Abstract
The substrate specificity of the mitochondrial metallopeptidase proteinase 1 (MP1) was investigated and its mitochondrial targeting signal identified. The substrate specificity of MP1 was examined with physiological peptides as substrates. Although the enzyme exhibits broad substrate specificity, there is a trend for peptides containing 13 or more residues to exhibit Km values of 2 μM or less. Three of four peptides containing 11 or fewer residues exhibited Km values above 10 μM. Similarly, peptides containing 13 or more residues exhibited kcat values below 10 min−1, while three of four peptides containing 11 or fewer residues exhibited kcat values above 30 min−1. Many of the peptide cleavage sites of MP1 resemble that of the mitochondrial processing protease (MPP), however MP1 does not process the precursor form of citrate synthase. The enzyme does however cleave the released prepeptide from precitrate synthase. A mitochondria localization was shown in MP1 transfected NT2 and HepG2 cells. Deletion of the N-terminal 15 amino acids caused MP1 to be mislocalized to the cytoplasm and nucleus. Furthermore, when fused to GFP, this 15-amino acid N-terminal sequence directed the fusion protein to the mitochondria.
Zinc-dependent peptidases of the M16 family possess the active site motif HEXXH in which the two histidine residues coordinate a zinc atom and the glutamate facilitates attack of a water molecule on the scissile bond. A subgroup of the M16 family of metallopeptidases contains an inverted Zn-binding motif HXXEH and these peptidases are referred to as inverzincins (1). The first identified inverzincin was pitrilysin protease III (2), a gene product of the ptr gene in E. coli.
Besides protease III (EC 3.4.24.55), several mammalian inverzincins have been identified and classified as M16A family members (3). These include insulysin (insulin degrading enzyme, IDE, EC 3.4.24.56) and nardilysin (NRDc, EC 3.4.24.61). Both protease III and insulysin exhibit a broad substrate specificity cleaving a variety of peptides. The structures of pitrilysin protease III and insulysin (4) are very similar and appear to be “clam-shaped” with the insulysin structure in a closed conformation and the protease III structure in an open conformation. The closed conformation tightly locks down the substrate in the interior of the enzyme, requiring a conformational change to permit product release. This conformational change is likely the ratelimiting step for the insulysin reaction. Both of these enzymes have an active site pocket that can accommodate and interact with a variety of peptides, explaining in part their broad substrate specificity. In contrast, nardilysin has a more defined substrate specificity cleaving peptides primarily at or between dibasic pairs (5).
In terms of cellular functions, insulysin has been implicated in regulating insulin and amyloid β peptide levels and thus has been linked to diabetes and Alzheimer’s disease (6,7). Furthermore, insulysin is up-regulated in activated macrophages (8) and some T cells (9) where its ability to cleave β-endorphin suggests a role in modulating immune function. Nardilysin has been implicated in regulating reproduction (10), cell cycling (11), and increasing the efficiency of energy metabolism (12). It has also been described as a surface receptor (13) and as an interacting protein for the PIP4 binding protein (14). The function of protease III remains unclear since there is no discernable phenotype detected in mutant ptr.
The mitochondrial processing peptidase (MPP, EC 3.4.24.64) has been classified as an M16B subfamily member. MPP plays an important role in the processing of nuclear encoded proteins as they enter the mitochondria. The cleavage of the presequence signal of the mitochondrial precursor proteins by MPP is an important step preceding folding and subsequent maturation of the precursor polypeptides. Except in plants, MPP is a heterodimeric enzyme with the Zn-binding motif located in β-MPP. As for the cleavage specificity of MPP, Arg is often found 2 or 3 residues at the amino side of the scissile bond. This specificity has been referred to as R-2 or R-3 motifs, respectively (15, 16). However, it is unclear how MPP makes a single and specific cleavage on preproteins that have very dissimilar presequences.
Mammalian metallopeptidase 1 (MP1, (17) and pre-sequence peptidases (PrePs) in Arabidopsis thaliana are the newest inverzincin family members and are included in an M16C subfamily. PrePs are located in both mitochondria and chloroplast (18). The function of PrePs is thought to involve the clearance of presequences that are derived from mitochondria precursor protein cleavage by MPP. Evidence that MP1 is also localized in mitochondria has been described (19, 20); however, a mitochondrial targeting sequence has not been identified or characterized. Based on sequence similarity and subcellular localization, MP1 may have a similarl function as the PrePs. MP1 was originally described on the basis of its ability to cleave the T13-R14 bond of leumorphin (dynorphin B-29); however there have not been any further studies on its substrate specificity. Furthermore, MP1 may have additional functions associated with tissue remodeling, as the expression of MP1 is up-regulated upon nuclear transplantation in mouse ES cells (21). The present study was designed to further characterize MP1 in terms of its substrate specificity and mitochondrial targeting.
Experimental Procedures
Materials
An antibody against the sequence (753AEMTDIKPILRKLPRIKK) of human MP1 was raised in a rabbit by Bethyl Lab. (Montgomery, Texas). The monoclonal anti-flag antibody M2 was purchased from Sigma-Aldrich. Anti-Lamp1, anti-calnexin, and anti-cytosolic thiolase were generously provided by Dr. S. W. Whiteheart (University of Kentucky). Polyclonal rabbit anti-mitochondrial malate dehydrogenase was a generous gift from Dr. Arnold W. Strauss (Vanderbilt University). Secondary florescent antibodies to mouse and rabbit were purchased from Vector Laboratories (Burlingame, CA). Poly-L-lysine was from Sigma-Aldrich. Recombinant yeast MPP was expressed in E. coli and purified as previously described (22). The citrate synthase presequence peptide (MALLTAAARLFGAKNASCLVLAARHAS-NH2) was synthesized by the W.M. Keck Foundation Biotechnology Resource Laboratory (New Haven, CT). Dynorphin B, and β-endorphin were purchased from neoMPS (San Diego, CA); dynorphin A and Leu-enkaphalin from Bachem (Torrance, CA), and the rest of the peptides used in this study were from California Peptide Research (Napa, CA).
cDNA subcloning
A cDNA clone for human MP1 (accession no. BC005025) was purchased from Invitrogen. The cDNA was subcloned into the pFastbac 1 cloning vector (Invitrogen) and used in the Bac-to-Bac expression system to generate baculovirus. The 5′ 416 bps were amplified and introduced into a Bam H1 site of pFastbac 1 by PCR using the following forward and reverse primers, respectively: 5′-TCT GGA TCC CAC CAT GTG GCG CTG C -3′ (BamH1 site underlined) and 5′-TTC ATG AAC GTG GAG AGG GAC -3′. This PCR product contains a Bam H1 and an internal Age I site. The remaining cDNA fragment was excised from the original clone with 5′-Age I and 3′-Xho1. These two fragments were then ligated into the pFastbac 1 vector.
