Abstract
PDZK1 (also known as CAP70, NHERF3, NaPi-Cap1) is a scaffolding protein composed of four PDZ (Post-Synaptic Density-95, Discs Large, Zonula Occludens-1) domains followed by a short carboxyl-terminal tail. This scaffold acts as a mediator of localization and expression levels of multiple receptors in the kidney, liver and endothelium. Here, we characterize the self-association properties of the protein. PDZK1 can undergo modest homo-dimerization in vivo and in vitro through self-association involving its third PDZ domain. In addition, the tail of PDZK1 interacts in an intramolecular fashion with the first PDZ domain, but this interaction does not contribute to dimer formation. The interaction between the tail of PDZK1 and its first PDZ domain induces the protein to adopt a more compact conformation. A head-to-tail association has also been reported for EBP50/NHERF1, a two PDZ domain member of the same scaffolding protein family as PDZK1, and shown to regulate binding of target proteins to the EBP50 PDZ domains. As opposed to EBP50, the association of PDZK1 with specific ligands for its PDZ domains is unaffected by the intramolecular association, establishing a different mode of interaction among these two members of the same scaffolding family. However, the tail of PDZK1 interacts with the PDZ domains of EBP50, and this interaction is negatively regulated by the intramolecular association of PDZK1. Thus, we have uncovered a regulated association between the two PDZ-containing scaffolding molecules, PDZK1 and EBP50.
An essential aspect of cell organization is that proteins have to be properly localized through specific and tightly regulated associations. One of the major contributors to achieving this state is through scaffold proteins, which are modular molecules often made up of multiple protein-protein interaction domains that serve to precisely link other molecules together to regulate their functions. Among the most common protein-protein interaction modules in scaffold proteins is the PDZ (Post-Synaptic Density-95, Discs Large, Zonula Occludens-1) domain. These domains are primarily known for their ability to interact with carboxyl-terminal tails of target proteins, although some internal binding motifs have also been found (1–3). By interacting with the tails of proteins, PDZ-containing molecules can mediate the localization of their binding partners to specific subcellular destinations (2). Further, scaffold proteins are themselves regulated in various ways, such as by phosphorylation, degradation or targeting to specific locations. In addition, PDZ motifs can undergo hetero- and homo-dimerization or oligomerization, which may contribute to their functions by allowing the formation of larger, more extensive, scaffolds (4, 5).
There is a growing awareness of regulation through intramolecular associations, wherein a motif in a protein interacts with another domain within that same molecule (6, 7). Well-studied examples of regulation by intramolecular associations include the tyrosine kinase Src, the membrane-cytoskeletal linking Ezrin-Radixin-Moesin (ERM) proteins, and the Arp2/3 complex activator Wiskott Aldrich Syndrome protein (WASP) (7, 8). In these and many other cases the intramolecular interaction maintains the protein in an autoinhibited state, which is relieved by some regulatory stimulus. Intramolecular associations may be regulated by binding of outside elements, such as binding of active Cdc42 to WASP, or by modification via covalent linkages, such as ubiquitination, which induces an intramolecular association in the EPS15 endocytic adaptor protein (9, 10). Phosphorylation is also a common regulator of intramolecular associations, where it may serve to decrease such an association, as in the ERM proteins, or stimulate them, as in phosphorylation of Src at tyrosine 527 (7, 11).
PDZ-containing proteins have also been found to be regulated by an intramolecular association, specifically between their own tail and one of their PDZ domains. In the case of EBP50/NHERF1, a scaffolding protein with two PDZ domains and a carboxyl-terminal tail that ends in a PDZ-binding motif, the tail interacts with the second PDZ domain, and in so doing reduces access of target molecules to both PDZ domains (12). Another example is the X11α/Mint1 scaffold protein (13).
In this study we have analyzed the in vitro properties of PDZK1 (also called CAP70, NHERF3, or NaPi-Cap1), a scaffolding protein with 4 tandem PDZ domains. PDZK1 is a physiologically important molecule implicated in tissue-specific regulation of expression and localization of membrane-associated receptors and directly linked to maintenance of serum cholesterol levels (14–16). We show that PDZK1 is able to undergo both an intermolecular and an intramolecular association. Furthermore, the intramolecular association regulates the ability of its tail to interact with EBP50, thereby regulating the association between these two scaffold proteins.
Experimental Procedures
DNA constructs
Xpress-tagged PDZK1 constructs were generated by PCR amplification of human PDZK1 cDNA (Open Biosystems) and ligation into pCDNA3.1HisC (Invitrogen). For GFP-tagging, PDZK1 constructs were subcloned into pEGFP-C1 (Clontech). The vector pGEX-6P-1 (GE Healthcare Life Sciences) was used for expression of PDZK1 constructs as GST fusions. Mutants were generated using the Quikchange Site-Directed Mutagenesis Kit (Stratagene). PDZ-GEF1 and OCTN1 cDNAs were purchased from Open Biosystems and the tails were subcloned into pGEX-6P-1 for expression as GST fusions. The construct expressing the GST-tagged tail of rabbit NHE3 was a gift from Peter S. Aronson (Yale University School of Medicine; (17)). The GST-tagged EBP50 PDZ domains and pcDNA3.1 His/Xpress-EBP50 construct were as previously described (18, 19).
Antibodies and Materials
Omni-probe monoclonal and polyclonal antibodies were purchased from Santa Cruz Biotechnology. Protein A Sepharose, glutathione agarose and ExtrAvidin HRP were purchased from Sigma. Glutaraldehyde was obtained from Electron Microscopy Sciences. Monoclonal antibodies versus E-cadherin and PDZK1 were purchased from BD Transduction Laboratories. Goat anti-mouse IRDye 800CW infrared-labeled secondary antibody was purchased from LI-COR Biosciences. Polyclonal antibody against Green Fluorescent Protein was generated in rabbits by Abraham Hanono using standard protocols. The polyclonal antibody versus EBP50 was as previously described (20).
Cell Culture and Transfections
JEG3 cells were obtained from the American Type Culture Collection and maintained in a 5% CO2 humidified atmosphere at 37°C in MEM (Invitrogen) with glutamine, penicillin, streptomycin and 10% fetal bovine serum. Transfections were performed with polyethylenimine (PEI) as described (18).
Binding Assays and Electrophoresis
Biotin-labeled PDZK1 constructs were synthesized by in vitro transcription/translation in rabbit reticulocyte lysate using the TNT Coupled Reticulocyte Kit and Transcend tRNA (Promega). GST fusion proteins were prepared according to standard protocols. Cleavage of GST from fusions was performed by an overnight incubation of the fusions with the 3C protease at 4°C in cleavage buffer (50 mM HEPES pH 7.0, 1 mM EDTA, 150 mM NaCl, 1mM dithiothreitol).
For crosslinking experiments, soluble proteins were incubated in 0.1% glutaraldehyde for five minutes at room temperature then quenched with 100 mM Tris pH 7.4 for ten minutes, boiled in SDS sample buffer and analyzed by SDS-PAGE and Coomassie staining using 4–15% Precast Gels (BioRad). Controls were treated identically, but with the addition of phosphate buffered saline as a vehicle control without glutaraldehyde.
