Abstract
A new method that uses immobilized trypsin concomitant with ultrasonic irradiation results in ultra-rapid digestion and thorough 18O labeling for quantitative protein comparisons. The reproducible and highly efficient method provided effective digestions in <1 min with a minimized amount of enzyme required compared to traditional methods. This method was demonstrated for digestion of both simple and complex protein mixtures, including bovine serum albumin, a global proteome extract from the bacteria Shewanella oneidensis, and mouse plasma, as well as 18O labeling of such complex protein mixtures, which validated the application of this method for differential proteomic measurements. This approach is simple, reproducible, cost effective, rapid, and thus well-suited for automation.
INTRODUCTION
Recent advances in high-resolution liquid chromatography and mass spectrometry have enabled high-throughput and sensitive global proteome analyses. As a result, mass spectrometry–based proteomics has become a preferred analytical tool for obtaining quantitative protein comparisons. In typical proteomic workflows, the largest time bottlenecks arise out of tedious and/or time-consuming sample preparation protocols. Digestions alone can take several hours to complete and this time can be doubled if performing an additional isotopic labeling step [e.g., ICAT1, SILAC2, and 18O3, 4] for quantitative proteomics. Of all the current isotopic labeling methods 18O labeling is arguably one of the most affordable, facile and versatile of all current labeling strategies. Qian et al recently demonstrated 18O labeling on a universal pooled reference sample, which was added to 18 severe burn patient plasma samples5. Using this strategy, both label-free quantitation was attainable simultaneously with precise peptide abundance ratios on a limitless number of samples. The label-free and labeled ratios together provided more precision and a greater number of protein abundance changes than either method alone demonstrating the utility of the approach.
In a typical 18O labeling protocol, proteins are first digested with trypsin for several hours, dried down, and then labeled overnight by incubation with H218O in the presence of trypsin. After quenching the trypsin with formic acid or thermal denaturation to stop the reaction, the two samples are combined prior to LC-MS analysis. In addition to long digestion times, traditional 18O labeling processes are challenged by trypsin autodigestion, back-exchange, and sample clean-up steps that can result in significant sample losses. In response to these challenges, we have investigated the use of immobilized trypsin in conjunction with high intensity focused ultrasound (HIFU) to produce a more robust digestion, as well as a quick and easy 18O labeling procedure. Immobilized trypsin is advantageous in that it is less susceptible to autolytic digestion, which leads to higher stability than free trypsin6, and can readily be removed once the reaction is complete, thereby helping in the avoidance of back-exchange7. The catalytic activity of trypsin is improved by immobilization due to an increased enzyme concentration. Since the enzyme concentration is directly proportional to the reaction rate (according to the Michaelis-Menten equation), a higher trypsin concentration results in shorter incubation periods. HIFU also increases the reaction kinetics by producing micro cavitation events, i.e., quick and centralized bursts of increased temperature and pressure8. Additionally, the use of ultrasonic energy aids in mixing and solubilizing proteins, which enhances enzymatic digestions9–11.
Herein, we report the evaluation of a HIFU-immobilized trypsin method for quantitative proteomic applications. The method was evaluated by digesting a diverse array of protein mixtures that ranged in protein complexity (i.e., BSA, Shewanella onedensis proteome, and mouse plasma), followed by 18O labeling of some samples to demonstrate the rapid HIFU-immobilized trypsin technique for determination of changes in protein abundances between samples. The entire process, including denaturation, digestion, and labeling was reduced to <5 min, making this method one of the most efficient of all current quantitative LC-MS proteomic technologies.
EXPERIMENTAL PROCEDURES
Materials and reagents
Immobilized trypsin was purchased from Applied Biosystems (Foster City, CA), Tris[2-carboxyethyl]phosphine (TCEP) and a BCA protein assay kit was purchased from Pierce (Rockford IL, USA). Bovine serum albumin (BSA), urea, iodoacetamide (IAA), ammonium bicarbonate, formic acid, 18O water, mouse plasma, and HPLC grade solvents were purchased from Sigma-Aldrich (St. Louis, MO).