For subcellular localization studies the full length MP1 cDNA was subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen) with the following modification. The stop codon was removed and an oligonucleotide encoding a flag epitope sequence followed by a stop codon (5′-TGT CCT CGA GTC ACT TAT CGT CGT CAT CCT TGT AAT CTC GGA TGA TCC AG -3′) (Xho1 site and stop codon underlined) was added to the 3′-end of the cDNA. To determine the mitochondrial targeting signal, two N-terminal truncation mutants MP1Δ15 and MP1Δ38 were constructed by PCR using the following forward primers, 5′-CAT AGG ATC CAT GAG CGG CGG ACA -3′ (MP1Δ15), and 5′-CAT AGG ATC CAT GCA GTA TAA ACT A-3′ (MP1Δ38), which contain a start codon preceded by a BamH 1 site. The reverse primer and the subsequent cloning procedures were the same as the above-mentioned method of cloning MP1 into pFastbac 1.
Expression and purification of MP1
Baculovirus expressing MP1 was prepared according to the manufacturer’s instructions and used to infect Sf9 cells. Three days post transfection, cells were collected by centrifugation at 2,000g for 10 min., and then frozen at −80°C until use. The cell paste was suspended in 10 volumes of 20 mM Tris buffer, pH 7.5 and sonicated with a Branson Sonifier 450 (Output control at 4, Duty cycle 30%, 10 sec for 3 times). The homogenate was centrifuged 30 min at 30,000g and the supernatant applied to Waters Acell Plus QMA anion exchange column (15 mL) in 20 mM Tris buffer, pH 7.5. Proteins were eluted with a 0 to 0.5 M linear NaCl gradient with MP1 eluting at ~0.2 M NaCl. The eluted enzyme was concentrated and applied to a Pharmacia Superdex 200 column (1.6cm × 60cm) equilibrated and run with 20 mM Tris buffer, pH 7.5. Lastly the enzyme was chromatographed on a 1 ml Pharmacia Mono Q column in the 20 mM Tris buffer and eluted with a linear salt gradient. The active MP1 fractions were pooled, desalted and concentrated with a Millipore Centricon concentrator and stored in 20% glycerol at −80°C. The purified enzyme appeared essentially homogeneous as judged by SDS-PAGE (figure 1a). The form of MP1 isolated by this method is processed at its N-terminus as determined by its molecular size and its mitochondrial location.
Figure 1. Recombinant human MP1 from Sf9 insect cells is processed.
(a) Purity of recombinant human MP1 derived from Sf9 cells. Recombinant MP1 (2 μg) purified as described in Experimental Procedures, was analyzed by SDS-PAGE on a 7.5% gel. Left lane shows the All Blue (BioRad) molecular weight standard. (b) Molecular weight comparison between purified MP1 from Sf9 cells and from E. coli. Lane 1 - purified MP1 from Sf9 cells; Lane 2. - purified MP1 from Sf9 cells and from E. coli mixed; Lane 3. - purified MP1 from E. coli. The recombinant precursor form of MP1 (p) is processed to the mature form (m) in Sf9 cells. (c) Subcellular localization of MP1 in Sf9 cells MP1 was detected with a rabbit anti-MP1 antibody and colocalized with MitoTracker (Invitrogen) (see Experimental Procedures).
MP1 was also expressed in E. coli as a GST fusion protein using the pGEX-5X expression vector. A TEV protease cleavage site followed by a BamH1 restriction site in frame with the MP1 cDNA was subcloned into the pGEX-5X vector. The construct pGEX-GST-TEV-MP1 was transformed into E. coli strain BL-21(DE3) and cells were grown at 37°C until an OD600 of 0.5 to 0.7 was reached. Expression was induced by the addition of isopropy-β-D-thiogalactoside to a final concentration of 0.5 mM. Induction was carried out overnight at room temperature. Cells were harvested and washed with PBS. Cell pellets were suspended in 25 mM potassium phosphate buffer, pH 7.5 containing 1 mM phenylmethylsulfonylfluride and 10 μM E-64, broken with a French press, and centrifuged at 30,000g at 4°C for 30 min. The supernatant was passed through a glutathione agarose column (Invitrogen) and washed with the same buffer without protease inhibitors. The fusion protein was eluted with 15 mM glutathione and cleaved with TEV protease (23). MP1 and GST were separated on an S-200 gel filtration column. The MP1 formed by this process contains the full-length unprocessed form of MP1 with two additional amino acids, Gln-Pro, at its N-terminus.
Assay of MP1 activity
MP1 activity was routinely assayed in 25 mM potassium phosphate buffer, pH 7.5 using the fluorogenic peptides Abz-RRQFKVVTRSQ-EDDnp and Abz-GGYRRGQ-EDDnp (Abz/Dnp peptides) as described (5). Hydrolysis of physiological peptides was determined by following their disappearance at 214 nm by reverse phase HPLC on a Phenomnex C18 column (5.0 × 4.6 mm). Peptides were separated by a linear gradient from 0.1% trifluoroacetic acid in 90% water/10% acetonitrile to 0.1% trifluoroacetic acid in 50% water/50% acetonitrile. The kcat values were determined based on the rate observed at 5% or less hydrolysis of a physiological peptide at a saturating concentration and using a molecular weight of 117 kDa for MP1. The Km of physiological peptides was determined by using them as alternate substrate inhibitors of Abz-GGYRRGQ-EDDnp hydrolysis over the range of 0.5Km to 2Km with 5 to 7 peptide concentrations used. Data was analyzed by Dixon plots with Graphpad Prism software (Graphpad, San Diego). Other kinetic parameters were determined by fitting to the Michaelis- Menton equation with the same software. Standard errors were 10% or less.
Identification of peptide cleavage products
Physiological peptides were incubated with MP1 for various times to establish their order of appearance. Cleavage products were separated by reverse phase HPLC on a Vydac C4 column (5). In the case of atrial natriuretic peptide and somatostatin the peptides were reduced and carboxymethylated prior to HPLC analysis. Peptides or their reaction products were collected manually, freeze-dried, and identified by mass spectrometry using a Bruker Autoflex time-of-flight mass spectrometer with alpha-cyano-4-hydroxycinnamic acid as the matrix or on a ThermoFinnigan LCQ, with sample introduction by direct infusion. These analyses were performed at the University of Kentucky Mass Spectrometry Core Facility.