To perform in vitro binding studies, biotin-labeled proteins prepared by in vitro transcription/translation reactions were incubated with GST fusions for 2 hours in binding buffer (20 mM Tris pH 7.4, 100 mM NaCl, 0.1% Triton X-100, 5% glycerol and 0.1% β-mercaptoethanol), then washed in the same buffer and boiled in SDS sample buffer. Complexes were resolved by SDS-PAGE and analyzed by Western blotting with ExtrAvidin HRP. Alternatively, binding assays were also performed between GST-fusion proteins bound to glutathione agarose and soluble proteins purified from bacteria. These experiments were performed similarly except that binding was detected by SDS-PAGE and Coomassie staining. In vitro binding studies from lysates were performed by standard protocols. In short, transfected JEG3 cells were lysed in binding buffer (20 mM Tris pH 7.4, 100 mM NaCl, 1% Triton X-100, 5% glycerol, 0.1% β-mercaptoethanol and Complete Protease Inhibitor (Roche)), spun to remove cellular debris, incubated with the GST-fusion proteins for 2 hours, washed in wash buffer (identical to binding buffer but with Triton X-100 adjusted to 0.1%) and boiled in SDS sample buffer. The resulting complexes were analyzed by Western blotting. For immunoprecipitations, transfected JEG3 cells were lysed in immunoprecipitation buffer (20 mM Tris pH 7.4, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5% glycerol and Complete Protease Inhibitor (Roche)) and centrifuged to remove cellular debris. Supernatants were incubated with Omni-probe antibody and Protein A Sepharose for 3 hours rotating at 4°C, washed in immunoprecipitation buffer with the Triton X-100 adjusted to 0.1% and then the immune complexes were solubilized in SDS sample buffer and boiled. Results were analyzed by Western blotting.
For the determination of binding constants, a limiting amount of resin-bound probe was used to precipitate soluble PDZK1 or PDZK1ΔDTEM. This was performed throughout a range of concentrations for the soluble PDZK1 constructs and analyzed by Western blotting. The Western blotting was quantified using an Odyssey Infrared Imaging system (LI-COR Biosciences). A best-fit curve was then established for the results and used to estimate the dissociation constants.
Native polyacrylamide gel electrophoresis was performed as previously described (21). In short, samples were run at 4°C on 8% polyacrylamide gels prepared without SDS and without stacking gel layers. Samples were not boiled and SDS, bromophenol blue and β-mercaptoethanol were excluded from the sample buffer. The results were analyzed by Coomassie staining.
The PDZK1 tail interacts directly with PDZK1
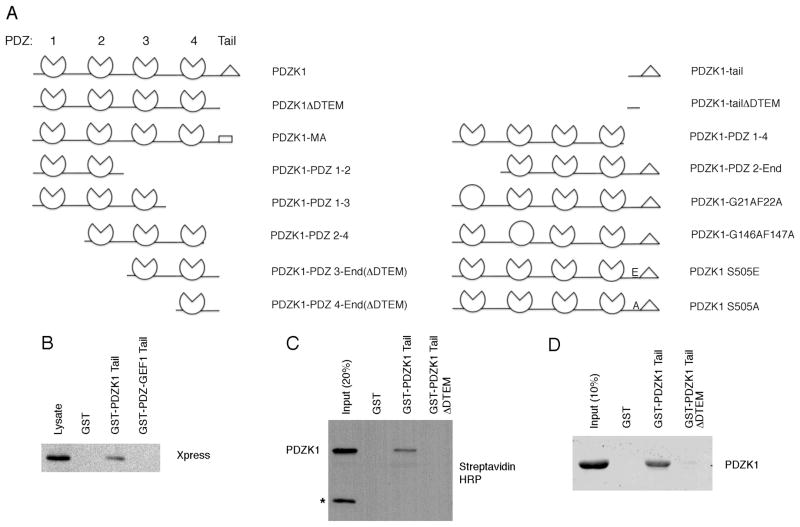
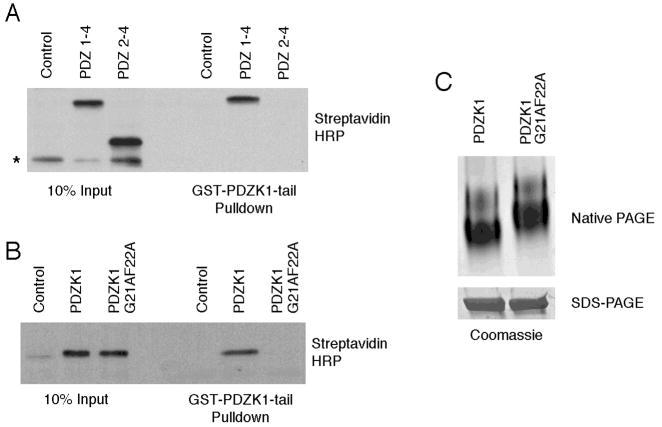
PDZK1 is a 519-residue protein containing four PDZ domains and ending in the sequence aspartic acid, threonine, glutamic acid and methionine (or DTEM) that we identified as a possible ligand for PDZ domains (Figure 1A shows all PDZK1 constructs used in this study). As some PDZ-containing proteins undergo an intramolecular interaction in which the carboxyl-terminal tail interacts with a PDZ domain, we therefore explored whether such an intramolecular interaction might exist in PDZK1. Recombinant protein consisting of GST fused to the 62-carboxyl-terminal residues of PDZK1 (GST-PDZK1-tail) was used as an affinity resin to recover binding proteins from JEG3 cells that had been transfected to express Xpress-tagged PDZK1. Xpress-PDZK1 was specifically recovered on the GST-PDZK1-tail construct, but not by GST alone (Figure 1B). As a control for specificity, the carboxyl-terminal tail of PDZ-GEF1 (amino acids 1432–1486), which ends in a strong consensus PDZ binding motif (valine, serine, alanine and valine, or VSAV), when fused to GST failed to precipitate PDZK1 (Figure 1B).
Figure 1. The PDZK1 tail interacts directly with PDZK1.
A. PDZK1 constructs used in this study. Pie shapes are used to represent functional PDZ domains while circles indicate PDZ domains that are no longer functional. The wedge shape represents a carboxyl-terminal tail containing the DTEM motif that is competent to bind PDZ domains. In the case of the PDZK1-MA mutant, the rectangle replaces the wedge at the carboxyl-terminus to illustrate that the tail is no longer able to bind PDZ domains. B. JEG3 cells were transfected with Xpress-tagged PDZK1 and subjected to pull-down assays with GST, GST-PDZK1-tail or GST-PDZ-GEF1-tail. Results were analyzed by Western blotting and indicated that the GST-PDZK1-tail specifically precipitates PDZK1. C. PDZK1 expressed as a biotin-labeled protein in an in vitro transcription/translation rabbit reticulocyte system was used in binding assays with GST, GST-PDZK1-tail or GST-PDZK1-tail ΔDTEM. The in vitro translated PDZK1 was found to interact directly with the PDZK1 tail and this was dependent on the presence of the PDZ binding motif DTEM on the carboxyl-terminus of the GST-PDZK1-tail. Note the presence of a background band at approximately 47 kDa that is not precipitated by the GST fusion proteins (denoted by an asterisk). D. Soluble PDZK1, purified from bacteria, was subjected to pulldowns with the indicated fusion proteins and analyzed by Western blotting. The results are consistent with those found in panel C.
To determine if the observed interaction is direct, the GST-PDZK1-tail fusion was also used to precipitate PDZK1 synthesized as a biotin-labeled protein in an in vitro transcription/translation rabbit reticulocyte system. The GST-PDZK1-tail fusion precipitated PDZK1, indicating a direct interaction between PDZK1 and its tail (Figure 1C). Further, the interaction requires the carboxyl-terminal four amino acids (DTEM), as a GST-PDZK1-tail construct lacking the terminal four residues (GST-PDZK1-tail ΔDTEM) failed to precipitate PDZK1 (Figure 1C). Similar experiments were performed using soluble PDZK1 purified from bacteria with identical results (Figure 1D).