Digestion Procedures
BSA was used to evaluate the method under several different conditions. First, equal aliquots of 100 µg of BSA were denatured in 8 M urea, reduced, and then alkylated simultaneously with 5 mM TCEP and 50 mM iodoacetamide in 25 mM ammonium bicarbonate (pH 8.25) for 3 min under ultrasonic irradiation. The reduced and alkylated samples were diluted 4-fold with 25 mM ammonium bicarbonate to decrease urea, TCEP, and IAA concentrations. 1 to 10 µL of immobilized trypsin slurry (as supplied by the manufacturer) was added to a final volume of 100 µL, and the solutions were sonicated for 15, 30, 60 or 120 s at 2% of the maximum ultrasound amplitude, which yielded 1–3 W of power at the end of the tip. All of the experiments employed a Sonicator 3000 instrument and 3.2 mm Microtip that were obtained from Mixonix (Farmingdale, NY, USA). The slurry was washed three times with 25 mM ammonium bicarbonate to eliminate non-volatile preservatives before adding it to the reaction tube. Finally, the enzymatic digests were filtered to remove the trypsin beads using 0.45 µm cut-off spin filters from Beckman Coulter, Inc. (Fullerton, CA) and the filtered peptide mixtures were transferred to new centrifuge tubes, acidified, and frozen with liquid N2 to stop the reaction. The samples were then dried by centrifugal evaporation and stored at −20 ºC.
The Shewanella oneidensis proteome was prepared as described previously12, and the soluble portion of the proteome was aliquoted as 50 µg samples and dried down. Dried proteins were resuspended in 8 M urea in 25 mM ammonium bicarbonate, 5mM TCEP and 50 mM IAA and then subjected to 3 min of ultrasonic irradiation. The samples were diluted and 5 µL of previously washed immobilized trypsin slurry was added after which the samples were sonicated for 30 to 90 s. A control was digested in parallel for 4 h in a thermo-mixer at 37 °C with shaking at 1500 rpm. Two replicate digestions were performed for each time point. After digestion, samples were acidified, and then immobilized trypsin was filtered out using again a 0.45 µm cut-off. Finally, digestion products were snap-frozen in liquid nitrogen and dried by centrifugal evaporation.
Mouse plasma was diluted 5 fold using 8 M urea in 25 mM ammonium bicarbonate and then reduced, alkylated, and digested in the same way as described above. In this case, no technical replicates were performed.
Post-digestion 18O labeling
Plasma samples were prepared as previously described with minor modifications13. Briefly, dried peptides were cleaned by loading them onto a 1-mL SPE C18 Discovery solid phase extraction (SPE) column (Supelco, Bellefonte, PA). Each sample was washed with 4 mL of 0.1% trifluoroacetic acid (TFA)/5% acetonitrile (ACN). Peptides were eluted from the SPE column with 1 mL of 0.1% TFA/80% ACN and then lyophilized. 100 µg aliquots of the desalted peptides were dried by centrifugal evaporation in a 0.6-mL centrifuge tube and the peptides were re-dissolved in 50 µL of 100 mM ammonium acetate (pH 6.75) with 5 µL of immobilized trypsin. After drying again by centrifugal evaporation, the samples were dissolved in 100 µL of either H216O or H218O (95%, Sigma-Aldrich) and sonicated for 30 to 90 s. Labeling was stopped by adding formic acid to 1% and the immobilized trypsin was filtered out. Samples were then mixed in a 1-to-1 ratio and prepared for LC-MS analysis.
As a control experiment to evaluate the complete proteomic workflow two identical aliquots of Shewanella oniedensis were subjected to reduction and alkylation, digestion, and labeling (either 18O or 16O). In order to avoid sample clean-up for post digestion labeling we incorporated a trifluoroethanol (TFE) assisted digestion, previously developed in our laboratory14 . Dried extracts were solubilized in a 50:50 TFE:50 mM ammonium bicarbonate solution and reduced and alkylated by adding TCEP and IAA to a final concentration of 5 mM and 50 mM respectively, followed with a 3 min sonication. Samples were subsequently diluted 10 times, and 5 µL of immobilized trypsin was added. The samples were again sonicated at 2% intensity for 30 s to perform the digestion. The enzymatic activity was stopped by acidification of the samples and physical removal of the immobilized trypsin by filtration. Filtrates were dried down by centrifugal evaporation. By using TFE as an organic co-solvent acting as a chaotrope there was no need for a separate sample cleanup. Post-digestion enzymatic labeling was performed as explained above. Once the labeling process was finished, samples were combined in a 1-to-1 ratio and submitted for LC-MS analysis.