Cell culture
HepG2 cells were grown in F12/MEM medium containing 10% iron-supplemented bovine calf serum (Atlanta Biological, Atlanta, GA). NT2 cells (ATCC# CRL-1973) were grown in DMEM medium containing 10% fetal calf serum, 4.5 g of glucose per liter, and 1% glutamine (Gibco). Penicillin/streptomycin (Gibco) (1%) was included in all media. Cells were kept at 37°C and 5% CO2. The FuGENE 6 reagent (Roche) was used for cell transfections. The DNA to reagent ratio was 1 μg per 3 μL of FuGENE 6 using a total of 6 μg DNA per 10-cm culture dish. Cells were harvested 48 hr after transfection. For Sf9 cells, baculovirus containing the full length MP1 was used. Virus obtained from the second round of amplification was diluted 10 times with Sf-900 II SFM (Invitrogen) and applied directly to Sf9 cells. Immunocytochemistry was performed after 2–3 days incubation.
Immunocytochemistry
Cells were plated on polylysine coated coverslips and transfected 12 to 24 h later. Cells were washed with PBS 24 h after transfection, and then fixed with 4% paraformaldehyde in PBS for 20 min, and permeablized with acetone/methanol (1:1) for 15 min at room temp. Fixed cells were blocked with 10% horse serum in PBS for 20 min and incubated with the appropriate antibody at a 1:200 dilution. The same blocking solution was used for washing before incubating with secondary antibodies. MitoTracker (Invitrogen) was used according to the manufacturer’s instruction. Coverslips were mounted with Vectashield/Dapi (Vector Laboratories) and examined with a Nikon Eclipse E600 epifluorescence microscope.
Subcellular localization by differential centrifugation
Harvested cells were washed two times with PBS, and transferred to 1 mM HEPES buffer, pH 7.4, on ice for 5 min. Cells were then adjusted to 10 mM HEPES buffer, pH 7.4 containing 250 mM sucrose and homogenized with a Balch-style ball homogenizer (24). Subcellular fractions were collected by differential centrifugation employing a modification of the method of Lutsenko and Cooper (25). Briefly, the P1 fraction containing unbroken cells and large cell fragments was obtained by centrifugation for 5 min at 500g; a P2 fraction containing mitochondria was obtained by centrifugation for 10 min at 3,000g; and finally a P3 fraction containing other light vesicles was obtained by centrifugation for 45 min at 20,000g. The supernatant of the P3 fraction (S3) contains soluble proteins.
Electrophoresis and Immunoblot Analysis
SDS-PAGE was performed in a Bio-Rad Protean system following the protocol of Laemmli (26). Resolved proteins were then transferred according to Towbin (27) onto a polyvinyldifluoride (PVDF-F) membrane (Millipore). Citritate synthase presequence peptide was resolved in a 10–20% Tricine gel using an Xcell SureLock Mini-gel system (Invitrogen). Specific proteins were detected and quantified using a Li-COR Odyssey Imaging system. The BioRad Precision Plus All Blue Protein standard was used for estimating molecular weights of protein bands.
Protein assay
Protein concentration was determined using Coomassie Plus (Pierce Chemical Company). Bovine serum albumin provided with the reagent was used as the protein standard.
In vitro Translation and processing
An MP1 cDNA and a mitochondrial citrate synthase cDNA (Accession no. BC010106, obtained from Invitrogen) were subcloned into the pBluescript vector by PCR. Proteins were expressed using a coupled transcription/translation system (Promega) and radiolabeled with Tran [35S] (20 μCi/50μL; MP Biomedicals). For studying preprotein processing the protocol of Cavadini et al. (28) was used except that the incubation temperature was 30°C. Briefly, one microliter of translation mixture was added to a reaction buffer containing 10 mM HEPES-KOH, pH 7.4, 1 mM DTT, 1 mM MnCl2, 1 mM ZnCl2, and varied amounts of processing enzymes. Processed products were analyzed by 10% SDS-PAGE and detected with a GE Typhoon system.
Results
Kinetics
MP1 expressed in Sf9 cells is processed as evidenced by the finding that it exhibits a lower molecular weight than the enzyme derived from E. coli, figure 1b. The latter form of MP1 was expressed as a fusion protein containing an internal TEV cleavage site such that cleavage by TEV generated the full length unprocessed MP1 with two additional N-terminal amino acids. Furthermore the Sf9 form of MP1 was shown to have a mitochondrial localization by immunofluorescence, figure 1c. For kinetic studies we used the mature processed enzyme.
Since MP1 was previously shown to cleave leumorphin at a monobasic site we initially characterized the enzyme using two intramolecularly quenched substrates containing either a part of the leumorphin sequence (Abz-RRQFKVVTRSQ-EDDnp) or a substrate containing a dibasic pair (Abz-YGGLRRGQ-EDDnp). The Km and kcat values for these substrates are nearly equivalent, being 1.07 μM and 4.8 min−1 for Abz-RRQFKVVTRSQ-EDDnp and 0.5 μM and 6.8 min−1 for Abz-YGGLRRGQ-EDDnp. The presence of one or two basic residues does not seem to affect the kinetics with these substrates.
We next studied the reaction of MP1 with a number of physiologically active peptides. The Km for each peptide was determined by using it as an alternate substrate inhibitor of Abz-GGYRRGQ-EDDnp hydrolysis and thus would reflect primarily if not totally the intact peptide, while kcat was determined from a linear initial rate at a saturating substrate concentration and it too would reflect reaction of the intact peptide. As shown in Table 1, MP1 exhibits a broad substrate specificity. There appears to be a trend that peptides containing 13 or more residues exhibit Km values of 2 μM or less. Of the four peptides containing 11 or fewer residues that were studied, three exhibited Km values above 10 μM. Similarly, peptides containing 13 or more residues exhibited kcat values below 10 min−1, while three of the four peptides containing 11 or fewer residues exhibited kcat values above 30 min−1. As a consequence kcat/Km values tend to vary little amongst substrates with the exception of human leumorphin and ANP, which exhibited the highest kcat/Km values. Of the substrates tested insulin exhibited the lowest kcat/Km, due to its high Km and low kcat values.
Table 1.