PDZK1 dimerizes in vitro and in vivo
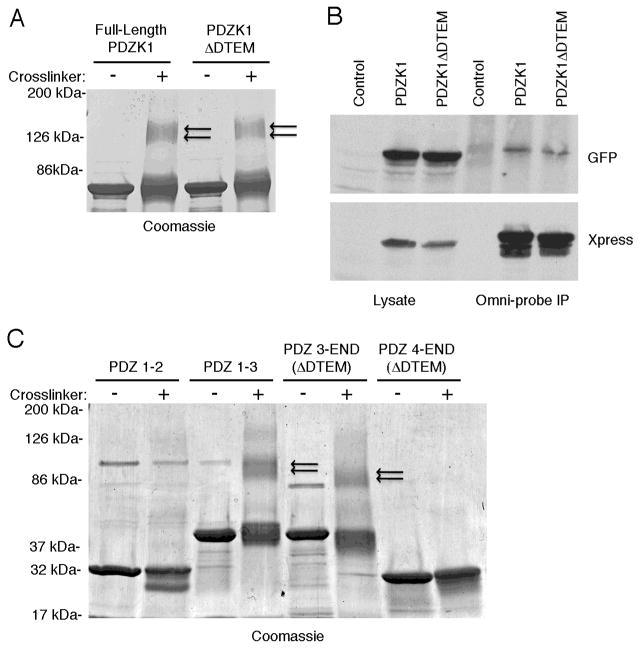
The ability of PDZK1 to interact with its own tail suggested the possibility that this association might mediate dimerization or oligomerization of the molecule. Purified recombinant PDZK1 was subject to chemical crosslinking to explore if dimers or higher oligomeric species exist. Analysis of the crosslinked material by SDS gel electrophoresis revealed the presence of both a monomeric species and a slower migrating species that would be consistent with the formation of a dimer; no higher oligomeric species were detected (Figure 2A). We next explored whether the carboxyl-terminal DTEM sequence is required for dimerization by subjecting recombinant PDZK1 lacking these carboxyl-terminal residues to the same crosslinking protocol. The same degree of crosslinked species was seen, indicating that the presence of these species is independent of the DTEM sequence, and not enhanced by it (Figure 2A). To determine whether PDZK1 might exist as dimers in cells, JEG3 cells were cotransfected to express both Xpress- and GFP-tagged PDZK1, the Xpress-tagged-PDZK1 was immunoprecipitated, and the precipitated material probed for GFP-PDZK1. Using this strategy, a modest amount of GFP-PDZK1 co-immunoprecipitated with Xpress-PDZK1 (Figure 2B). In a similar experiment, equivalent coprecipitation was found when constructs deleted for the carboxyl-terminal DTEM sequence were employed (Figure 2B). These co-immunoprecipitation experiments were performed from JEG3 cells, as they do not contain endogenous PDZK1 that may have confounded the results of this experiment. Our results indicate that PDZK1 has the ability to form dimers, albeit weakly, with the possibility that dimerization may be enhanced in vivo in some fashion.
Figure 2. PDZK1 dimerizes in vitro and in vivo.
A. Soluble wild-type PDZK1 and PDZK1 ΔDTEM proteins were crosslinked with 0.1% glutaraldehyde (or vehicle control) for 5 minutes then quenched with 100 mM Tris pH 7.4. The resulting complexes were solubilized in sample buffer and subjected to SDS-PAGE and Coomassie staining. Both the wild-type PDZK1 and PDZK1 ΔDTEM were found to dimerize (double arrows) to an equal extent, indicating that PDZK1 dimerization does not involve the carboxyl-terminal tail. B. JEG3 cells were transfected with either control DNA or Xpress and GFP tagged wild-type PDZK1 or PDZK1 ΔDTEM then subjected to Omni-probe (for the Xpress tag) immunoprecipitations 48 hours later. GFP-tagged PDZK1 variants co-immunoprecipitated with the Xpress-tagged constructs, thus indicating that PDZK1 dimerizes in vivo, independent of the tail of PDZK1. C. The indicated soluble PDZK1 constructs were subjected to in vitro crosslinking assays with 0.1% glutaraldehyde as in A. Proteins containing the third PDZ domain were able to dimerize (double arrows) while those constructs that lacked this domain did not associate with themselves.
Since PDZK1 dimer formation does not involve the carboxyl-terminal DTEM sequence, it may be through a PDZ-PDZ interaction, a possibility suggested for EBP50 (5, 22). To explore this scenario, we determined the ability of PDZK1 truncations to homo-dimerize as assessed by chemical crosslinking. A construct composed of PDZ domains 1–2 (amino acids 1–226) did not dimerize, while a molecule composed of PDZ domains 1–3 (amino acids 1–351) was able to self-associate (Figure 2C). Similarly, an amino-terminally truncated construct composed of PDZs 3 and 4 to the end of the protein (amino acids 227–515) was able to dimerize while a construct composed of just PDZ 4 to the end of the protein (amino acids 352–515) was not (Figure 2C). These two amino-terminal truncations also lacked the final four amino acids (DTEM) to avoid any spurious binding that may be contributed by this sequence in the context of truncation mutants. Taken together, the results indicate that the third PDZ domain of PDZK1 is the sole PDZ domain required for dimerization of the molecule. Furthermore, the specific abilities of the constructs composed of PDZs 1–3 and PDZs 3–4 to homo-dimerize indicate that PDZ 3 is likely associating with itself, or its flanking regions, as these are the only constant regions in these two constructs.
The conformation of PDZK1 is regulated by an intramolecular association with its tail
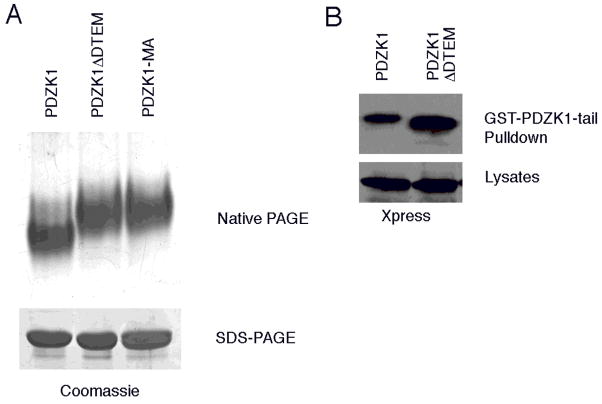
The ability of the GST-PDZK1-tail to bind PDZK1 (Figure 1), which is unrelated to the formation of dimers documented above (Figure 2), suggests the possibility of an intramolecular association. As an intramolecular interaction of this kind might affect the conformation of PDZK1, we compared the migration of PDZK1 and mutants lacking this interaction by native gel electrophoresis, which can reveal differences in molecular shape. Wild-type PDZK1, PDZK1 ΔDTEM and PDZK1-MA, a mutant in which an alanine was added after the carboxyl-terminal methionine as an alternate way to disrupt the PDZ binding motif (19), were analyzed by native PAGE. Although these constructs migrate identically on conventional denaturing SDS-PAGE, both the PDZK1 ΔDTEM and PDZK1-MA mutants exhibited a slower migration than wild-type on native gels (Figure 3A). Since all three of these constructs have similar molecular weights and isoelectric points, and more compact proteins migrate faster in native gels, the results indicate that the intramolecular interaction mediated by the tail induces a more compact conformation for PDZK1.
Figure 3. The conformation of PDZK1 is regulated by an intramolecular association with its tail.
A. Soluble PDZK1 wild-type, ΔDTEM and MA fusions were expressed, purified from bacteria and subjected to electrophoresis under native conditions or by standard SDS-PAGE. Both the PDZK1 ΔDTEM and MA variants ran more slowly than the wild-type PDZK1 under native conditions, a result indicative of a more open conformation adopted by these mutants. B. JEG3 cells were transfected with Xpress-tagged PDZK1 or PDZK1 ΔDTEM then allowed to express the constructs for 48 hours. The cells were then lysed and the lysates subjected to binding assays with the GST-PDZK1-tail fusion. Analysis of the precipitated proteins by Western blotting showed that the PDZK1 ΔDTEM protein interacts with the GST-PDZK1-tail fusion more efficiently than wild-type PDZK1 does.
