Abstract
The PRL (phosphatase of regenerating liver) phosphatases represent a distinct class of protein tyrosine phosphatases, which are implicated in tumorigenesis and metastasis processes. Accumulating evidence indicates that alteration in PRL1 expression affects cell motility and tumor metastasis, although the biochemical pathways regulated by PRL1 remain less well defined. We find that elevated expression of PRL1 increases the levels of the matrix metalloproteinases MMP2 and MMP9. We have studied whether MMP2 and MMP9 are regulated by PRL1 and participate in PRL1-dependent cell migration and invasion. To this end, knockdown or inhibition of MMP2 and MMP9 by either siRNA or a specific small molecule inhibitor blocks the PRL1-mediated cell migration and invasion. In addition, we report that up-regulation of PRL1 activates the Src kinase through increased Tyr416 phosphorylation, which culminates in the phosphorylation of the focal adhesion proteins FAK and p130Cas, as well as ERK1/2 activation. We provide evidence that both the Src and ERK1/2 pathways contribute to the increased motility of the PRL1 cells. We further demonstrate that Src and ERK1/2 activities are required for the PRL1-induced increase in MMP2 and MMP9, likely through activation of the transcription factors AP1 and Sp1. Accordingly, increased PRL1 expression results in activation of Src and ERK1/2, which stimulates MMP2 and MMP9 production, leading to increased cell migration and invasion.
Protein tyrosine phosphorylation plays an important role in regulating a wide array of signaling events essential for the proper function of many cellular processes, including proliferation, metabolism, differentiation, survival/apoptosis, as well as adhesion and motility (1). The level of tyrosine phosphorylation is modulated by the coordinated action of protein tyrosine kinases and phosphatases. Similar to the kinases, dysregulation of protein tyrosine phosphatases has been linked to several human diseases such as diabetes, autoimmune disorders and cancer (2). The PRL (phosphatase of regenerating liver) phosphatases represent a distinct class of protein tyrosine phosphatases, which are implicated in a number of tumorigenesis and metastasis processes (3). The PRL phosphatases include three members (PRL1, PRL2, and PRL3), which share a high degree (>75%) of amino acid sequence identity.
PRL1 was originally identified as an immediate early gene in regenerating liver (4). Subsequent studies revealed that PRL1 expression is elevated in several tumor cell lines, and cells expressing high levels of PRL1 exhibit enhanced proliferation and anchorage-independent growth (4-6). Interestingly, up-regulation of the related PRL2 and PRL3 also promotes cell growth (3). In addition to a role in cell proliferation, the PRLs are also involved in tumor metastasis. For example, the PRL3 message is amplified in colorectal cancer metastases, whereas its expression in primary tumors and normal colorectal epithelium is undetectable (7). CHO or HEK293 cells stably expressing PRL1 or PRL3 display enhanced motility and invasiveness (8-10), and CHO cells with elevated PRL1 or PRL3 induce metastatic tumor formation in nude mice (8). In addition, an increase in PRL1 or PRL3 expression promotes motility and invasion in colon adenocarcinoma cells (11), while transient reduction of PRL1 or PRL3 abrogates colon cancer cell motility and hepatic colonization (12). Finally, knockdown of endogenous PRL1 inhibits human A549 lung cancer cell invasion (13). Collectively, these studies reveal that an excess of PRL1 or PRL3 is a key contributing factor to the acquisition of metastatic properties of tumor cells.
Given their involvement in human malignancies, the PRL phosphatases have emerged as highly attractive targets for novel anticancer therapy. Unfortunately, although considerable evidence has now accumulated suggesting that the PRLs may play key causal roles in tumor metastasis, the underlying mechanisms by which these phosphatases promote cell migration and invasion remain poorly understood. Tumor metastasis is a multi-step process by which cancer cells disseminate from a primary tumor to distant secondary organs or tissues. Proteolysis and remodeling of the extracellular matrix (ECM) represent one of several initiating events that allow cancer cells to invade into the surrounding stroma. Matrix metalloproteinases (MMPs) are major hydrolytic enzymes targeting ECM during metastasis and there is a clear connection between MMPs, ECM degradation and cancer cell invasion (14).
In this report, we show that up-regulation of PRL1 activates Src and ERK1/2 pathways, and increases both MMP2 and MMP9 expression. Knockdown or inhibition of MMP2 and MMP9 by either siRNA or a specific small molecule inhibitor blocks the PRL1-mediated cell migration and invasion. In addition, we provide evidence that induction of MMP2 and MMP9 by PRL1 requires activation of the Src and ERK1/2 pathways. We further establish that the PRL1-induced MMP2 and MMP9 expression is driven by the ERK1/2-mediated activation of the transcriptional factors AP1 and Sp1. Finally, we demonstrate that lung cancer cell lines with higher migration and invasion potential also express elevated levels of PRL1 as well as MMP2 and MMP9, and display higher Src and ERK1/2 activities. Collectively, the results support a mechanism for the PRL1-induced cell migration and invasion that involves induction of MMP2 and MMP9 expression via the Src and ERK1/2 signaling pathways.
Materials and Methods
Materials
Anti-HA-antibody was purchased from Santa Cruz Biotechnology. Anti-phosphotyrosine (pY20), p130Cas, FAK, and actin antibodies were from BD Biosciences. Anti-Src, anti-Src-pY416, and anti-Src-pY527 antibodies were from Biosource. Polyclonal anti-ERK1/2 and anti-pERK1/2 (Thr-202/Tyr-204) antibodies were purchased from Cell Signaling (Beverly, MA). Src inhibitor SU6656, MEK1/2 inhibitor U0126 and MMP2/9 inhibitor (2R)-[(4-Biphenylylsulfonyl) amino]-N-hydroxy-3-phenylpropionamide (BiPS) were purchased from Calbiochem. Zeocin was purchased from Invitrogen. Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, penicillin, and streptomycin were from Invitrogen. The expression plasmid encoding PRL1 was described in (10).
Cell culture and stable clone selection
HEK293 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (50 units/ml), and streptomycin (50 μg/ml) under a humidified atmosphere containing 5% CO2. HEK293 cells were seeded at 40% confluency in antibiotic-free medium and grown overnight. Transfection was performed using Fugene 6 (Roche, Indianapolis) according to the manufacturer’s recommendations. 24 h after transfection, stable cell lines were established by selection with Zeocin (500 μg/ml). Lung cancer cell lines are from ATCC and cultured according to ATCC cell culture conditions.
mRNA extraction and RT-PCR
mRNA from different cell lines was prepared using Trizol reagent (Invitrogen). mRNA was treated by DNase and quantified by absorbance at 260 and 280 nm. RT-PCR was performed using the Invitrogen SuperScript one-step RT-PCR kit according to manufacture’s instruction. Equal amounts of RNA (200 ng) were used as templates in each reaction. The sequences of specific primers were as follows: PRL1 sense, 5′-CCAGCTCCTGTGGAAGTCAC-3′, and antisense: 5′-CCATCATCAAAAGGCCAATC-3′; 18S ribosome RNA sense, 5′-CGCCGCTAGAGGTGAAATTC-3′, and antisense, 5′-TTGGCAAATGCTTTCGCTC-3′; MMP2 sense, 5′-ACCTGGATGCCGTCGTGGAC-3′, and antisense: 5′-TGTGGCAGCACCAGGGCAGC-3′; MMP9 sense, 5′-CACTGTCCACCCCTCAGAGC-3′, and antisense, 5′-GCCACTTGTCGGCGATAAGG-3′. The PCR products were separated by 1% agarose gel and visualized by ethidium bromide staining. Representative results from two independent experiments are shown.