Kinetics of hydrolysis of physiological peptides by MP1.
| Peptide | Peptide Size (no. residues) | Km (μM) | kcat* (min−1) | kcat/Km (M × min−1 × 105) |
|---|---|---|---|---|
| Leu-enkephalin | 5 | 20.8 ± 0.9 | 39.0 ± 3.9 | 18.7 ± 2.1 |
| Bradykinin | 9 | 16.2 ± 0.8 | 35.4 ± 0.9 | 21.8 ± 1.2 |
| Angiotensin I | 10 | 1.7 ± 0.1 | 7.6 ± 0.4 | 44.6 ± 4.3 |
| Substance P | 11 | 13.8 ± 1.0 | 32.7 ± 2.6 | 23.6 ± 2.5 |
| Dynorphin B | 13 | 1.9 ± 0.2 | 8.9 ± 0.1 | 46.3 ± 6.8 |
| Dynorphin A | 17 | 0.8 ± 0. 1 | 4.8 ± 0.4 | 62.6 ± 6.8 |
| Endothelin 1 | 21 | 0.7 ± 0.1 | 1.1 ± 0.0 | 15.8 ± 4.6 |
| Somatostatin | 28 | 0.1 ± 0.0 | 0.6 ± 0.0 | 41.7 ± 3.8 |
| Atrial natriuretic peptide | 28 | 0.2 ± 0.0 | 4.5 ± 0.4 | 199.2 ± 18.6 |
| Leumorphin (human) | 29 | 0.4 ± 0.0 | 5.9 ± 0.4 | 140.6 ± 10.7 |
| Leumorphin (porcine) | 29 | 0.8 ± 0.0 | 4.9 ± 0.2 | 63.5 ± 4.5 |
| β-endorphin | 31 | 0.9 ± 0.0 | 5.8 ± 0.1 | 62.6 ± 5.1 |
| Calcitonin | 32 | 2.4 ± 0.0 | 3.8 ± 0.1 | 15.4 ± 0.6 |
| β-Amyloid (1–40) | 40 | 1.7 ± 0.1 | 3.3 ± 0.1 | 19.5 ± 1.1 |
| Growth Hormone Releasing factor | 40 | 0.5 ± 0.0 | 1.4 ± 0.1 | 29.4 ± 3.4 |
| Insulin | 51 | 54.5 ± 5.5 | 0.2 ± 0.0 | 0.03 ± 0.00 |
Since most peptides are cleaved at multiple sites kcat could reflect cleavages at more than one site.
Km values were determined by using the indicated peptide as an alternate substrate inhibitor of 2.2 μM Abz-GGFYRRVGQ-EDDnp hydrolysis, (Km = 1.07 μM). Under the assay conditions employed little if any of the inhibitory peptide would have been cleaved.
Cleavage sites
MP1 dependent peptide cleavage products were isolated by HPLC and identified by mass spectrometry. This analysis showed that MP1 cleaved at multiple sites within its peptide substrates, and that some products which appear to be derived from the intact peptide, as judged by identification of both N and C-terminal fragments, appeared at latter time points. In addition the secondary cleavage of some initial products was evident based on the analysis of product appearance as a function of time. For example, in the case of β-endorphin, MP1 initially cleaves this peptide at Ala21-Ile22 generating fragments YGGFMTSEKSQTPLVTLFKNA (b, Figure 2) and IIKNAYKKGE. The latter peptide was identified near the void volume of the column (not shown). Fragment b is subsequently cleaved by MP1 at Gly3-Phe4 at the N-terminal region generating FMTSEKSQTPLVTLFKNA (c, Figure 2). We also found that MP1 acts on FMTSEKSQTPLVTLFKNA (fragment c) by removing the N-terminal Phe (F). Additionally, MP1 cleaves the C-terminal region of fragment c at Leu17-Phe18, Thr16-Leu17 and to a lesser extent Pro13-Leu14 and Leu14-Val15.
Figure 2. HPLC chromatogram of β-endorphin and its fragments produced by hydrolysis by MP1.
β-endorphin (30 μM) was cleaved by 2 μg of MP1 for the indicated time in 25 mM Tris buffer, pH 7.5. The reaction was stopped by the addition of trichloroacetic acid to 0.1% and subjected to reverse phase HPLC on a Vydac C4 column as previously described (5). Peaks were collected manually and subjected to mass spectral analysis for identification.
One of the substrates of MP1, dynorphin 29 also called leumorphin (YGGFLRRQFKVVTRSQQDPNAYSGQLFDA), was reported to be cleaved at the T-R bond generating dynorphin B and dynorphin 14–29 (17). Although we indeed detected the MP1 dependent cleavage of porcine dynorphin 29 at the T-R bond (Table 2), this turned out to be a minor cleavage site as it was not observed until latter time points. Instead the initial cleavage sites were at D18-P19 and L26-F27. On the other hand, the initial cleavage site of human dynorphin 29, which differs by five residues (E17=Q, Y22=S, E23=G, E24=Q, and V29=A), is at the T-R bond, showing the influence of C-terminal residues on the cleavage site. This may also reflect differences in conformation of the bound peptide. The product dynorphin B was subsequently hydrolyzed at the K-V, F-K and Y-G sites, which are the same cleavages observed when dynorphin B was the substrate.
Table 2.
Cleavage sites of human pitrilysin MP1 on physiological peptides.
| Peptide | Cleavage Sites |
|---|---|
| Angiotensin I | DRVY▼IHP▼FHL |
| Atrial natriuretic peptide (ANP) | SLRRSSCFGGRMD▼RIGAQSGLGCNS▼FRY |
| Bradykinin | RPPG▼FSPFR |
| Calcitonin | CGNLSTC▼MLG▼TYTQDFNK↓FHTFPQTAIGVGAP |
| Dynorphin A | Y↓GGFLRR↓IRP▼KLKWDNQ |
| Dynorphin B | Y↓GGFLRRQF↓K▼VVT |
| Growth Hormone Releasing Factor (GRF) | Y↓ADAIFTNSYRKVLG▼QLS▼ARKL↓QDIMQQGESNQERGA |
| Leu-enkephalin | Y▼GGFL |
| Leumorphin (human) | Y↓GGFLRRQF↓K▼VVT▼RSQQDPNAYSGQLFDA |
| Leumorphin (porcine) | Y↓GGFLRRQFK↓VVT↓RSQED▼PNAYYEEL▼FDV |
| Substance P | RPKPQQF↓FG▼LM |
| β Amyloid (1–40) | DAEFRHDSGYEVHHQKLVFF▼AEDVGSNKGAIIGL↓MVGGVV |
| β-endorphin | YGG↓F↓MTSEKSETPLV↓T↓L▼FKNA▼IIKNAYKKGE |
Peptide hydrolysis was followed at several time points to determine initial and secondary or slower cleavage sites. Filled triangles indicate initial cleavage sites, while arrows indicate secondary cleavage sites that were either derived from initial products or appeared at latter time points.