The data suggest that in wild-type PDZK1, the tail may be bound to a PDZ domain in the molecule. If the tail occupies this site, wild-type PDZK1 might be expected to bind less efficiently to the GST-PDZK1-tail than a PDZK1 ΔDTEM construct. Accordingly, JEG3 cells expressing either tagged wild-type PDZK1 or PDZK1 ΔDTEM were lysed and the ability of the tagged protein to be recovered on immobilized GST-PDZK1-tail was analyzed. The GST PDZK1-tail fusion was found to be more efficient at recovering the PDZK1 ΔDTEM construct than the wild-type PDZK1 protein (Figure 3B). Interestingly, this effect is pronounced in pulldowns performed from cell lysates, but is not readily apparent when the GST PDZK1-tail fusion is used to precipitate PDZK1 and PDZK1 ΔDTEM purified from bacteria (data not shown and Figure 6D). Thus, it is likely that some additional regulation of this conformation occurs in mammalian cells that is absent from the in vitro system. Possibilities include an as yet undetermined post-translational modification such as phosphorylation or possibly the binding of some cytosolic factor.
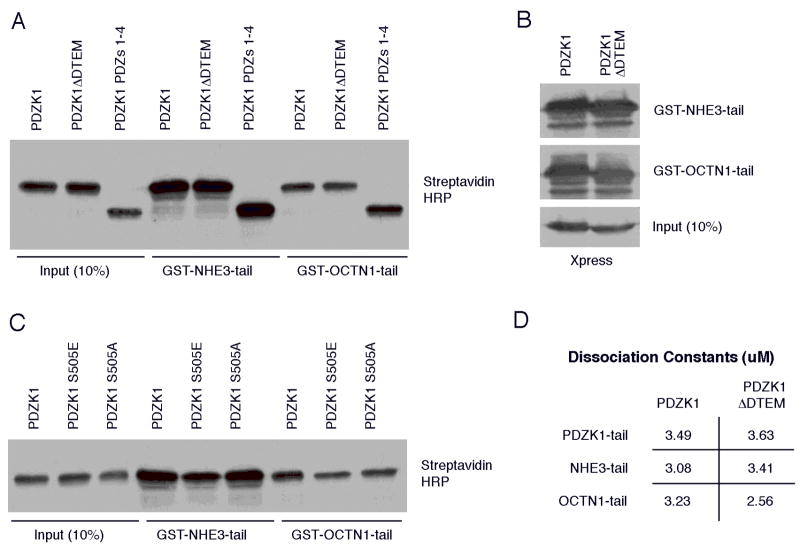
Figure 6. The presence or absence of the PDZK1 tail does not appreciably affect binding to PDZ 1 or PDZ 2.
A. PDZK1 or the indicated mutants were expressed as biotin-labeled proteins using a rabbit reticulocyte in vitro transcription/translation system then subjected to pulldowns with the either the GST-tagged tail of NHE3 or OCTN1 conjugated to glutathione agarose. The complexes were then analyzed by SDS-PAGE and Western blotting using Streptavidin HRP to detect biotin-labeled proteins. The PDZK1 constructs were each able to efficiently interact directly with the GST-fusion proteins. B. The indicated GST fusion proteins were used to precipitate either full-length PDZK1 or PDZK1 ΔDTEM from cell lysates. Similar to the in vitro results observed in panel A, both of these PDZK1 constructs bound similarly to the resin-bound probes. C. In a similar protocol to that used in A, the PDZK1 S505E exhibited a marginal loss of binding to the NHE3 tail as compared to wild-type PDZK1, but overall the phospho-mimetic mutants bound similarly to the wild-type protein. D. Estimated dissociation constants for the indicated interactions as detailed in Experimental Procedures.
The PDZK1 tail interacts with the first PDZ domain of PDZK1
Our findings that the presence of the PDZK1 carboxyl-terminal DTEM sequence is necessary for the tail to bind PDZK1 (Figure 1C) and affects the protein’s conformation (Figure 3A), but does not contribute to dimerization (Figure 2A), suggested a canonical intramolecular PDZ interaction. To further test this, a truncation approach was used to determine which PDZ domain of PDZK1 might be able to interact with the PDZK1 tail. In vitro binding experiments were performed between in vitro transcription/translation reaction products of PDZK1 truncations and the GST-PDZK1-tail fusion. A biotin-labeled PDZK1 protein composed of PDZ domains 1–4 was found to be precipitated by the GST-PDZK1-tail fusion while a protein consisting of PDZ domains 2–4 was not, suggesting that the first PDZ domain of PDZK1 might be the domain responsible for tail binding (Figure 4A). As an alternate means of testing this finding, a construct with a non-functional first PDZ domain was generated to test binding to the GST-PDZK1-tail fusion. The mutations glycine 21 to alanine and phenylalanine 22 to alanine (G21AF22A) were introduced into the GLGF carboxylate-binding loop of the first PDZ domain of full length PDZK1 (23). This mutant was unable to bind the GST-PDZK1-tail fusion, indicating that the tail binds the first PDZ of PDZK1 using the same binding pocket generally used by target proteins of PDZ domains (Figure 4B). If this model is correct, the PDZK1-G21AF22A mutant protein is predicted to have a more extended conformation than wild-type PDZK1. Native gel electrophoresis of these proteins shows that indeed the PDZK1-G21AF22A mutant migrates more slowly than wild-type PDZK1 and is therefore more extended (Figure 4C).
Figure 4. The PDZK1 tail interacts with the first PDZ domain of PDZK1.
A. The indicated constructs were expressed as biotin-labeled proteins using a rabbit reticulocyte in vitro transcription/translation system. A GST-PDZK1-tail fusion protein was used to precipitate proteins and complexes were then analyzed by SDS-PAGE and Western blotting using Streptavidin HRP to detect biotin-labeled proteins. The results showed that the first PDZ of PDZK1 is required for interaction with the tail of PDZK1. The asterisk indicates a background band in the lysates that is not precipitated by the GST fusion. B. In vitro transcription/translation reactions were performed with the indicated constructs then binding assays were performed and analyzed as in panel A. A functional PDZ 1 is required for full-length PDZK1 to interact with the tail of PDZK1, as mutation of the carboxylate-binding loop of PDZ 1 (G21AF22A) inhibited the association. C. Soluble wild-type PDZK1 and the G21AF22A mutant were subjected to gel electrophoresis under native conditions to test for a conformational switch. The mutant exhibited slower migration than the wild-type protein, confirming that the first PDZ was involved in an intramolecular association with the tail to regulate the conformation of PDZK1. Both the wild-type PDZK1 and PDZK1 G21AF22A ran identically under standard SDS-PAGE conditions.