Immunoblotting and immunoprecipitation
Cells were grown to 70% confluency, washed with ice-cold phosphate-buffered saline, and lysed on ice for 30 min in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl, 10 mM sodium phosphate, 10 mM sodium fluoride, 1 mM sodium pervanadate, 1 mM benzamidine, 1% Triton X-100, 10 μg/ml leupeptin, and 5 μg/ml aprotinin). Cell lysates were cleared by centrifuging at 15,000 rpm for 10 min. Lysate protein concentration was estimated using BCA protein assay kit (Pierce). For immunoprecipitation, 3 μg of antibody was added to 1 mg of cell lysate and incubated at 4 °C for 4 hr with protein A/G-agarose beads. After extensive washing, protein complex was boiled with sample buffer, separated by SDS-PAGE, transferred electrophoretically to nitrocellulose membrane, and immunoblotted with appropriate antibodies followed by incubation with horseradish peroxidase-conjugated secondary antibodies. The blots were developed by the enhanced chemiluminescence technique (ECL kit, Amersham Biosciences). Representative results from at lease 2 independent experiments are shown. Densitometry was performed using ImageJ software.
Src kinase assay
Cell lysate from each cell line was incubated with anti-Src antibody for 4 h at 4 °C together with 20 μl of protein A/G. Half of the immunocomplex was subjected to immunoblotting to determine the levels of total Src protein. The other half was used to assay for Src kinase activity using enolase as a substrate as described previously (9). The immunoprecipitate was washed three times with the lysis buffer and twice with the kinase buffer (40 mM HEPES, pH 7.4, 10 mM MgCl2, 3 mM MnCl2). The immunoprecipitate was then incubated with 10 μCi of [γ-32P]ATP, 50 μM ATP, 1 mM dithiothreitol, and 5 μg of acid-treated enolase in a 30-μl volume at 30 °C for 10 min. The reaction was terminated by adding 10 μl of 4x SDS loading buffer. The sample was analyzed by SDS-PAGE with 10% gel. 32P-Labeled enolase was visualized by autoradiography. Densitometry was performed using ImageJ software.
Cell motility assay
Cell migration was measured as described (8) with some modifications. The assay was performed with gelatin-coated Transwells (6.5 mM diameter; 8 μM pore size polycarbonate membrane) obtained from Corning (Costar, Acton, MA). Cells (3.75 × 105) in 1.5 ml serum-free medium were placed in the upper chamber, whereas the lower chamber was loaded with 2.6 ml medium containing 10% FBS. After incubation at 37°C with 5% CO2 for 24 h, the total numbers of cells that migrated into the lower chamber was counted by hemacytometer. Experiments were performed in duplicates.
Cell invasion assay
Transwells (Biocoat Matrigel 24-well invasion chamber) with filters coated with extracellular matrix (ECMatrix gel) on the upper surface were purchased from BD Biosciences. The experiments were performed according to the manufacturer’s protocol. Cells (2.5 × 104) were added to the upper chamber in serum-free medium, and the total invasive cells in the lower chamber were stained and counted after 24 h of incubation at 37°C with 5% CO2. Experiments were performed in duplicates.
siRNA knockdown
Duplex siRNAs were provided by Proligo at 50 μM concentration. The siRNA sequences were: human MMP2-1, sense 5′-GCAACCUGUUUGUGCUGAATT, antisense 5′-UUCAGCACAAACAGGUUGCTT, target position 498; MMP2-2, sense 5′-AGCGUGAAGUUUGGAAGCATT, antisense 5′-UGCUUCCAAACUUCACGCUTT, target position 2243; MMP2 scramble, sense 5′- CGCAACGUUGUUGUUACGATT, antisense 5′-UCGUAACAACAACGUUGCGTT; human MMP9-1, sense 5′-ACCACAACAUCACCUAUUGTT, antisense 5′-CAAUAGGUGAUGUUGUGGUTT, target position 372; MMP9-2, sense 5′-GCAUAAGGACGACGUGAAUTT, antisense 5′-AUUCACGUCGUCCUUAUGCTT, target position 1312; MMP9 scramble, sense 5′-ACACCAUUAACUCCUAACGTT, antisense 5′-CGUUAGGUGUUAAUGGUGUTT, human PRL1-1, sense 5′-CAACCAAUGCGACCUUAAATT, antisense 5′-UUUAAGGUCGCAUUGGUUGTT, target position 1224; PRL1-2, sense 5′-CUGGUUAAGUCUUGUGAAATT, antisense 5′- UUUCACAAGACUUAACCAGTT, target position 1399; PRL1 scramble, sense 5′-AACACUAAGCGCAUCAUACTT, antisense 5′-GUAUGAUGCGCUUAGUGUUTT. siRNAs were transfected by RNAiFect (Qiagen) and the knockdown of targeted genes were verified after 72 hr by either RT-PCR or Western blotting analysis.
Gelatin zymography
PRL1 cells were incubated in serum-free DMEM for 24 h. The conditioned medium was collected, concentrated and 30 μg of total protein was resolved in 7.5% polyacrylamide gels containing 0.1% gelatin. After electrophoresis, the gels were washed for 1 h in 2.5% (v/v) Triton X-100 to remove SDS and then incubated overnight at 37 °C in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM ZnCl2 and 10 mM CaCl2 to allow proteolysis of the gelatin substrate. Bands corresponding to activity were visualized by negative staining using 0.5% Coomassie brilliant blue R-250 (Bio-Rad, CA, USA). Representative results from two independent experiments are shown. Densitometry was performed using ImageJ software and band density was normalized to the non-specific band staining on the gel.
Nuclear extraction and gel shift assay
Nuclear extracts were prepared from cells as described (15). PRL1 cells were resuspended in 1 ml of cold hypotonic RSB buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2) supplemented with 0.25% NP-40 and protease inhibitors. Following a 12-min incubation on ice, the cells were lysed with a Dounce homogenizer. Nuclei were resuspended in 2 packed nuclear volumes of extraction buffer C (420 mM KCl, 20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA, 20% glycerol) supplemented with protease inhibitors and incubated on ice for 30 min. Protein concentrations were determined with the Bio-Rad protein assay. The consensus sequences of the double-stranded oligonucleotides used in the gel shift assay were: AP1: 5′-CGC TTG ATG AGT CAG CCG GAA-3′ and 3′-GCG AAC TAC TCA GTC GGC CTT-5′; Sp1: 5′-CAG AGA GGG GCG GGC CCG AGT G-3′ and 5′- CAC TCG GGC CCG CCC CTC TCT G-3′. For the binding reaction, 32P-labeled DNA fragments (20,000 to 25,000 cpm), 5 μg of nuclear extract, and 2.5 μg of poly(dI-dC), as a nonspecific competitor, were added to a solution containing 10 mM HEPES [pH 7.9], 4 mM dithiothreitol, 0.5% Triton X-100, 100 mM KCl, and 2.5% glycerol in a final assay volume of 25 μl. The binding assay was carried out at room temperature for 30 min, and DNA-protein complexes were separated by gel electrophoresis. Where indicated, excess unlabeled competitor DNA fragments corresponding to the consensus sequences for the AP1 and Sp1 binding sites were added to the assay mixture to ascertain binding specificity. Representative results from two independent experiments are shown.