As noted in the reaction with Leu-enkephalin, MP1 was able to cleave an N-terminal tyrosine, which was also detected in the reactions with dynorphin B, dynorphin A, and GRF (Table 2) and as noted above an N-terminal Phe. Judging from the cleavage products listed in Table 2, it is unlikely that this activity can be attributed to contamination by an aminopeptidase since only an N-terminal Tyr or Phe were cleaved. However, to rule out aminopeptidase activity we carried out the hydrolysis of β-endorphin, dynorphin A and leumorphin in the presence of 10 μM bestatin, a broad spectrum aminopeptidase inhibitor. We observed no effect on the rate of peptide hydrolysis or the distribution of cleavage products. Additionally, we used an anti-MP1 antibody to immunodeplete MP1 and measure the reaction of the immunodepleted sample with Leu-enkephalin (YGGFL). As the Y-G bond is the only cleavage in Leu-enkephalin, it is clear that elimination of MP1 by immunodepletion effectively eliminated the N-terminal cleavage activity toward Leu-enkephalin, Figure 3.
Figure 3. MP1 can cleave an N-terminal tryosine residue.
Leu-enkephalin (500 μM) was cleaved by 1 μg of MP1 in 25 mM phosphate buffer, pH 7.5. The reaction was stopped and analyzed by reverse phase HPLC as described in figure 2. Peaks were identified by comparison to authentic standards. Upper panel – MP1 incubated with rabbit IgG on ice for 1 hr was then absorbed with protein A Sepharose. Lower panel - MP1 incubated with anti-MP1 on ice for 1 hr and then absorbed with protein A Sepharose.
Since MP1 can cleave a peptide at multiple sites and secondary cleavages were demonstrated, we focused on the initial cleavage sites and some notable secondary cleavages as shown in Table 2. Examination of the residues at the P1′ and P1 sites (based on the nomenclature of Schechter and Berger (29)) did not reveal a clear pattern of cleavage preference for particular a residue. However, two cleavage patterns were observed. The first became apparent when the cleavage sites were aligned. Here we note that many, but not all, of the cleavage sites occur two to five residues C-terminal to an arginine residue, Table 3. This resembles the cleavage preference of mitochondrial processing protease (MPP). Another cleavage preference is seen at the amino side of Phe when it is located within 3–5 residues to the C-terminus. Examples in Table 2 are angiotensin I, atrial natriuretic peptide, bradykinin, porcine leumorphin, and β-endorphin (fragment c conversion to fragment e, Fig. 2).
Table 3.
MP1 cleavage patterns
| a. MP1 cleavage motifs mimic that of mitochondrial processing proteases. | ||
|---|---|---|
| Peptide | Cleavage alignment | Motif |
| Angiotensin I | DRVY▼IHPFHL | R-3 |
| ANP | RRSSCFGGRMD▼RIGAQSGLGCNSFRY | R-3 |
| Bradykinin | RPPG▼FSPFR | R-4 |
| Dynorphin A | YGGFLRRIRP▼KLKWDNQ | R-2, R-4 or R-5 |
| Dynorphin B | YGGFLRRQF▼KVVT | R-3 or R-4 |
| Dynorphin B | YGGFLRRQFK▼VVT | R-4 or R-5 |
| GRF | ..QLSARKL▼LQDIMSRQQGESNQE | R-3 |
| GRF | IFTNSYRKVLG▼QLS | R-5 |
| b. Primary cleavage sites that did not contain an R-2 to R-5 motif | |
|---|---|
| Peptide | Cleavage site |
| Calcitionin | CNS▼FRY |
| Calcitonin | STC▼MLG |
| ANP | MLG▼TYT |
| Leu-enkephalin | Y▼GGFL |
| β-amyloid 1–40 | VFF▼A |
| β-endorphin | KNA▼IIK |
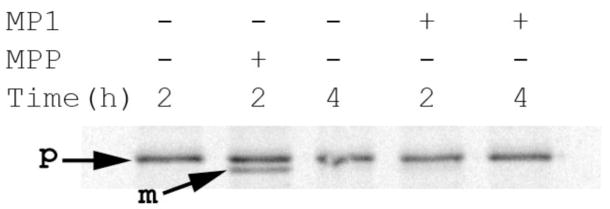
Since one of the cleavage patterns of MP1 resembles that of the mitochondrial processing protease, we determined whether or not MP1 is able to actually process a mitochondrial precursor protein. We tested radiolabeled in vitro translated full-length citrate synthase (CS), which contains the presequence (MALLTAAARLFGAKNASCLVLAARHAS), as a substrate. In the presence of 0.2 μg of yeast mitochondrial processing protease (yMPP) full length citrate synthase is processed to its mature form within a 2 hr incubation period, Figure 4. However; MP1 did not generate mature CS even with a five times higher amount of enzyme (1 μg) and longer incubation time (4 hrs) suggesting it does not behave as a protein processing enzyme, but rather a true peptidase cleaving peptides, but not proteins.
Figure 4. Precitrate synthase is processed by yMPP, but not by MP1.
The mitochondrial precursor form of CS was generated by in vitro translation in the presence of 35S Met/Cys. From this translation mixture one microliter was incubated with MP1 (1 μg) or yMPP (0.2 μg), and then analyzed by SDS-PAGE. P = precursor form; M = mitochondrial processed form.
To determine whether MP1 could be involved in the clearance of presequence peptides we synthesized the citrate synthase presequence peptide and tested it as a substrate for MP1. As shown in Figure 5a, MP1 can in fact cleave the presequence peptide. We determined the kinetic parameters for this 27 amino acid peptide and observed a Km of 0.17 μM, a kcat of 1.1 min−1, and a kcat/Km of 6.7 × 105 M × min−1. The cleavage sites were also determined using mass spectrometry to detect peptide fragments generated by MP1 cleavage (Figure 5b). Only the initial cleavage sites are shown in this figure. In each case both the N- and C-terminal complementary fragments were identified, showing that MP1 cleaves primarily at sites in the middle of the peptide.
Figure 5. MP1 cleaves the presequence peptide of citrate synthase.