The tails of NHE3 and OCTN1 are specific binding partners of PDZK1’s PDZ 1 and PDZ 2, respectively
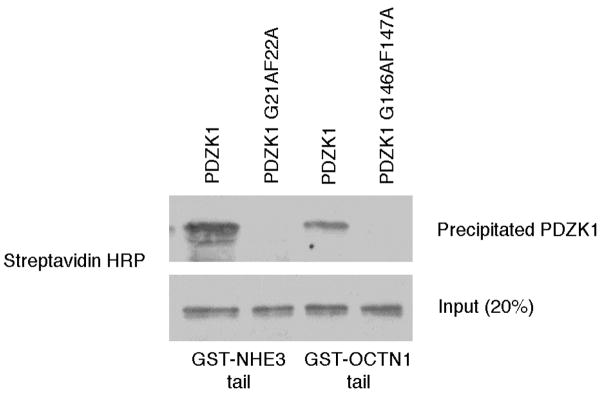
We next wanted to determine if the intramolecular association of PDZK1 affected interactions with its PDZ domain ligands. To do this, we sought specific binding partners of the PDZ domains. The first PDZ domain has a distinctive specificity with the tails of binding partners ending in the following sequences: -STHM (NHE3), -STPM (MAP17), and –DTEM (PDZK1 tail), indicating that its preferred binding motif is X-T-X-M (with X representing a variable residue) (24–26). The second PDZ domain also has a binding partner, OCTN1, with an atypical tail, ending in an –ITAF motif (26). To test the specificity of these interactions, in vitro binding assays were performed between the tails of NHE3 and OCTN1 fused to GST and either wild-type PDZK1 or full-length constructs with mutated binding pockets in PDZs 1 (G21AF22A) or 2 (G146AF147A). The PDZK1 G21AF22A mutant failed to bind NHE3 and the PDZK1 G146AF147A mutant failed to bind OCTN1, thus confirming that these are the sole PDZ domain binding sites for these proteins and establishing them as specific probes for the binding properties of the first 2 PDZ domains of PDZK1 (Figure 5). It should be noted that the G21AF22A mutant conformationally opens PDZK1 (Figure 4), and could thus affect binding to NHE3, but data to be presented below (Figure 6) show that this is not the case. Further, NHE3 has been previously reported to be a specific binding partner for PDZ 1 (26), and shares a related tail sequence with other PDZ 1 binding partners, as mentioned above.
Figure 5. The tails of NHE3 and OCTN1 are specific binding partners of PDZK1’s PDZ 1 and PDZ 2, respectively.
PDZK1 or the indicated mutants were expressed as biotin-labeled proteins using a rabbit reticulocyte in vitro transcription/translation system then subjected to pulldowns with the indicated GST constructs. The complexes were then analyzed by SDS-PAGE and Western blotting using Streptavidin HRP to detect biotin-labeled proteins. The G21AF22A and G146AF147A mutations inhibited binding to the tails of NHE3 and OCTN1, respectively.
Conversely, PDZs 3 and 4 share a common binding motif: X-T-K/R-L, and we were unable to find specific binding partners for these PDZs (data not shown) (26). Therefore, the remainder of this study focuses on regulation of binding of the first 2 PDZ domains. It should also be noted that although the binding partners of PDZ 3 and 4 that we tested bind without complete specificity in vitro, this may not be the case in vivo.
The PDZK1 intramolecular association does not appreciably affect binding to PDZ 1 or PDZ 2
The tail of PDZK1 interacts with the first PDZ in an intramolecular association, regulating the conformation of the molecule. Other molecules that undergo an intramolecular interaction, such as the Ezrin-Radixin-Moesin proteins, have been proven to exhibit altered binding to target proteins based on conformation (8, 27, 28). This suggests a similar possibility for PDZK1. Accordingly, we tested the abilities of PDZK1, PDZK1 ΔDTEM or a construct consisting of just PDZs 1–4 and lacking the entire tail, to interact with the tails of NHE3 and OCTN1. The interactions were tested using pulldowns of the soluble PDZK1 constructs generated in rabbit reticulocyte and each interacted equivalently with tails of OCTN1 and NHE3 fused to GST (Figure 6A). As the tail of PDZK1 is able to precipitate the PDZK1 ΔDTEM variant more efficiently than wild-type PDZK1 from cell lysates (Figure 3B), pulldowns using the tails of NHE3 and OCTN1 fused to GST were performed from JEG3 cells transfected with either full-length PDZK1 or the PDZK1 ΔDTEM mutant. However, similar to the results from rabbit reticulocyte (Figure 6A), each PDZK1 variant bound both fusions equally well (Figure 6B). It may be that preferential binding exists between these PDZK1 variants in vivo for some binding partners. However, for the binding partners tested here, PDZK1’s intramolecular association does not markedly affect ligand binding to PDZ 1 or PDZ 2.
The phosphorylation of serine 509 in the tail of rat PDZK1 (equivalent to serine 505 in the human isoform used here) is the only documented post-translational modification for the protein. This phosphorylation is involved in regulation of total levels of the Scavenger Receptor β1, a PDZ 1 binding partner of PDZK1 (29). We therefore generated both phosphomimetic (S505E) and non-phosphorylatable (S505A) constructs to determine if they exhibited altered binding for PDZ 1 or 2. To do this, wild-type PDZK1 and the two phospho-mutants were expressed in rabbit reticulocyte and pulldowns were performed with GST-NHE3 tail and GST-OCTN1 tail. The phosphomimetic S505E mutant exhibited a very mildly reduced binding to a PDZ 1 binding partner, but not to a partner of PDZ 2 (Figure 6C). Thus, the phosphorylation of the PDZK1 tail does not appear to be a major regulator of the affinity of its PDZ domains for exogenous ligands under the conditions tested.
To more thoroughly access the possible role of the PDZK1 tail in regulating the molecule’s association with binding partners we estimated the dissociation constants for the interactions between full-length PDZK1 and PDZK1ΔDTEM and the tails of NHE3, OCTN1 and PDZK1. To do this, a limiting amount of the indicated resin-bound tail (NHE3, OCTN1 or PDZK1) was used to precipitate PDZK1 over a wide concentration range that included saturating amounts of the soluble proteins. Results were then quantitated using an Odyssey Infrared Imaging System and fit to curves to determine representative binding dissociation constants (Figure 6D). The data illustrate that there are no appreciable differences between the ability of PDZK1 and PDZK1ΔDTEM to bind each of these probes. Furthermore, PDZK1 binds nearly equivalently to each of these proteins (Figure 6D).
The PDZK1 tail interacts with EBP50
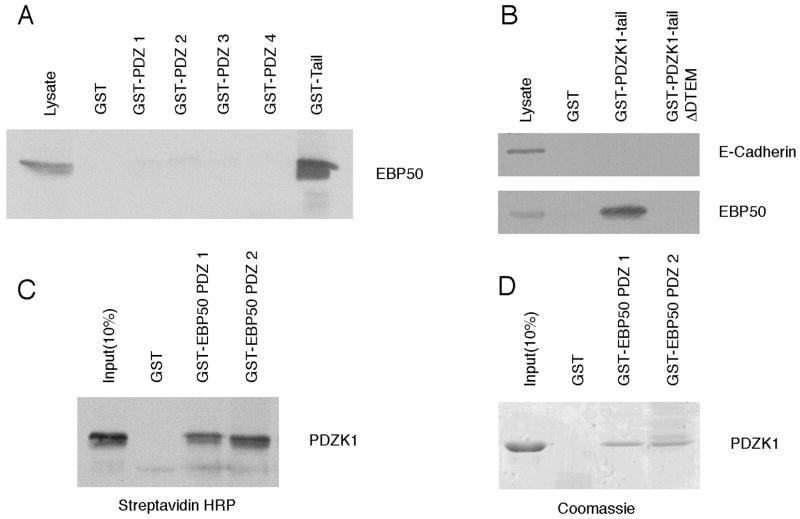
As PDZK1’s intramolecular association had no significant effect on the interactions between its PDZ domains and their ligands, we sought to determine if it may affect binding of the PDZK1 tail to other proteins. There are no known binding partners for this region of PDZK1. However, it has been published that a construct consisting of PDZ 4 to the end of PDZK1 interacts with the PDZ-domain containing protein EBP50 in a yeast 2-hybrid screen, possibly through a PDZ-PDZ interaction (26). An alternative possibility is that the tail of PDZK1 can interact with the PDZ domains of EBP50. To test this, pulldowns with either the tail or PDZ domains of PDZK1 fused to GST were performed from JEG3 lysates. Indeed, it was found that the tail of PDZK1 was able to precipitate endogenous EBP50, while the PDZ domains did not (Figure 7A). EBP50 binding to the PDZK1 tail depends on the DTEM motif (Figure 7B). To confirm that this interaction is direct and to determine which of EBP50’s PDZ domains is involved, pulldowns were performed from rabbit reticulocyte lysate expressing biotin-labeled PDZK1. Surprisingly, it was found that the first and second PDZ domains of EBP50 were each able to precipitate PDZK1 to an equal, yet modest extent (Figures 7C and 7D).