LC-MS(/MS) analysis
For the BSA analyses, 500 fmol of the protein digest were analyzed by LC-MS/MS using an Agilent HPLC-Chip system coupled to a MSD XCT Ultra ion trap (Agilent Technologies, Santa Clara, CA). Separations were performed on a chip that contained a 40-nL enrichment column and a 43 mm × 75 µm analytical column, both packed with 5 µm ZORBAX 300SB C18 particles. A flow rate of 1 µL/min was employed for loading and enriching the sample. Peptides were eluted at 600 nL/min, using either a 1) 5 min gradient from 5% to 90% B (Solvent B: 0.5% formic acid in water:acetonitrile 10:90; Solvent A: 0.5% formic acid in water:acetonitrile 97:3) with a separation window of ∼2 min and total analysis time of 12 min, or 2) 55 min gradient from 5% to 90% B.
For the S. oneidensis proteome and mouse plasma analyses, 2 µg of the digested peptides were analyzed using an in-house developed capillary LC system coupled online to a linear ion trap mass spectrometer (Thermo-Fisher, San Jose, CA) with an in-house developed ESI source12 . Reversed-phase capillary columns were prepared by slurry packing 5-µm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 150 µm × 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) that utilized a 2-µm stainless steel retaining screen within a stainless steel union (Valco Instruments Co., Houston, TX). The mobile phase consisted of 0.2% acetic acid and 0.05% TFA in water (A) and 0.1% TFA in 90% acetonitrile/10% water (B). Mobile phases were degassed online using a vacuum degasser (Jones Chromatography Inc., Lakewood, CO), and the HPLC system was equilibrated at 10,000 psi with 100% mobile phase A for the initial starting conditions. After loading 2.5 µg of peptides onto the column, the mobile phase was held at 100% A for 20 min. Exponential gradient elution was performed by increasing the mobile-phase composition from 0 to 77% B over ∼100 min, using a stainless steel mixing chamber, followed by column washing at 100% B for 10 min. Data were collected in a data-dependent mode (m/z 400–2000) where the ten most abundant ions were selected for tandem MS after a full scan. A dynamic exclusion time of 1 min and normalized collision energy of 35% were used for the entire run. A potential of 2.2 kV was applied to the union to form the electrospray. Quantitative analyses were carried out using the same chromatography system but in this case coupled on line to a hybrid LTQ-Orbitrap mass spectrometer. Data were collected in a data-dependent mode where the six most abundant ions were selected for tandem MS in the ion trap after a full, high-resolution survey scan.
Protein identification and validation
Peptide identification was performed using SEQUEST15 to search MS/MS spectra against an IPI Mus musculus or Shewanella oneidensis databases downloaded from NCBI. Database search parameters included a dynamic modification for Met oxidation and for carbamidomethylation of Cys. Peptide identification statistics that included estimating random match probabilities and false discovery rates were performed using the two-variable Gaussian method16 with some modifications as described elsewhere17. For application of the accurate mass and time (AMT) tag strategy, the high-resolution LC-MS spectra were processed using Decon-2LS software (publicly available at http://omics.pnl.gov/software/) to obtain peak lists that contained the monoisotopic mass and observed charge. A reference AMT tag database was generated from peptides previously identified by LC-MS/MS, using MTDB creator software (publicly available at http://omics.pnl.gov/software/). Once generated, high resolution MS datasets were matched to the reference database as described below.
Quantitative proteomic strategy
LC-MS datasets were analyzed using VIPER software (publicly available at http://omics.pnl.gov/software/). Details of the quantitation strategy are described elsewhere17. Briefly, data processing ratios were calculated across the chromatographic peak, taking into account the labeling efficiency18, which corrects the ratio as a function of the remaining unlabelled peptide from the labeled sample. Then the ratio calculated for each pair is averaged from several ratios calculated across the chromatographic peak and an error in terms of the standard deviation is used to discriminate between poorly matched peaks and real pairs. Pairs were then identified by comparing the mass and the elution time of the pairs with the mass and the normalized elution time (NET) of the peptides in the AMT tag database19 (match tolerance of ±1.4 ppm and ± 0.02 LC NET). Finally a statistical analysis was carried out in the same way as described before4, i.e., fitting a histogram of the ratios to a Gaussian distribution and using the average and standard deviations to discriminate between pairs that represented a real differential expression event.