(a) The reaction mixture contained 3.6 μg of purified MP1 and 8.6 μg of peptide in 50 mM Tris-HCl buffer, pH 7.5. The molar ratio of enzyme to peptide was 1:100. Aliquots were removed at the times indicated and analyzed on a 10–20% Tricine gel stained with Coomassie blue. The last lane to the right shows a peptide alone control. (b). Cleavage sites were determined by detecting peptide fragments in the reaction mixture by mass spectrometry. Secondary structure was predicted with the Prof Prediction program (www.proteinpredict.org). Helical structures are showed in red and random coil in green.
Effects of metal ions
AtPreP, the plant homolog of MP1, requires Mg+2 or Ca+2 for full activity. We thus examined the effect of metal ions on MP1 activity. As shown in Table 4 the enzyme is inhibited by nickel and zinc and slightly activated by manganese, which parallels that observed with nardilysin (39). Other metals were without effect.
Table 4.
Effect on Metal Ions on hMP1
| Salt | % Activity Remaining * |
|---|---|
| CaCl2, 1 mM | 98 ± 0 |
| KCl, 2 mM | 77 ± 1.9 |
| MgCl2, 1 mM | 102 ± 0.9 |
| MnCl2, 1 mM | 143 ± 6.5 |
| NaCl, 2 mM | 83 ± 0 |
| NiCl2, 1 mM | 18 ± 2.8 |
| ZnCl2, 1 mM | 0 |
MP1 was diluted in 20 mM Tris-HCl, pH 7.5 in the presence of metal salts at the concentration indicated. MP1 activity was assayed in duplicate with 2.7 μM Abz-YGGLRRGQ-EDDnp as substrate under the same conditions. Experiments were conducted in duplicate and average values are presented with the range given.
Subcellular Localization
To determine the subcellular localization of MP1, we employed two approaches, namely differential centrifugation and immunocytochemistry in a neuronal NT2 cell line and in a non-neuronal HepG2 cell line. Differential centrifugation showed that MP1 is enriched in the pellet derived from centrifugation at 3,000g (P2 fraction) and co-sediments with mitochondrial malate dehydrogenase (Figure 6). Although not shown, fractionation of HepG2 cells produced the same result. Sonication of the P2 fraction released the enzyme demonstrating that MP1 is in the soluble fraction of the mitochondrion in HepG2 cells. MP1 was also shown to colocalize with the soluble mitochondrial protein mMDH by immunocytochemistry (Figure 7).
Figure 6. Subcellular localization of MP1.
NT2 cells were fractionated by differential centrifugation as described in Methods. (a) Thirty microgram of protein from each subcellular fraction was analyzed by Western blotting. The antibodies used are shown at the right column of the blots. (b) The intensity of the bands representing the fluorescence signal of each antibody were calculated from the data in figure 6a and expressed relative to total fluorescence for each antibody.
Figure 7. Subcellular localization of MP1 in NT2 and HepG2 cells.
NT2 or HepG2 were transfected with either full length Flag tagged MP1 (wtMP1-Flag) or an N-terminal truncated MP1-Flag. After 24 h culture, cells were fixed and probed with rabbit anti-mitochondrial malate dehydrogenase and mouse-anti Flag (see Experimental procedure).
The first 15 amino acids of MP1 contain basic and hydrophobic residues organized in groups when this sequence is analyzed on a helical wheel, suggesting this region might contain a mitochondrial targeting signal. To determine whether or not this presequence actually contains a functional mitochondrial-targeting signal, a truncation mutant MP1Δ15 was constructed. Additionally, as 22- (30) or 29- (31) amino acid long presquences are predicted to be cleaved during the translocation process; we also constructed another 38-residue truncation mutant (MP1Δ38) to ensure that the putative target signal is completely removed. We found that both of these truncation mutants, when expressed in NT2 or HepG2 cells, are mislocalized to cytoplasmic compartments and the nucleus. This is shown in Figure 7 for MP1Δ15. Next, we generated a construct in which the N-terminal 15 amino acid putative MP1 mitochondrial targeting sequence (MWRCGGRRGLCVLRR-) was fused to GFP. When transfected into NT2 or HepG2 cells this construct produced mitochondrial GFP, Figure 8.
Figure 8. N-terminal sequence of MP1 targets GFP to mitochondria.
NT2 and HepG2 cells are transfected with a GFP expression vector to which a 15-residue N-terminal MP-1 is fused (pEGFP-N15). Cells were incubated for 24 hrs and then stained and imaged as described in Experimental procedures.
Discussion
In this study we have examined the properties of mammalian pitrilysin MP1 in terms of its intracellular targeting and its substrate specificity. Although other metallopeptidases of the inverzincin family localize to both the cytosol and mitochondria (i.e. insulysin (32) and neurolysin (33)), we demonstrate here that MP1 is primarily, if not exclusively, a mitochondrial protein with its mitochondrial targeting signal sequence located within a 15-residue presequence (MWRCGGRRGLCVLRR). Removal of this presequence mislocalizes MP1 in cells, while this sequence is sufficient to target a non-mitochondrial protein (GFP) into mitochondria. It is generally accepted that mitochondrial presequences are amphipathic helices in which basic and hydrophobic residues are localized to different sides on a helical cylinder (34), suggesting an initial interaction with multiple proteins at the mitochondrial membrane. In the case of the presequence of MP1, Arg residues at positions 3, 7, and 14 can be clustered on the same side of a helical wheel. Interestingly, MP1 exists as two variants. The MP1 clone (Accession no. BC005025) used in the present study contains an Arg in the 8th position. There is a more predominant variant containing Gln at this position due to a variation of the 8th codon CGG (8Arg) versus CAG (8Gln). The 8Arg variant exists at a frequency of ~20% based on 100 EST clones examined. However, it is likely that 8Arg is not important in mitochondrial targeting since it faces away from the basic cluster. The MP1 mitochondrial sequence is functionally conserved as mammalian MP1 is targeted to insect mitochondria.
MP1 was initially described as a metallopeptidase cleaving the opioid peptide leumorphin, but its specificity was not investigated. We have further characterized MP1 with a series of opioid and other bioactive peptides that have been studied as substrates for other metallopeptidases. MP1 generally exhibits a preference for peptides of 13 or more amino acids in terms of lower Km values, whilst kcat values show an inverse relationship with peptides of 13 or more amino acids exhibiting lower kcat values. Although the structure of MP1 has not been determined, its exhibits 54% sequence similarity (30% sequence identity) to insulysin suggesting it has a similar structure. The long extended substrate-binding site of insulysin, if conserved in MP1 would account for the lower Km values generally seen with larger peptides. At the same time these extended interactions may contribute to the lower kcat values if product dissociation or opening of a closed conformation were rate limiting.