Figure 7. The PDZK1 tail interacts with EBP50.
A. The indicated domains of PDZK1 were expressed as GST fusion proteins and used to perform pulldowns from JEG3 lysates. The tail of PDZK1 was able to efficiently precipitate endogenous EBP50. The lysate lane was equivalent to 10% of the protein used per pulldown. B. In a similar experiment, the carboxyl-terminal DTEM motif of the PDZK1 tail is needed for its interaction with EBP50, with E-Cadherin serving as a negative control. C. PDZK1 was expressed as a biotin-labeled protein in rabbit reticulocyte and subjected to pulldowns against either GST, or the GST-conjugated PDZ domains of EBP50. Both the first and second PDZ domains of EBP50 were able to interact directly with PDZK1. D. Soluble PDZK1, purified from bacteria, was subjected to pulldowns with the indicated fusion proteins and analyzed by SDS-PAGE and Coomassie staining. The results are consistent with those found in panel C.
PDZK1’s intramolecular interaction regulates its association with EBP50
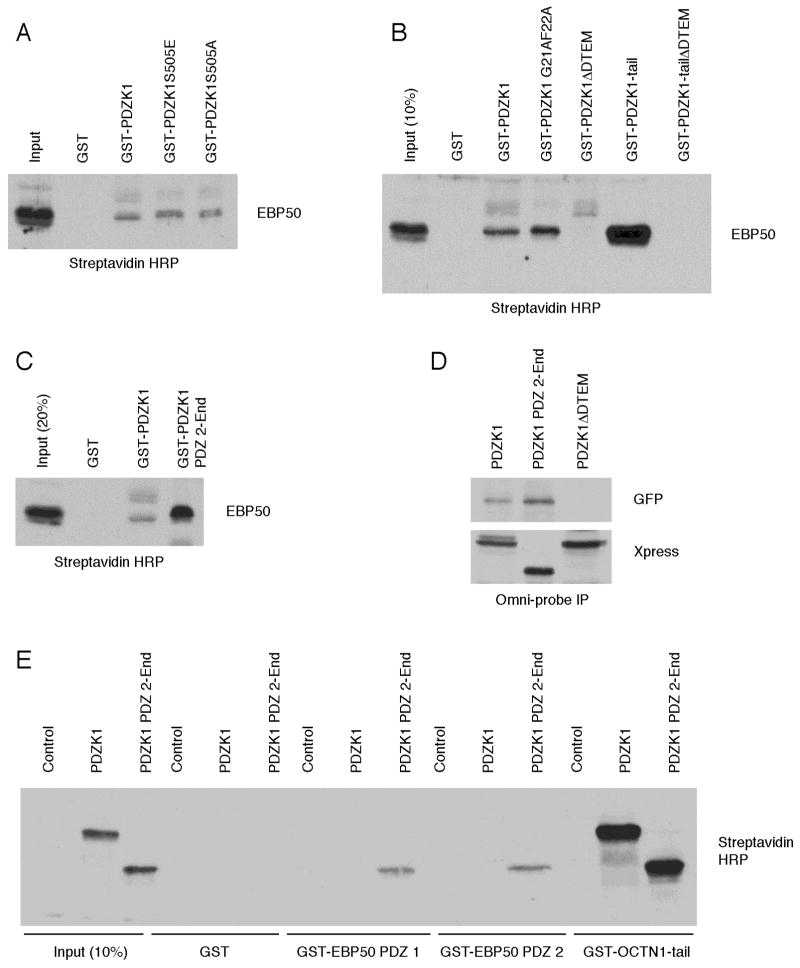
As a first step toward investigating possible regulation of the interaction between PDZK1 and EBP50, we tested the ability of the phospho-mutants of PDZK1 to interact with EBP50. EBP50 was synthesized in the rabbit reticulocyte system and subjected to pulldowns by wild-type or phospho-mutant full-length PDZK1 GST fusion proteins coupled to glutathione agarose. Each of the PDZK1 mutants interacted equivalently with EBP50 in vitro, indicating that phosphorylation of serine 505 is unlikely to regulate the interaction between PDZK1 and EBP50 (Figure 8A).
Figure 8. PDZK1’s intramolecular interaction regulates its association with EBP50.
A. EBP50 was expressed as a biotin-labeled protein in rabbit reticulocyte and subjected to pulldowns versus either GST, as a control, wild-type PDZK1 or the phospho-mimetic PDZK1 mutants, S505E and S505A. The complexes were then analyzed by SDS-PAGE and Western blotting using Streptavidin HRP to detect biotin-labeled proteins. Each of the PDZK1 fusions was able to interact equivalently with EBP50. B. GST pulldowns were performed as in A with the indicated constructs and analyzed via Western blotting with Streptavidin HRP. Note that the amounts of GST fusions used per pulldown were adjusted to ensure that the same amount of PDZK1 tail was included in each. Full-length PDZK1 was less efficient than the PDZK1 tail at precipitating EBP50, indicating an intramolecular regulation of PDZK1’s ability to bind EBP50. The absence of the DTEM motif causes loss of the interaction. C. The indicated GST-fusions were utilized to precipitate EBP50 that had been generated in vitro. The loss of the first PDZ domain of PDZK1 increased the binding to EBP50. D. GFP-EBP50 was co-expressed with the indicated Xpress-tagged PDZK1 constructs in JEG3 cells followed by Omni-probe (anti-Xpress) immunoprecipitations. Similar to in vitro results, the PDZK1 PDZ 2-End construct co-precipitated EBP50 more efficiently than the wild-type PDZK1 did, while the PDZK1 construct lacking the DTEM motif did not interact with EBP50. E. Full-length PDZK1, PDZK1 PDZ 2-End or non-coding control DNA were used to express biotin-labeled proteins in the rabbit reticulocyte system. The generated proteins were then subjected to pulldowns with the indicated GST constructs. As in previous experiments, the truncated PDZK1 protein interacted more efficiently with the EBP50 PDZ domains than full-length PDZK1 did. A GST-OCTN1 construct was able to precipitate both PDZK1 constructs efficiently, indicating that the increased binding of PDZK1 PDZ 2-End as opposed to wild-type PDZK1 is not general but instead specific to binding partners of the PDZK1 tail.