Luciferase assay
Luciferase assay were modified as previously described (16). Cells were transfected by various human MMP2 or MMP9 promoter-luciferase constructs kindly provided by Dr. Benveniste (University of Alabama, Birmingham, AL) (17, 18) and Dr. Boyd (University of Texas, MD Anderson Cancer Center) (19, 20). Cells were harvested and lysed 48 h after transfection. Luciferase activities were determined using the luciferase assay system (Promega, Madison, WI) according to the manufacturer’s specifications. Individual assays were normalized by internal Renilla luciferase activity. Experiments were performed in triplicate and repeated two or three times with similar results. The statistics were done using Student’s t-test, with P < 0.05 considered significant.
Results
PRL1 promotes cell migration and invasion
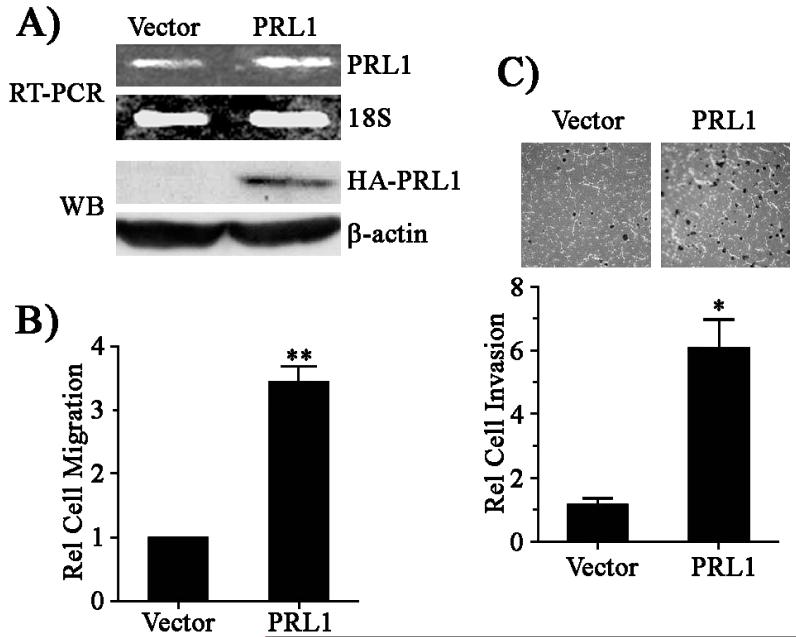
To define the mechanism by which PRL1 promotes cell migration and invasion, we established HEK293 cell lines overexpressing PRL1. The level of ectopically expressed PRL1 in the pool of stably transfected cells was 2 fold higher than that of the endogenous PRL1 in vector control cells (Figure 1A). Using Transwell assay, we found that the PRL1 cells exhibited a 3.5-fold increase in cell migration as compared to the vector control (Figure 1B). We also assessed the effect of PRL1 expression on cell invasion. In comparison with the control cells, the PRL1 cells displayed a 6-fold increase in cell invasion as determined by the Matrigel assay (Figure 1C). These results are in complete agreement with previous observations from a number of cell lines, including HEK293 (8, 10, 11), that ectopic expression of PRL1 enhances cell motility and invasion. Acquisition of these motile and invasive properties in PRL1 cells is consistent with PRL1’s involvement in tumor metastasis.
Figure 1.
PRL1 promotes cell migration and invasion. (A) RT-PCR analysis of PRL1 expression in stable cell lines using 18S rRNA as a control. The expression of the HA-tagged PRL1 protein was measured by Western blot. (B) Cell migration was quantitated over 24 h using the Transwell assay as described in Materials and Methods. Relative cell migration was normalized to vector control. Results were presented as mean ± S.D. **, p<0.01. (C) Cell invasion was measured over 24 h using the Matrigel method as described in Materials and Methods. Relative cell invasion was normalized to vector control. Results were presented as mean ± S.D. *, p<0.05.
PRL1 activates Src and ERK1/2 pathways
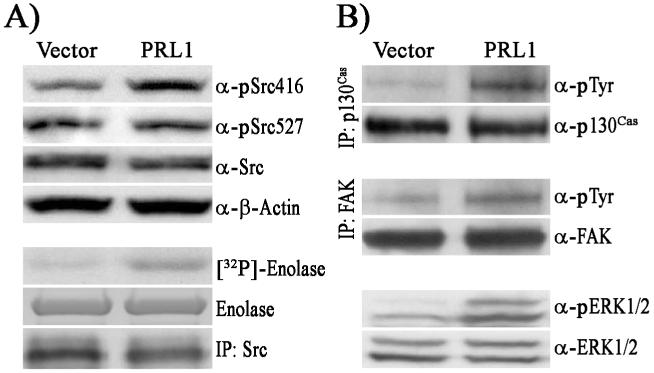
Little is known about the biochemical mechanisms underlying the PRL phosphatases-mediated cell invasion and tumor metastasis, although the PRLs have been linked to several signaling molecules, including Src, ERK1/2, PI3K/Akt, and the Rho family of GTPases (9, 11, 13, 21). Since Src kinase is intimately involved in cell migration and invasion, we set out to determine whether Src is activated in PRL1 expressing cells. Src activity is regulated by phosphorylation at two distinct tyrosine residues. Autophosphorylation of Tyr416 in the kinase domain activates Src, while phosphorylation of Tyr527 in the C-terminal tail by the C-terminal Src kinase (Csk) blocks Src activity. Src phosphorylation levels at Tyr416 and Tyr527 were determined using phosphospecific antibodies directed toward pSrc416 and pSrc527, respectively. Western blotting analysis showed that Src Tyr416 phosphorylation level was 1.9-fold higher in PRL1 cells, while no significant change in pTyr527 was observed (Figure 2A). The results suggest that Src is activated as a result of increased PRL1 expression. To directly confirm Src activation, Src protein was immunoprecipitated from cell lysates with anti-Src antibodies and assayed for kinase activity using enolase and [32P]-ATP as substrates. As shown in Figure 2A, Src kinase activity was 3.4-fold higher in PRL1 cells than that in vector control cells.
Figure 2.
PRL1 activates Src and ERK1/2. (A) Cell lysates (50 μg) were resolved by SDS-PAGE and subjected to Western blotting to detect pSrc527, pSrc416, and Src. Actin was used as a loading control. Kinase activity of the immunoprecipitated Src was measured with enolase and [32P]ATP as substrates. (B) Activation status of p130Cas, FAK, and ERK1/2. p130Cas and FAK were immunoprecipitated from cell lysates (1000 μg) and resolved by SDS-PAGE. Tyrosine phosphorylation levels were detected by Western blot using anti-pY20 antibody. ERK1/2 and pERK1/2 were detected by Western blot from total cell lysates (50 μg).