There are however exceptions in terms of the trends observed between substrate size and Km. Most notable is insulin with its two chains linked by disulfide bonds. It may be too large to optimally fit into the active site of MP1. Another exception is angiotensin I which is a 10 residue peptide that has a lower Km and lower kcat that other similar sized peptides. This apparent anomaly likely reflects a sequence that interacts favorably with the active site.
When comparing the residues at the P1 and P1′ cleavage sites of physiological peptides, the cleavage preference of MP1 appears unique as it does not follow any known pattern of other inverzincins. For example, insulysin prefers hydrophobic or basic residue at either the amino side (35) or carboxyl side (36) of the scissile bond. PreP, a plant homolog of MP1, prefers P1 to be a basic residue and P1′ to be uncharged residue (18). However, P1 can be a basic, acidic, small or hydrophobic residue for MP1. Two consequences result from these differences. First, MP1 shares substrates, but not cleavage sites with other inverzincins. For example, except for the T-R bond in leumorphin, there are no overlapping cleavage sites for this substrate between bacterial pitrilysin (37) and MP1. Second, MP1 exhibits even broader substrate specificity than other M16 metallopeptidases. This is exemplified by the fact that angiotensin I and bradykinin are not cleaved by protease III (37), insulysin (38) or nardilysin (39), but are substrates of MP1. Additionally, the ability to cleave an N-terminal Phe or Tyr residue is unique to MP1.
It is noted that the multiple cleavage sites for MP1 within the citrate synthase presequence peptide are clustered in the middle of the peptide. Structural analysis predicts that this central region is an unstructured one flanked by two helical structure regions of the peptide. Thus in this case the open accessibility of this central region could contribute as a cleavage site determinant.
Based solely on its mitochondrial localization, MP1 has been postulated to be a peptidase that clears free presequences inside the mitochondria (20). We have provided in vitro evidence that MP1 can in fact degrade a mitochondrial presequence peptide, that of mammalian citrate synthase. The cleavage of the citrate synthase presequence exhibited similar kinetics to other peptides examined, indicating MP1 is indeed a potential candidate for such a function. If in fact MP1 functions to hydrolyze released mitochondrial presequence peptides, then its broad substrate specificity may serve this function well as mitochondrial presequence peptides are rather diverse in their sequences. In the yeast intermembrane space, a metallopeptidase MoP112 (40) belonging to M16 pitrilysin family has recently been shown to cleave peptides along with another metallopeptidase saccharolysin (PRD1). Presequence peptides were detected in the double mutant Δmop112Δprd1, but not found in wild type yeast. Interestingly, although fewer presequences were detected in mutants of either Δmop112 or Δprd1, the identity of these peptides differed indicating that these two enzymes have overlapping but not identical specificity. It is tempting to speculate that MP1 may serve a similar function as MoP112 or PRD1 in mammalian mitochondria.
Based on the ability of MP1 to degrade a number of physiological peptides, it is also likely that the enzyme may degrade other peptides in the mitochondria. Besides presequence peptides, mitochondrial peptides are derived from the degradation of misfolded proteins by ATP- dependent proteases in the inner mitochondrial membrane (41). These peptides are subsequently either degraded by oligopeptidases or exported out of the mitochondria.
Acknowledgments
This research was supported by grants from the National Institute on Drug Abuse, grant DA 02243 and from the National Center for Research Resources (NCRR), grant P20RR02017.
We thank NIDA for providing dynorphin B and β-endorphin through NeoMPS, Inc. We would also like to thank Drs Sydney Whiteheart and Becky Dutch for technical advice on immunochemistry and radiolabeling experiments, respectively. We thank Dr. Carol Beach for conducting the mass spectrometry analysis at the University of Kentucky Proteomics Core facility, which is supported in part by a National Center for Research Resources grant P20RR02017.
Abbreviations used
- Abz
2-aminobenzoyl
- ANP
Atrial natriuretic peptide
- EDDnp
ethylenediamine-2,4-dinitrophenyl
- Fmoc
9-fluorenylmethoxycarbonyl
- EST
expressed sequenced tag
- GFP
Green fluorescent protein
- GRF
Growth Hormone Releasing Factor
- MALDI-TOF
Matrix Assisted Laser Desorption Ionization-Time Of Flight
References
- 1.Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;345:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 2.Finch PW, Wilson RE, Brown K, Hickson ID, Emmerson PT. Complete nucleotide sequence of the Escherichia coli gene encoding protease III. Nucleic Acids Res. 1979;14:7695–7703. doi: 10.1093/nar/14.19.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow KM, Csuhai E, Juliano MA, Pyrek JS, Juliano L, Hersh LB. Studies on the subsite specificity of rat nardilysin (N-arginine dibasic convertase) J Biol Chem. 2000;275:19545–19551. doi: 10.1074/jbc.M909020199. [DOI] [PubMed] [Google Scholar]
- 6.Hersh LB. Peptidases, proteases and amyloid beta-peptide catabolism. Curr Pharm Des. 2003;9:449–454. doi: 10.2174/1381612033391676. [DOI] [PubMed] [Google Scholar]
- 7.Qiu WQ, Folstein MF. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer’s disease: review and hypothesis. Neurobiol Aging. 2006;2:190–198. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Sarada B, Thiele D, Dang T, Safavi A, Hersh LB, Cottam GL. Increased expression of an endopeptidase (gamma-EGE/IDE) hydrolyzing beta-endorphin during differentiation and maturation of bone marrow macrophages. J Leukoc Biol. 1997;62:753–760. doi: 10.1002/jlb.62.6.753. [DOI] [PubMed] [Google Scholar]
- 9.Sarada B, Thiele D, Dang T, Lee J, Safavia A, Hersh LB, Cottam GL. Anti-CD3 activation of human CD4+ T cells increases expression of the intracellular beta-endorphin endopeptidase (IDE/gamma-EpGE) J Neuroimmunol. 1998;85:59–68. doi: 10.1016/s0165-5728(97)00268-3. [DOI] [PubMed] [Google Scholar]