Since PDZK1 undergoes an intramolecular association between its first PDZ domain and its tail, it seemed likely that the ability of the tail to bind EBP50’s PDZ domains might be influenced by the intramolecular association. Thus, the ability of in vitro-generated EBP50 to bind a variety of GST-coupled PDZK1 constructs was tested (Figure 8B). The amount of each of the GST fusion proteins was adjusted to normalize the amount of PDZK1 tail present per pulldown. The tail of PDZK1 is far more competent than the full-length protein to precipitate EBP50 (compare lanes 3 and 6, Figure 8B), suggesting that the intramolecular association present in the full-length PDZK1 protein reduces the ability of the tail to interact with the PDZ domains of EBP50. As expected, loss of the DTEM motif eliminates the ability of PDZK1 constructs to interact with EBP50 (Figure 8B). Curiously, however, the PDZK1 G21AF22A mutant, which has a defective PDZ 1 and therefore the tail should be free to bind EBP50, exhibits similar binding to EBP50 as wild-type PDZK1 (Figure 8B). Thus, loss of binding between the tail of PDZK1 and the carboxylate-binding loop of PDZ 1 does not fully release the tail to bind other proteins, although PDZK1 is in a more open conformation (Figure 4C). This means that other residues in PDZK1 are also involved in limiting the accessibility of the PDZK1 tail. To define the area of PDZK1 involved in inhibition of tail binding, mutants were made that contained amino-terminal truncations of the PDZ domains. Interestingly, removal of the first PDZ domain from PDZK1 completely released the inhibition of tail binding (Figure 8C). Thus, there may be secondary interactions between the first PDZ domain of PDZK1 and its tail in addition to the traditional GLGF motif-mediated interaction. This result was reproduced in vivo through co-immunoprecipitations from JEG3 cells, where the PDZK1 PDZ 2-End construct was more efficient than the full-length protein at precipitating EBP50 (Figure 8D). As expected, a PDZK1 mutant lacking the DTEM motif does not precipitate EBP50 (Figure 8D). We then wished to determine if the tail-PDZ 1 interaction that regulates the binding to EBP50 also affects PDZ ligands for PDZK1. We therefore assessed the ability of GST-EBP50 PDZ 1, GST-EBP50 PDZ 2 and the GST-OCTN1-tail, which binds the second PDZ of PDZK1, to bind in vitro synthesized full length PDZK1 and the PDZ 2-End construct. The results confirm that PDZK1 PDZ 2-End interacts better than wild-type PDZK1 with the PDZ domains of EBP50 (Figure 8E, note that at this length exposure the full-length PDZK1 is not readily apparent in the GST-EBP50 PDZ pulldowns, but appears on a longer exposure (data not shown)). Importantly, both of the PDZK1 constructs were efficiently recovered by the GST-OCTN1-tail, illustrating that the regulation of binding between the PDZK1 tail and PDZ domains of EBP50 is a phenomenon specific to the tail that does not extend to the PDZ domains of PDZK1 (Figure 8E).
Discussion
In this study we present the first molecular characterization of the scaffolding protein PDZK1. We first show that PDZK1 is able to homo-dimerize in a manner that is not dependent upon its carboxyl-terminal tail. Second, we show that the tail of PDZK1 undergoes an intramolecular association with the first PDZ. This interaction induces the molecule to adopt a more compact conformation. Interestingly, we found no evidence for head-to-tail anti-parallel dimerization, suggesting that dimers in which the tail of each molecule associates with the first PDZ domain of the other either cannot form, or are much less stable than intramolecularly associated monomers. We also find that the tail of PDZK1 interacts with the PDZ scaffolding protein EBP50 and that this interaction is regulated by the presence of the first PDZ domain of PDZK1.
The ability of the PDZK1 tail to interact with PDZ 1 suggests the possibility that there may be an effect on the ability of PDZK1 to bind its various PDZ ligands. This is especially important considering the critical role of PDZ 1 in regulation of the scavenger receptor, class B, type I in the liver (30). Such a method of regulation is seen in EBP50, which has been shown to undergo an intramolecular self-association between its tail and second PDZ domain (12). This interaction was found to reduce binding to both PDZ domains of EBP50, as an EBP50 ΔSNL (analogous to the PDZK1 ΔDTEM mutant) mutant was better able to bind both PTEN and β-catenin, known binding partners of PDZ 1 and 2, respectively (12). However, we were unable to find a similar regulation of binding of the tails of ligands to the PDZ domains in PDZK1, indicating that this molecule has an alternate mode of regulation to that reported for EBP50. Among the possible alternatives are trivial explanations, such as regulated binding to PDZK1 is dependent on specific binding partners that are not represented in the tested ones. However, it seems equally likely that our results point to an alternative mode of regulation.
One interesting possible function for the conformational change of PDZK1 is that it may regulate the spacing of binding proteins in relation to one another without significantly affecting their binding affinity for PDZK1. For example, PDZ domains of PDZK1 have been shown to bind disparate signaling molecules, such as the Ste20-Related Kinase SLK, Ubiquitin-conjugating enzyme 9, the cystic fibrosis transmembrane conductance regulator (CFTR) and the somatostatin receptor (26, 31, 32). There may be functional crosstalk in signaling pathways, such as sumoylation of receptors or activation of kinases by extracellular ligation of receptors, that is dependent on PDZK1’s role as a scaffold protein. For instance, the closed conformation of PDZK1 may bring a receptor into contact with a kinase, while the open conformation separates these molecules. Indeed, PDZK1 has recently been shown to be an important component of the functional crosstalk between the CFTR and the multidrug resistance protein 4 (MRP4) in the gut epithelium (33).
By contrast, the intramolecular association of PDZK1 clearly regulates its interaction with the microvillus protein EBP50. An interesting aspect of this finding involves the fact that EBP50 has also been found to undergo a head-to-tail intramolecular interaction to regulate its interaction with its PDZ binding partners, as well as being involved with its association with the microvillar ERM proteins (12). Furthermore, the ERM proteins were among the first molecules found to undergo an intramolecular interaction involving their amino-terminal FERM (band 4.1, Ezrin, Radixin, Moesin) domains and their carboxyl-terminal C-ERMAD (Carboxyl-terminal ERM Association Domain) domains, which regulates their association with EBP50, the plasma membrane and filamentous-actin (8, 27, 28). Taken together with the results presented herein, there may be a macromolecular complex found in microvilli consisting of PDZK1, EBP50, Ezrin and filamentous-actin, as well as membrane-associated proteins, such as NHE3. The core components of this complex, PDZK1, EBP50, and Ezrin, can each undergo intramolecular conformational changes wherein they open and then interact more strongly with one another (Figure 9). For example, one could envision NHE3 interacting with the first PDZ domain of PDZK1, thus releasing its tail to interact with EBP50, leading to a release of the EBP50 tail to interact with Ezrin, which could then stabilize the active form of Ezrin allowing its carboxyl-terminus to bind filamentous-actin, thereby creating a domino-effect of conformational change that nucleates the assembly of a large structural complex. Alternatively, this complex could be nucleated with Ezrin as the upstream component, as illustrated in Figure 9.
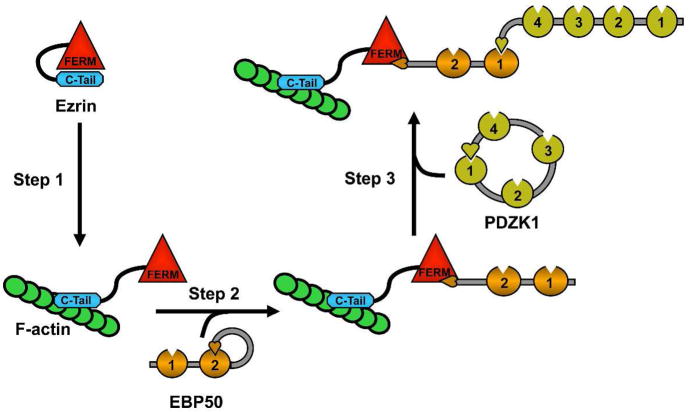
Figure 9. A potential mechanism for the formation of a macromolecular complex composed of F-actin, Ezrin, EBP50 and PDZK1.
The ERM protein Ezrin is well characterized for its ability to exist in an auto-inhibited state wherein its amino-terminal FERM domain is associated with its carboxyl-terminus (C-Tail) in an intramolecular association. Upon activation by phosphorylation, this intramolecular association is released and the carboxyl-terminus is able to bind F-actin and the FERM domain is free to associate with EBP50 (Step 1). EBP50 exists in an auto-inhibited state wherein its tail is bound intramolecularly to its second PDZ domain (PDZ domains are denoted by pie shapes), thus inhibiting binding of target molecules to both PDZs. This inhibition is released upon binding of the Ezrin FERM domain to the EBP50 tail (Step 2). The PDZ domains of EBP50 are then free to bind the tail of PDZK1, thereby releasing the intramolecular association between the first PDZ of PDZK1 and the carboxyl-terminal tail (Step 3), although the functional consequences of the opening of PDZK1 have yet to be resolved. Thus, there is likely to exist a progressive mechanism whereby the opening of one of these molecules may stabilize the opening of the next, thus nucleating the assembly of a large, macromolecular, structural complex.