To further corroborate that Src is activated in PRL1 cells, we next examined the status of a number of signaling molecules downstream of Src, including the focal adhesion kinase (FAK), the adaptor protein p130Cas, and ERK1/2. It is well established that Src regulates cell motility by targeting FAK, p130Cas, and the ERK1/2 pathway (22, 23). Both FAK and p130Cas are direct substrates of Src kinase and they are important mediators of focal adhesion turnover and cell migration. Upon Src activation, FAK and p130Cas increase their tyrosine phosphorylation levels and relocate to focal adhesions to participate in cellular migratory processes. Src can also mediate activation of the ERK1/2 pathway, which is involved in several fundamental cellular processes in cell proliferation, survival, and motility. Immunoprecipitation experiments revealed that the levels of tyrosine phosphorylation of FAK and p130Cas in PRL1 cells were increased by 1.5- and 3.2-fold, respectively (Figure 2B). Moreover, ERK1/2 is also activated in PRL1 cells, with phospho-ERK1/2 level 2.6-fold higher than the vector control (Figure 2B). These results indicate that elevated PRL1 expression leads to Src activation, which promotes phosphorylation of focal adhesion proteins and ERK1/2 activation. The increase in phosphorylation of FAK and p130Cas are consistent with the pro-migratory actions of PRL1. Activation of ERK1/2 may also contribute to the PRL1 induced cell migration and invasion (see below).
Elevated PRL1 expression leads to MMP2 and MMP9 activation
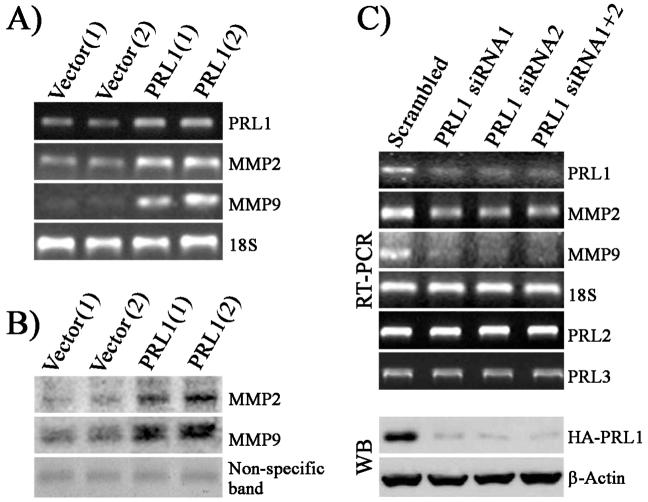
In addition to its role in cell growth and survival, the ERK1/2 pathway also promotes tumor invasiveness through up-regulation of MMPs (24), which play important roles in tumor metastasis (14). Among the MMP family members, MMP2 (gelatinase A) and MMP9 (gelatinase B) are most relevant to tumor invasion. Both MMP2 and MMP9 can degrade collagen of the basement membrane, which induces cell migration and is required for cells to break up the ECM and invade into the surrounding tissues (25, 26). Since ERK1/2 are activated in PRL1 cells and MMP2 and MMP9 are essential for cell invasion, we sought to determine whether PRL1 cell lines have elevated MMP2 and MMP9 expression. RT-PCR analysis revealed that MMP2 and MMP9 levels are 3.6 and 4-fold higher, respectively, in two individual clones of PRL1 cell lines (Figure 3A). Both MMP2 and MMP9 are synthesized as pro-enzymes in the cell. When secreted, they are cleaved and become catalytically activated (14). Therefore, we further investigated whether the PRL1 cells also produce more secreted and active forms of MMP2 and MMP9. As shown in Figure 3B, PRL1 significantly increased mature and active MMP2 (3.7x) and MMP9 (3x) production, as measured by gelatin zymography. To confirm that the enhanced MMP2 and MMP9 expression is mediated by PRL1, we knocked down PRL1 expression in PRL1 cells using siRNAs, and measured the levels of MMP2 and MMP9 by RT-PCR. As shown in Figure 3C, the PRL1 message and protein levels were reduced by nearly 80-90% in PRL1 knockdown cells. No changes were observed in the message levels of the related PRL2 and PRL3, demonstrating the specificity of the siRNAs for PRL1. As expected, the reduction in PRL1 resulted in ∼70% decrease in MMP2 and MMP9 expression (Figure 3C). Taken together, the data indicate that elevated PRL1 expression leads to MMP2 and MMP9 activation and a decrease in PRL1 suppresses MMP2 and MMP9 production.
Figure 3.
PRL1 cell lines have increased levels of MMP2 and MMP9 production. (A) PRL1, MMP2, and MMP9 mRNA levels were determined by RT-PCR using 18S rRNA as a control. (B) Aliquots of cell culture medium from control and PRL1 cell lines containing the same amount of protein were subjected to gelatin zymography. Staining of a non-specific protein band was used as loading control. (C) Knockdown of PRL1 using siRNA reduces MMP2 and MMP9 expression. PRL1, PRL2, PRL3, MMP2, and MMP9 levels were determined by RT-PCR.
PRL1 stimulates MMP2 and MMP9 expression through Src and ERK1/2 mediated AP1 and Sp1 activation
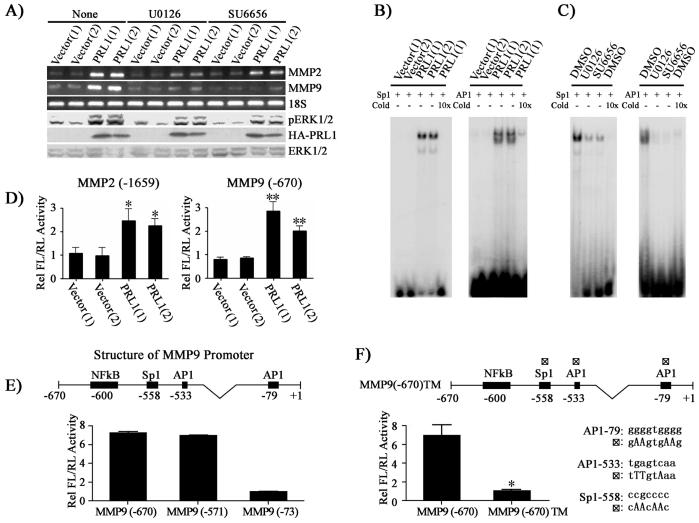
We have shown that Src and ERK1/2 pathways are activated in PRL1 cells. To determine whether Src and ERK1/2 activities are required for the increased MMP2 and MMP9 production in PRL1 cells, we examined the effects of inhibiting Src or ERK1/2 pathways on MMP2 and MMP9 levels using the Src specific inhibitor SU6656 and a specific MEK1/2 inhibitor U0126. As shown in Figure 4A, treatment of the cells with U0126 decreased ERK1/2 activity by 3.2 fold and reduced the expression levels of MMP2 and MMP9 by 8.1- and 8.8-fold, respectively. Similarly, inhibition of Src by SU6656 led to a 3.5 fold reduction in ERK1/2 activity and 2.9 and 11-fold decrease in MMP2 and MMP9 expression respectively. These results support that PRL1 activates MMP2 and MMP9 expression via Src and ERK1/2 signaling pathways.
Figure 4.