- 10.Chesneau V, Prat A, Segretain D, Hospital V, Dupaix A, Foulon T, Jegou B, Cohen P. NRD convertase: a putative processing endoprotease associated with the axoneme and the manchette in late spermatids. J Cell Sci. 1996;109:2737–2745. doi: 10.1242/jcs.109.11.2737. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Wang X, Hockman S, Snow EC, Hersh LB. Subcellular localization of nardilysin during mouse oocyte maturation. Arch Biochem Biophys. 2005;434:187–194. doi: 10.1016/j.abb.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 12.Chow KM, Ma Z, Cai J, Pierce WM, Hersh LB. Nardilysin facilitates complex formation between mitochondrial malate dehydrogenase and citrate synthase. Biochim Biophys Acta. 2005;1723:292–301. doi: 10.1016/j.bbagen.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Nishi E, Prat A, Hospital V, Elenius K, Klagsbrun M. N-arginine dibasic convertase is a specific receptor for heparin-binding EGF like growth factor that mediates cell migration. EMBO J. 2001;20:3342–3350. doi: 10.1093/emboj/20.13.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stricker R, Chow KM, Walther D, Hanck T, Hersh LB, Reiser G. Interaction of the brain-specific protein p42IP4/centaurin-alpha1 with the peptidase nardilysin is regulated by the cognate ligands of p42IP4, PtdIns(3,4,5)P3 and Ins(1,3,4,5)P4, with stereospecificity. J Neurochem. 2006;98:343–354. doi: 10.1111/j.1471-4159.2006.03869.x. [DOI] [PubMed] [Google Scholar]
- 15.Ito A. Mitochondrial processing peptidase: Multiple-site recognition of precursor proteins. Biochem Biophys Res Comm. 1999;265:611–616. doi: 10.1006/bbrc.1999.1703. [DOI] [PubMed] [Google Scholar]
- 16.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 17.Mzhavia N, Berman YL, Qian Y, Yan L, Devi LA. Cloning, expression, and characterization of human metalloprotease 1: a novel member of the pitrilysin family of metalloendoproteases. DNA Cell Biol. 1999;18:369–380. doi: 10.1089/104454999315268. [DOI] [PubMed] [Google Scholar]
- 18.Ståhl A, Nilsson S, Lundberg P, Bhushan S, Biverståhl H, Moberg P, Morisset M, Vener A, Mäler L, Langel U, Glaser E. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J Mol Biol. 2005;349:847–60. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nature biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 20.Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Eneqvist T, Tjernberg L, Ankarcrona M, Glaser E. Degradation of the amyloid beta - protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281:29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- 21.Suemizu H, Aiba K, Yoshikawa T, Sharov AA, Shimozawa N, Tamaoki N, Ko MS. Expression Profiling of Placentomegaly Associated with Nuclear Transplantation of Mouse ES Cells. Dev Biol. 2003;253:36–53. doi: 10.1006/dbio.2002.0870. [DOI] [PubMed] [Google Scholar]
- 22.Gakh O, Obsil T, Adamec J, Spizek J, Amler E, Janata J, Kalousek F. Substrate binding changes conformation of the alpha-, but not the beta-subunit of mitochondrial processing peptidase. Arch Biochem Biophys. 2001;385:392–6. doi: 10.1006/abbi.2000.2167. [DOI] [PubMed] [Google Scholar]
- 23.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Prot Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Xu W, He P, Medrano EE, Whiteheart SW. Gaf-1, a gamma-SNAP-binding protein associated with the mitochondria. J Biol Chem. 2001;276:13127–13135. doi: 10.1074/jbc.M009424200. [DOI] [PubMed] [Google Scholar]
- 25.Lutsenko S, Cooper MJ. Localization of the Wilson’s disease protein product to mitochondria. Proc Natl Acad Sci USA. 1998;95:6004–6009. doi: 10.1073/pnas.95.11.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavadini P, Gakh O, Isaya G. Protein import and processing reconstituted with isolated rat liver mitochondria and recombinant mitochondrial processing peptidase. Methods. 2002;26:298–306. doi: 10.1016/S1046-2023(02)00035-X. [DOI] [PubMed] [Google Scholar]
- 29.Schechter I, Berger A. On the size of the active site in proteases. I Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 30.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 32.Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, Guarente L, Selkoe DJ. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–446. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato A, Sugiura N, Saruta Y, Hosoiri T, Yasue H, Hirose S. Targeting of endopeptidase 24.16 to different subcellular compartments by alternative promoter usage. J Biol Chem. 1997;272:15313–15322. doi: 10.1074/jbc.272.24.15313. [DOI] [PubMed] [Google Scholar]
- 34.Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 35.Song ES, Mukherjee A, Juliano MA, Pyrek JS, Goodman JP, Jr, Juliano L, Hersh LB. Analysis of the subsite specificity of rat insulysin using fluorogenic peptide substrates. J Biol Chem. 2001;276:1152–1155. doi: 10.1074/jbc.M008702200. [DOI] [PubMed] [Google Scholar]
- 36.Dieter Muller, Christian Schulze, Hans Baumeister, Friedrich Buck, Dietmar Richter. Rat insulin-degrading enzyme: cleavage pattern of the natriuretic peptide hormones ANP, BNP, and CNP revealed by HPLC and mass spectrometry. Biochemistry. 1992;31:11138–11143. doi: 10.1021/bi00160a026. [DOI] [PubMed] [Google Scholar]
- 37.Cornista J, Ikeuchi S, Haruki M, Kohara A, Takano K, Morikawa M, Kanaya S. Cleavage of various peptides with pitrilysin from Escherichia coli: kinetic analyses using beta-endorphin and its derivatives. Biosci Biotechnol Biochem. 2004;68:2128–2137. doi: 10.1271/bbb.68.2128. [DOI] [PubMed] [Google Scholar]
- 38.Safavi A, Miller BC, Cottam L, Hersh LB. Identification of gamma-endorphin-generating enzyme as insulin-degrading enzyme. Biochemistry. 1996;35:14318–14325. doi: 10.1021/bi960582q. [DOI] [PubMed] [Google Scholar]
- 39.Csuhai E, Safavi A, Hersh LB. Purification and characterization of a secreted arginine-specific dibasic cleaving enzyme from EL-4 cells. Biochemistry. 1995;34:12411–12419. doi: 10.1021/bi00038a039. [DOI] [PubMed] [Google Scholar]
- 40.Kambacheld M, Augustin S, Tatsuta T, Stefan Muller S, Langer T. Role of the Novel Metallopeptidase MoP112 and Saccharolysin for the Complete degradation of Proteins Residing in Different Subcompartments of Mitochondria. J Biol Chem. 2005;280:20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- 41.Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996;15:4218–4229. [PMC free article] [PubMed] [Google Scholar]