We have also found that PDZK1 is able to homo-dimerize, although to a limited extent. It may be that the amount of dimerization observed in vitro, or in vivo in non-stimulated cells, is only indicative of a low, baseline level of self-association that may be regulated in vivo by some stimulus. As the tail of PDZK1 does not mediate this, it involves a PDZ-PDZ interaction. Thus, it is expected that the dimeric form of the scaffold protein may not interact with some PDZ-domain binding partners, thereby providing a functional difference between monomeric and dimeric PDZK1 in vivo.
The work reported here shows that PDZK1 is more than a simple scaffolding protein as it can participate in both intramolecular and intermolecular self-associations. Further, the ability to undergo an intramolecular association regulates its ability to directly interact with the microvillar scaffold protein EBP50. Future studies investigating the physiological role of this protein will need to take into account these newly discovered properties.
Acknowledgments
This work was supported by the National Institutes of Health Grant GM036652 to AB and the National Research Service Award Postdoctoral Fellowship GM080847 to DPL.
The authors thank Peter S. Aronson for supplying pGEX-6P-2 rabbit NHE3 tail and Abraham Hanono for generating polyclonal antibody versus GFP.
Abbreviations
- EBP50
Ezrin Binding Phosphoprotein 50
- ERM
Ezrin Radixin Moesin
- GFP
Green Fluorescent Protein
- HRP
Horseradish Peroxidase
- NHERF
Sodium Hydrogen Exchanger Regulatory Factor
- PDZ
Post-Synaptic Density-95, Discs Large, Zonula Occludens-1
- SDS PAGE
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
References
- 1.Christopherson KS, Hillier BJ, Lim WA, Bredt DS. PSD-95 assembles a ternary complex with the N-methyl-D-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J Biol Chem. 1999;274:27467–27473. doi: 10.1074/jbc.274.39.27467. [DOI] [PubMed] [Google Scholar]
- 2.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 3.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 4.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 5.Fouassier L, Yun CC, Fitz JG, Doctor RB. Evidence for ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) self-association through PDZ-PDZ interactions. J Biol Chem. 2000;275:25039–25045. doi: 10.1074/jbc.C000092200. [DOI] [PubMed] [Google Scholar]
- 6.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14:690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 8.Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- 9.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 10.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 11.Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- 12.Morales FC, Takahashi Y, Momin S, Adams H, Chen X, Georgescu MM. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol Cell Biol. 2007;27:2527–2537. doi: 10.1128/MCB.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long JF, Feng W, Wang R, Chan LN, Ip FC, Xia J, Ip NY, Zhang M. Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem. Nat Struct Mol Biol. 2005;12:722–728. doi: 10.1038/nsmb958. [DOI] [PubMed] [Google Scholar]
- 14.Kocher O, Pal R, Roberts M, Cirovic C, Gilchrist A. Targeted disruption of the PDZK1 gene by homologous recombination. Mol Cell Biol. 2003;23:1175–1180. doi: 10.1128/MCB.23.4.1175-1180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kocher O, Yesilaltay A, Cirovic C, Pal R, Rigotti A, Krieger M. Targeted disruption of the PDZK1 gene in mice causes tissue-specific depletion of the high density lipoprotein receptor scavenger receptor class B type I and altered lipoprotein metabolism. J Biol Chem. 2003;278:52820–52825. doi: 10.1074/jbc.M310482200. [DOI] [PubMed] [Google Scholar]
- 16.Silver DL, Wang N, Vogel S. Identification of small PDZK1-associated protein, DD96/MAP17, as a regulator of PDZK1 and plasma high density lipoprotein levels. J Biol Chem. 2003;278:28528–28532. doi: 10.1074/jbc.M304109200. [DOI] [PubMed] [Google Scholar]
- 17.Thomson RB, Wang T, Thomson BR, Tarrats L, Girardi A, Mentone S, Soleimani M, Kocher O, Aronson PS. Role of PDZK1 in membrane expression of renal brush border ion exchangers. Proc Natl Acad Sci U S A. 2005;102:13331–13336. doi: 10.1073/pnas.0506578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reczek D, Bretscher A. Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J Cell Biol. 2001;153:191–206. doi: 10.1083/jcb.153.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bretscher A, Gary R, Berryman M. Soluble ezrin purified from placenta exists as stable monomers and elongated dimers with masked C-terminal ezrin-radixin-moesin association domains. Biochemistry. 1995;34:16830–16837. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- 22.Shenolikar S, Minkoff CM, Steplock DA, Evangelista C, Liu M, Weinman EJ. N-terminal PDZ domain is required for NHERF dimerization. FEBS Lett. 2001;489:233–236. doi: 10.1016/s0014-5793(01)02109-3. [DOI] [PubMed] [Google Scholar]
- 23.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 24.Kocher O, Comella N, Gilchrist A, Pal R, Tognazzi K, Brown LF, Knoll JH. PDZK1, a novel PDZ domain-containing protein up-regulated in carcinomas and mapped to chromosome 1q21, interacts with cMOAT (MRP2), the multidrug resistance-associated protein. Lab Invest. 1999;79:1161–1170. [PubMed] [Google Scholar]
- 25.Kocher O, Comella N, Tognazzi K, Brown LF. Identification and partial characterization of PDZK1: a novel protein containing PDZ interaction domains. Lab Invest. 1998;78:117–125. [PubMed] [Google Scholar]
- 26.Gisler SM, Pribanic S, Bacic D, Forrer P, Gantenbein A, Sabourin LA, Tsuji A, Zhao ZS, Manser E, Biber J, Murer H. PDZK1: I a major scaffolder in brush borders of proximal tubular cells. Kidney Int. 2003;64:1733–1745. doi: 10.1046/j.1523-1755.2003.00266.x. [DOI] [PubMed] [Google Scholar]
- 27.Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117:1547–1552. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- 28.Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T, Shibata N, Nishimoto-Shibata T, Feng D, Ikemoto M, Motojima K, Iso ON, Tsukamoto K, Tsujimoto M, Arai H. Regulation of SR-BI protein levels by phosphorylation of its associated protein, PDZK1. Proc Natl Acad Sci U S A. 2005;102:13404–13409. doi: 10.1073/pnas.0506679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenske SA, Yesilaltay A, Pal R, Daniels K, Rigotti A, Krieger M, Kocher O. Overexpression of the PDZ1 domain of PDZK1 blocks the activity of hepatic scavenger receptor, class B, type I by altering its abundance and cellular localization. J Biol Chem. 2008;283:22097–22104. doi: 10.1074/jbc.M800029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Yue H, Derin RB, Guggino WB, Li M. Accessory protein facilitated CFTR-CFTR interaction, a molecular mechanism to potentiate the chloride channel activity. Cell. 2000;103:169–179. doi: 10.1016/s0092-8674(00)00096-9. [DOI] [PubMed] [Google Scholar]
- 32.Wente W, Stroh T, Beaudet A, Richter D, Kreienkamp HJ. Interactions with PDZ domain proteins PIST/GOPC and PDZK1 regulate intracellular sorting of the somatostatin receptor subtype 5. J Biol Chem. 2005;280:32419–32425. doi: 10.1074/jbc.M507198200. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Krishnamurthy PC, Penmatsa H, Marrs KL, Wang XQ, Zaccolo M, Jalink K, Li M, Nelson DJ, Schuetz JD, Naren AP. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell. 2007;131:940–951. doi: 10.1016/j.cell.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]