PRL1 increases MMP2 and MMP9 expression through Src and ERK1/2 mediated AP1 and Sp1 activation. (A) Inhibition of Src (2.5 μM SU6656) or ERK1/2 (1 μM U0126) lead to reduced MMP2 and MMP9 expression. MMP2 and MMP9 expression was measured by RT-PCR, and pERK1/2, ERK1/2 and HA-PRL1 were determined by Western blot analysis. (B) AP1 and Sp1 transcriptional factors are involved in the PRL1-induced MMP2 and MMP9 expression. Nuclear extracts of control and PRL1 cell lines were prepared. Electrophoretic mobility shift assays were performed with 5 μg of nuclear extract with 20,000 cpm of labeled Sp1 or AP1 probe. Competition analysis was performed in the presence of a 10-fold molar excess of unlabeled probes. (C) Sp1 and AP1 activation requires Src and ERK1/2 activity. Cells were preincubated with either Src (SU6656, 2.5 μM) or MEK1/2 (U0126, 1 μM) inhibitor, and the DNA binding activity of AP1 and Sp1 were determined using gel shift assays. (D, E, F) Requirement of AP1 and Sp1 for PRL1-induced MMP2 and MMP9 expression. Control and PRL1 cell lines were co-transfected with either 200 ng of the MMP2 or the MMP9 luciferase reporter constructs (D) or MMP9 deletion mutant luciferase reporter constructs (E) or a MMP9 luciferase reporter construct (MMP9 (-670)TM) with all three AP1 and Sp1 sites mutated (changes in nucleotides were made according to the consensus sequences and published data (38, 39) and depicted in the figure) (F) together with 10 ng Renilla-TK luciferase reporter as an internal control. After 48 hr, cells were lysed and dual luciferase measurements were performed. Firefly luficerase values were normalized against Renilla luciferase values. Relative luciferase activity was normalized to vector control cell (D) or to MMP9 (-73) (E) or to MMP9 (-670)TM (F). Results were presented as mean ± S.D. *, p<0.05 and **, p<0.01.
To further define the mechanism for the increased MMP2 and MMP9 levels in PRL cells, we determined the status of the transcription factors Sp1 and AP1, which regulate MMP2 and MMP9 expression (17, 18, 27, 28). Interestingly, ERK1/2 activation has been shown to be required for full activity of Sp1 and AP1. To this end, ERK1/2 directly phosphorylates Sp1 on Thr453 and Thr739. Mutation of these sites to alanines decreases the ERK1/2-dependent transcriptional activity of Sp1 (29). c-Jun, a critical component of the AP1 complex, can be phosphorylated by ERK1/2 on Ser63 and Ser73, and the phosphorylation of these sites are required for AP1 to become transcriptionally active (30). Since ERK1/2 activation appears to be responsible for the enhanced MMP2 and MMP9 expression in PRL1 cells, we ascertained whether this occurs through up-regulation of Sp1 and AP1 activity. We first determined whether Sp1 and AP1 are activated in PRL1 cells. Nuclear extracts from both the control and PRL1 cells were prepared and gel shift assays were performed using radioactive, double-stranded oligonucleotides corresponding to the consensus sequences of the Sp1 and AP1 binding sites. As revealed in Figure 4B, substantially more AP1- and Sp1-DNA complex formation was observed in PRL1 cells than in the control cells, indicating that both AP1 and Sp1 are activated as a result of increased PRL1 expression. In addition, AP1- and Sp1-DNA interactions were disrupted when an excess of unlabeled oligonucleotide was present in the assay mixture, which confirms the binding specificity of the 32P-labled probes with the transcription factors. We next evaluated whether Src and ERK1/2 activities are required for AP1 and Sp1 activation in PRL1 cells. The cells were incubated with either the Src inhibitor SU6656 or the MEK1/2 inhibitor U0126 before the nuclear extracts were prepared for gel shift assays. The results revealed that inhibition of the Src or ERK1/2 pathways abrogate the formation of the AP1- and Sp1-DNA complexes, indicating that Src and ERK1/2 are indeed essential for Sp1 and AP1 activation in PRL1 cells (Figure 4C).
To further confirm the importance of AP1 and Sp1 for the PRL1-mediated up-regulation of MMP2 and MMP9, we transfected control and PRL1 cell lines with luciferase reporter constructs of MMP2 and MMP9 promoter regions encompassing the AP1 and Sp1 sites. Consistent with the results from the gel shift assays, the luciferase reporter activities were increased 2-3 fold in PRL1 cell lines (Figure 4D). When luciferase reporters lacking the AP1 and Sp1 sites (-73) in MMP9 promoter region were used to transfect the PRL1 cells, approximately 7-fold decrease in luciferase activity was observed (Figure 4E). In contrast, no change in luciferase activity was observed when the NFκB site (-571) was removed from the MMP9 promoter. Furthermore, when AP1 (-79), AP1 (-533) and Sp1 (-558) sites were mutated, the luciferase activity for the triple mutant construct was 7 fold lower than that of the control (Figure 4F), thus validating the requirement of AP1 and Sp1 for PRL1 induced MMP2 and MMP9 expression. Taken together, our data suggest that PRL1 promotes the activation of the Src and ERK1/2 pathways, which drive the AP1 and Sp1-dependent transcriptional up-regulation of MMP2/MMP9.
Up-regulation of MMP2 and MMP9 is required for PRL1 induced cell migration and invasion
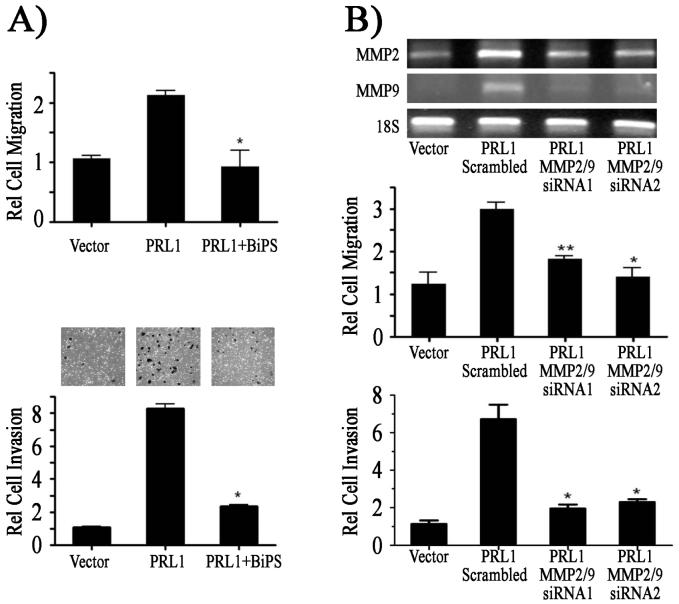
Up-regulation of MMP2 and MMP9 is often associated with increased cell migration, invasion and cancer metastasis (14, 31). Given the observed increase in MMP2 and MMP9 expression in PRL1 cells, we suspected that MMP2 and MMP9 might contribute to the enhanced migratory and invasive properties of the PRL1 cells. To test this hypothesis, we employed either a small MMP2/9 inhibitor (2R)-[(4-Biphenylylsulfonyl) amino]-N-hydroxy-3-phenylpropionamide (BiPS) (32) to inhibit the MMP2/9 activity or siRNAs to knockdown both MMP2 and MMP9 levels in PRL1 cells. The results showed that either inhibition of MMP2/9 catalytic activity or reduction of MMP2/9 protein levels abrogated the PRL1-mediated cell migration and invasion (Figure 5). Therefore, we conclude that the increased MMP2 and MMP9 expression are essential to the PRL1-induced cell migration and invasion.
Figure 5.
Enhanced MMP2 and MMP9 expression is required for the PRL1 induced cell migration and invasion. (A) Cells were treated with 0.5 μM of the MMP2/9 inhibitor BiPS and the rates of cell migration and invasion were quantitated over 24 h using Transwell and Matrigel assays respectively. Relative cell migration/invasion was normalized to vector control cells. (B) The levels of MMP2 and MMP9 in PRL1 cells were knocked down by siRNA and the rates of cell migration and invasion were quantitated over 24 h using the Transwell and Matrigel assays respectively. Relative cell migration/invasion was normalized to vector control cells. Results were presented as mean ± S.D. *, p<0.05 and **, p<0.01.
PRL1 overexpression is correlated with increased MMP2 and MMP9 expression in lung cancer cell lines
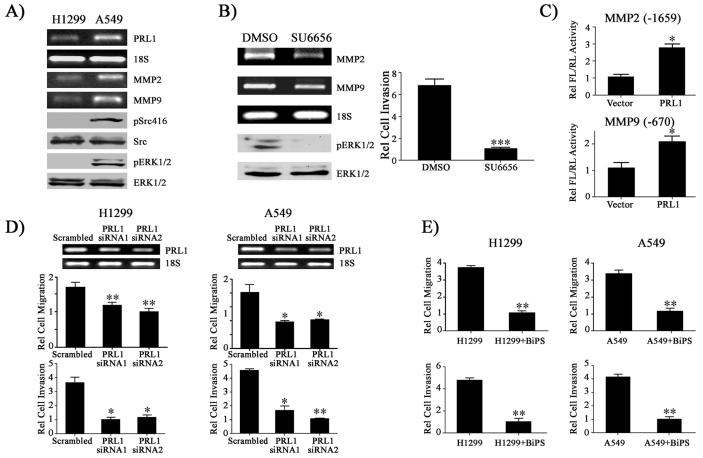
Lung cancer is the leading cause of cancer death in the United States. The major cause for lung cancer related fatality is the development of metastasis. PRL1 is expressed in normal lung bronchiolar epithelium and is overexpressed in many tumor cell lines, including lung cancer cells (5). Recent studies indicate that silencing of PRL1 in human A549 lung cancer cells inhibits cell invasion (13). To ascertain whether MMP2 and MMP9 expression is also elevated in cells that naturally overexpress PRL1, we examined two lung cancer cell lines, H1299 and A549. We determined the levels of PRL1, MMP2, and MMP9 as well as the activation status of Src and ERK1/2 in H1299 and A549 cells. As shown in Figure 6A, A549 cells, which possess strong invasive activity, express 2.5-fold higher PRL1 than H1299 cells, which have weaker invasive activity (33). RT-PCR analysis revealed that A549 cells also express correspondingly 2.2 and 3.2-fold higher MMP2 and MMP9, respectively. Moreover, consistent with our observations with the PRL1-expressing HEK293 cell lines, Src and ERK1/2 activities are also substantially higher in A549 cells than those in H1299 cells. Since Src activity is higher in A549 cells, we further determined whether Src activity is required for MMP2/9 expression and cell invasion. Similar to results obtained with PRL1 expressing HEK293 cells, treatment of A549 cells with SU6656 caused considerable reduction in ERK1/2 activity and significantly reduced MMP2 and MMP9 expression (Figure 6B). Additionally, Src inhibition by SU6656 also resulted in a 6.7-fold decrease in cell invasion as compared to the vehicle control. Thus, increased expression of PRL1 in lung cancer cells also correlates with increased Src and ERK1/2 signaling as well as MMP2 and MMP9 expression.
Figure 6.
PRL1 enhances the invasiveness of lung cancer cells by increasing MMP2/9 expression through the Src and ERK1/2 pathways. (A) An increase in PRL1 expression is correlated with Src and ERK1/2 activation as well as enhanced MMP2/9 expression in lung cancer cell lines. (B) Inhibition of Src by SU6656 in A549 cells decreases ERK1/2 activity and suppresses MMP2/9 expression and cell invasion. A549 cells were treated with Src inhibitor SU6656 (2.5 μM) for 4 hr. RNA was prepared and MMP2 and MMP9 expression was determined by RT-PCR. pERK1/2, ERK1/2 were determined by Western blot analysis. For cell invasion assay, A549 cells were treated with either DMSO or SU6656 and the rates of cell invasion were quantitated over 24 hr using the Matrigel assay. Relative cell invasion was normalized with the SU6656 treatment group. Results were presented as mean ± S.D. *** p<0.001. (C) PRL1 induces MMP2/9 expression in lung cancer cell. H1299 cells were co-transfected by 500 ng PRL1 together with either 200 ng of the MMP2 or the MMP9 luciferase reporter constructs with 10 ng Renilla-TK luciferase reporter as an internal control. After 48 hr, cells were lysed and dual luciferase measurements were performed. Firefly luficerase values were normalized against Renilla luciferase values. Relative luciferase activity was normalized to vector control cell. Results were presented as mean ± S.D. *, p<0.05 and **, p<0.01. Knockdown of MMP2 and MMP9 using siRNAs (D) or inhibition of MMP2 and MMP9 using small molecule inhibitor BiSP (0.5 μM) (E) in H1299 and A549 reduce lung cancer cell migration and invasion. Relative cell migration/invasion was normalized to the PRL1 siRNA2 group (D) or to the BiPS treated group (E). Results were presented as mean ± S.D. *, p<0.05 and **, p<0.01.
To determine whether PRL1 is also capable of inducing MMP2/9 expression in lung cancer cells, we cotransfected H1299 cells with a plasmid encoding PRL1 together with luciferase reporter constructs carrying the full-length MMP2 or MMP9 promoter. Compared with the vector control, luciferase reporter activities were increased 2-3 fold when PRL1 and MMP reporter constructs were coexpressed in H1299 cells (Figure 6C). These results support the notion that the PRL1-mediated expression of MMP2 and MMP9 may be a general mechanism responsible for the PRL1-induced cell invasion and migration. To address the potential biological relevance of PRL1 and MMP2/9 in lung cancer metastasis, we determined the effect of PRL1 knockdown or MMP2/9 inhibition on A549 and H1299 cell migration and invasion. As expected, knockdown of endogenous PRL1 with small interfering RNA in lung cancer cells attenuated both cell migration and invasion (Figure 6D). Moreover, inhibition of MMP2 and MMP9 with the small molecule inhibitor BiPS also blocked lung cancer cell migration and invasion (Figure 6E). The results support the conclusion that PRL1 enhances the invasiveness of lung cancer cells by increasing MMP2/9 expression through the Src and ERK1/2 pathways.
Discussion
The involvement of PRL1 in oncogenesis and metastasis has been widely documented. However, the signaling pathways regulated by PRL1 remain to be fully defined. Obtaining this knowledge is vital for the development of effective anticancer therapies targeting PRL1. To gain further insight into the cellular pathways regulated by PRL1, we have utilized HEK293 cell lines stably overexpressing PRL1 as model systems. Consistent with previous results from other cell lines, we find that an increase in PRL1 expression leads to enhanced cell migration and invasion. In addition, we show that ectopic expression of PRL1 in HEK293 cells results in Src activation. The activity of Src is regulated by phosphorylation on two tyrosine residues, Tyr416 in the kinase activation loop and Tyr527 in the C-terminal tail. Phosphorylation at Tyr416 activates Src whereas Tyr529 phosphorylation inhibits the Src kinase. Thus, depending on which tyrosine experiences changes in phosphorylation, Src could exhibit either increased or decreased activity. Our data indicate that the PRL1-induced Src activation occurs as a result of increased phosphorylation on Tyr416 in the kinase activation loop. Interestingly, ectopic expression of PRL3 in HEK293 cells also leads to Src activation (9). However, in the latter case, Src is activated through a reduction in phosphorylation of the inhibitory Tyr527 rather than an increase in Tyr416 phosphorylation. Thus, although PRL1 and PRL3 share high structural homology, they may promote Src activation through different mechanisms.
The exact mechanism by which PRL1 promotes Src activation remains to be established. Although both Src and PRL1 reside in plasma membrane, it is unlikely that PRL1 directly activates Src. First, we failed to detect direct binding between Src and PRL1 by immunoprecipitation experiments (data not shown). Second, the PRL1-mediated Src activation involves an increase in Tyr416 phosphorylation, which cannot be directly effected by a phosphatase. Given the requirement of phosphatase activity for PRL function (8, 9), it is likely that PRL1 dephosphorylates an inhibitory substrate(s) to promote cellular signaling. Identification of direct substrates of the PRL phosphatases will further our understanding of the biochemical mechanism of the PRL1-mediated Src activation. It may also be insightful to examine whether additional kinases are also involved in PRL1 signaling. In any event, it appears that elevated expression of either PRL1 or PRL3 can lead to Src activation, which is a hallmark for oncogenic transformation and tumor metastasis.
Elevated Src kinase activity drives focal adhesion turnover that is associated with cell motility and metastasis. To this end, activated Src binds and phosphorylates FAK leading to FAK activation and creation of additional binding sites for SH2-domaining containing molecules such as the Crk adaptor protein and paxillin into the focal adhesion complexes (22). Src kinase can further recruit and phosphorylate the SH-3-domain containing adaptor protein p130Cas in focal adhesions. Our results show that tyrosine phosphorylation of FAK and p130Cas, two direct substrates of Src, is indeed increased in PRL1 cells. The phosphorylation of FAK and p130Cas stimulates the localization of these phosphoproteins to newly formed focal adhesions, which should contribute to the pro-migratory actions of PRL1. In addition to regulating the dynamics of focal adhesion turnover, Src is also important for controlling the activity of the Rho family of small GTPases, including Rho, Rac, and Cdc42, which are critical regulators of actin cytoskeleton reorganization during cell migration. Interestingly, alteration in PRL1 or PRL3 expression has been reported to perturb the activity of the Rho family of GTPases (11, 13).
In addition to Src activation, ERK1/2 are also up-regulated in PRL1 cells. Aside from its role in regulating cell growth and survival, the ERK1/2 pathway has been implicated in the regulation of cell motility and tumor metastasis (34). A hallmark of metastatic cancer cells is the capacity for matrix degradation through the localized secretion of ECM-degrading MMPs. One suggested mechanism for ERK1/2 to promote invasiveness in tumor cells is through up-regulation of MMPs (e.g., MMP9) for extracellular matrix remodeling (24). We provide evidence that the levels of MMP2 and MMP9 are elevated in PRL1 cells. Both MMP2 and MMP9 have been shown to play critical roles in cancer metastasis (35-37). That MMP2 and MMP9 are essential for the PRL1 dependent cell migration and invasion is quite consistent with the known activities of these proteases. Thus, our finding that an increase in PRL1 expression promotes MMP2 and MMP9 activation offers a potential mechanism for the enhanced motility and invasive phenotypes of the PRL1 cells. We further demonstrate that MMP2 and MMP9 are up regulated by ERK1/2 through the transcription factors AP1 and Sp1, and participate in the PRL1-mediated cell migration and invasion.
Finally, to ascertain whether the PRL1-induced Src and ERK1/2 activation and the associated increase in MMP2 and MMP9 expression represent a general phenomenon, we also determined the levels of PRL1, phospho-Src416, phospho-ERK1/2, MMP2, and MMP9 in two lung cancer cell lines A549 and H1299. Similar to the results obtained with the HEK293 cell lines ectopically expressing PRL1, higher PRL1 expression is correlated with activation of the Src and ERK1/2 pathways and an increase in MMP2 and MMP9 levels. Moreover, increased MMP2 and MMP9 expression is also correlated with increased migration and invasion in lung cancer cells. Thus, it appears that a similar mechanism is also operative in lung cancer cells, namely PRL1 activates Src and ERK1/2, which promote focal adhesion turnover and augment MMP2 and MMP9 expression, leading to increased cell migration and invasion.
Abbreviation
- PRL
phosphatase of regenerating liver
- ECM
extracellular matrix
- MMP
matrix metalloproteinases
- FAK
focal adhesion kinase
- CSK
C-terminal Src kinase
Footnotes
This work was supported by National Institutes of Health Grant CA69202. Y. L. was supported in part by a fellowship from Lungs for Life, Inc.
References
- 1.Hunter T. The phosphorylation of proteins on tyrosine: its role in cell growth and disease. Philos. Trans. R. Soc. London, Ser. B. 1998;353:583–605. doi: 10.1098/rstb.1998.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;11:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 3.Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol. Cancer Ther. 2005;4:1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 4.Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol. Cell. Biol. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Kirby CE, Herbst R. The tyrosine phosphatase PRL1 localizes to the endoplasmic reticulum and the mitotic spindle and is required for normal mitosis. J. Biol. Chem. 2002;277:46659–46668. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- 6.Werner SR, Lee PA, DeCamp MW, Crowell DN, Randall SK, Crowell PL. Enhanced cell cycle progression and down regulation of p21(Cip1/Waf1) by PRL tyrosine phosphatases. Cancer Lett. 2003;202:201–211. doi: 10.1016/s0304-3835(03)00517-2. [DOI] [PubMed] [Google Scholar]
- 7.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St. Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V, Pallen CJ, Manser E, Hong W. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63:2716–2722. [PubMed] [Google Scholar]
- 9.Liang F, Liang J, Wang WQ, Sun JP, Udho E, Zhang Z-Y. PRL3 Promotes Cell Invasion and Proliferation by Down-regulation of Csk Leading to Src Activation. J. Biol. Chem. 2007;282:5413–5419. doi: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 10.Sun J-P, Luo Y, Yu X, Wang W-Q, Zhou B, Liang F, Zhang Z-Y. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. J. Biol. Chem. 2007;282:29043–29051. doi: 10.1074/jbc.M703537200. [DOI] [PubMed] [Google Scholar]
- 11.Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Res. 2006;66:3153–3161. doi: 10.1158/0008-5472.CAN-05-3116. [DOI] [PubMed] [Google Scholar]
- 12.Kato H, Semba S, Miskad UA, Seo Y, Kasuga M, Yokozaki H. High expression of PRL3 promotes cancer cell motility and liver metastasis in human colorectal cancer: a predictive molecular marker of metachronous liver and lung metastases. Clin. Cancer Res. 2004;10:7318–7328. doi: 10.1158/1078-0432.CCR-04-0485. [DOI] [PubMed] [Google Scholar]
- 13.Achiwa H, Lazo JS. PRL-1 tyrosine phosphatase regulates c-Src levels, adherence, and invasion in human lung cancer cells. Cancer Res. 2007;67:643–650. doi: 10.1158/0008-5472.CAN-06-2436. [DOI] [PubMed] [Google Scholar]
- 14.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 15.Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y, Kwon HJ, Montano S, Georgiadis M, Goebl MG, Harrington MA. Phosphorylation of SIMPL modulates RelA-associated NF-{kappa}B-dependent transcription. Am. J. Physiol. Cell Physiol. 2007;292:C1013–1023. doi: 10.1152/ajpcell.00456.2006. [DOI] [PubMed] [Google Scholar]
- 17.Qin H, Sun Y, Benveniste EN. The transcription factors Sp1, Sp3, and AP-2 are required for constitutive matrix metalloproteinase-2 gene expression in astroglioma cells. J. Biol. Chem. 1999;274:29130–29137. doi: 10.1074/jbc.274.41.29130. [DOI] [PubMed] [Google Scholar]
- 18.Bian J, Sun Y. Transcriptional activation by p53 of the human type IV collagenase (gelatinase A or matrix metalloproteinase 2) promoter. Mol. Cell Biol. 1997;17:6330–6338. doi: 10.1128/mcb.17.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gum R, Lengyel E, Juarez J, Chen JH, Sato H, Seiki M, Boyd D. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J. Biol. Chem. 1996;271:10672–10680. doi: 10.1074/jbc.271.18.10672. [DOI] [PubMed] [Google Scholar]
- 20.Yan C, Wang H, Boyd DD. KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha -induced block of p65/p50 nuclear translocation. J. Biol. Chem. 2001;276:1164–1172. doi: 10.1074/jbc.M008681200. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- 22.Avizienyte E, Frame MC. Src and FAK signalling controls adhesion fate and the epithelial-to-mesenchymal transition. Curr. Opin. Cell Biol. 2005;17:542–547. doi: 10.1016/j.ceb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 23.O’Neill GM, Fashena SJ, Golemis EA. Integrin signalling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10:111–119. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- 24.McCawley LJ, Li S, Wattenberg EV, Hudson LG. Sustained activation of the mitogen-activated protein kinase pathway. A mechanism underlying receptor tyrosine kinase specificity for matrix metalloproteinase-9 induction and cell migration. J. Biol. Chem. 1999;274:4347–4353. doi: 10.1074/jbc.274.7.4347. [DOI] [PubMed] [Google Scholar]
- 25.Stetler-Stevenson WG. Type-IV collagenases in tumor invasion and metastasis. Cancer Metastasis Rev. 1999;9:289–303. doi: 10.1007/BF00049520. [DOI] [PubMed] [Google Scholar]
- 26.Johnsen M, Lund LR, Romer J, Almholt K, Dano K. Cancer invasion and tissue remodeling: common themes in proteolytic matrix degradation. Curr. Opin. Cell Biol. 1998;10:667–671. doi: 10.1016/s0955-0674(98)80044-6. [DOI] [PubMed] [Google Scholar]
- 27.Kuo L, Chang HC, Leu TH, Maa MC, Hung WC. Src oncogene activates MMP-2 expression via the ERK/Sp1 pathway. J. Cell Physiol. 2006;207:729–734. doi: 10.1002/jcp.20616. [DOI] [PubMed] [Google Scholar]
- 28.Chung TW, Lee YC, Kim CH. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J. 2004;18:1123–1125. doi: 10.1096/fj.03-1429fje. [DOI] [PubMed] [Google Scholar]
- 29.Milanini-Mongiat J, Pouysségur J, Pagès G. Identification of Two Sp1 Phosphorylation Sites for p42/p44 Mitogen-activated Protein Kinases. J. Biol. Chem. 2002;277:20631–20639. doi: 10.1074/jbc.M201753200. [DOI] [PubMed] [Google Scholar]
- 30.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2002;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Zhang ZG, Zhang RL, Gregg SR, Hozeska-Solgot A, LeTourneau Y, Wang Y, Chopp M. Matrix metalloproteinase 2 (MMP2) and MMP9 secreted by erythropoietin-activated endothelial cells promote neural progenitor cell migration. J. Neurosci. 2006;26:5996–6003. doi: 10.1523/JNEUROSCI.5380-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobsen EJ, Mitchell MA, Hendges SK, Belonga KL, Skaletzky LL, Stelzer LS, Lindberg TJ, Fritzen EL, Schostarez HJ, O’Sullivan TJ, Maggiora LL, Stuchly CW, Laborde AL, Kubicek MF, Poorman RA, Beck JM, Miller HR, Petzold GL, Scott PS, Truesdell SE, Wallace TL, Wilks JW, Fisher C, Goodman LV, Kaytes PS, Ledbetter SR, Powers EA, Vogeli G, Mott JE, Trepod CM, Staples DJ, Baldwin ET, Finzel BC. Synthesis of a series of stromelysin-selective thiadiazole urea matrix metalloproteinase inhibitors. J. Med. Chem. 1999;42:1525–1536. doi: 10.1021/jm9803222. [DOI] [PubMed] [Google Scholar]
- 33.Su JL, Yang PC, Shih JY, Yang CY, Wei LH, Hsieh CY, Chou CH, Jeng YM, Wang MY, Chang KJ, Hung MC, Kuo ML. The VEGF-C/Flt-4 axis promotes invasion and metastasis of cancer cells. Cancer Cell. 2006;9:209–223. doi: 10.1016/j.ccr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 34.Vial E, Pouysségur J. Regulation of tumor cell motility by ERK mitogen activated protein kinases. Ann. N. Y. Acad. Sci. 2004;1030:208–218. doi: 10.1196/annals.1329.027. [DOI] [PubMed] [Google Scholar]
- 35.Itoh T, Tanioka M, Yoshida H, Yoshioka T, Nishimoto H, Itohara S. Reduced angiogenesis and tumor progression in gelatinase Adeficient mice. Cancer Res. 1998;58:1048–1051. [PubMed] [Google Scholar]
- 36.Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, Shipley JM, Senior RM, Shibuya M. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/s1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 37.Jodele S, Chantrain CF, Blavier L, Lutzko C, Crooks GM, Shimada H, Coussens LM, Declerck YA. The contribution of bone marrowderived cells to the tumor vasculature in neuroblastoma is matrix metalloproteinase-9 dependent. Cancer Res. 2005;65:3200–3208. doi: 10.1158/0008-5472.CAN-04-3770. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh D. Status of the transcription factors database (TFD) Nucleic Acids Res. 1993;21:3117–3118. doi: 10.1093/nar/21.13.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato H, Kita M, Seiki M. v-Src activates the expression of 92-kDa type IV collagenase gene through the AP-1 site and the GT box homologous to retinoblastoma control elements. A mechanism regulating gene expression independent of that by inflammatory cytokines. J. Biol. Chem. 1993;268:23460–23468. [PubMed] [Google Scholar]