RESULTS AND DISCUSSION
Evaluation of HIFU and immobilized trypsin digestion performance
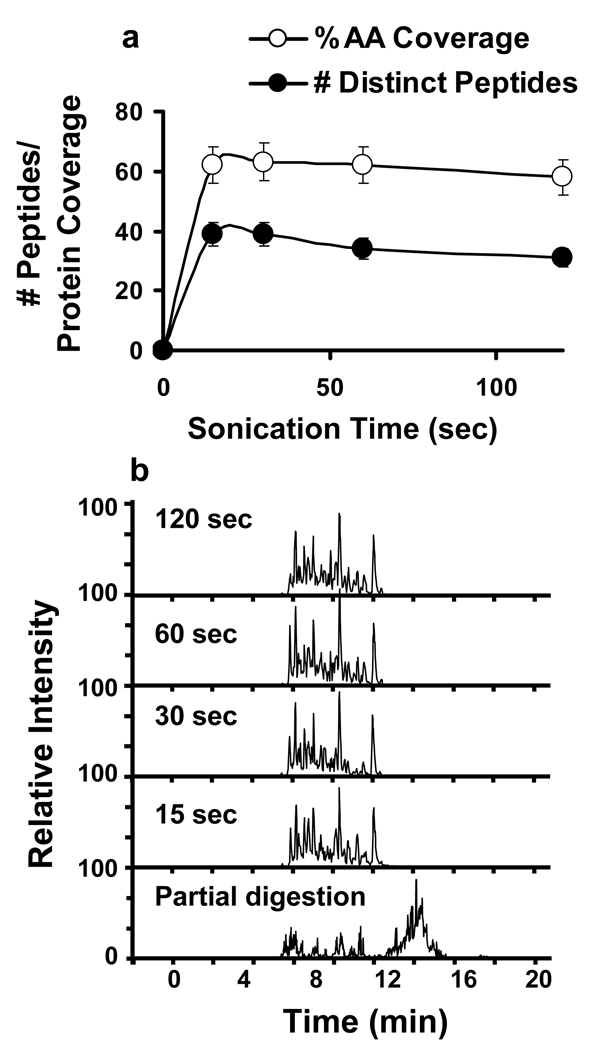
Protein digestion was initially evaluated using BSA as a standard protein. HIFU time varied from 15 s to 2 min for a sample volume of 100 µL that contained 100 µg total protein and 10 µL of washed immobilized trypsin slurry. Following digestion, the samples were acidified and the trypsin-coated beads were removed by filtering. The tryptic digests were then analyzed by LC-Chip-MS/MS in a completely automated fashion, using a 5 min gradient. Optimal digestion occurred within 15 s, and no significant differences in terms of coverage or number of peptides were observed for sonication times in the range of 15–120 s (Figure 1a). In addition, no traces of undigested protein where detected. Figure 1b shows the chromatograms of all the experiments in addition to a control BSA digestion under unfavorable conditions, where undigested or partially digested protein was observed. Protein coverage was typically >60% using the aforementioned fast 5 min gradient (Figure 1a). This coverage is equivalent to that of the control experiments performed with a typical overnight digestion (data not shown). Furthermore, this rapid digestion technique increased protein coverage by 2.3 fold compared to a similar technique that utilized a combination of microwave technology and magnetic silica microspheres, which only obtained 26% coverage20. The minimal amount of immobilized trypsin slurry necessary for maximal protein coverage was 2 µL (see supplementary figure), which suggests that costs can effectively be minimized to < $1 for immobilized trypsin per 100 µg of sample digested. While the price of 2 µg of free trypsin is similar, activity decreases more rapidly with free trypsin (especially when used in conjunction with ultrasound techniques), which may lead to incomplete digestions under certain circumstances. Moreover, free trypsin cannot be removed quickly and thoroughly from a protein mixture, which is important for 18O labeling strategies in order to avoid back exchange.
Figure 1.
a) Comparison of the number of uniquely identified peptides and BSA amino acid coverage at different irradiation times 15, 30, 60, and 120 s. Empty circles correspond to protein coverage. Filled circles correspond to number of identified peptides. b) Reprentative LC-MS/MS chromatograms for each of 15, 30, 60 and 120 s digestion experiments as well as for a control BSA “incomplete digestion”.
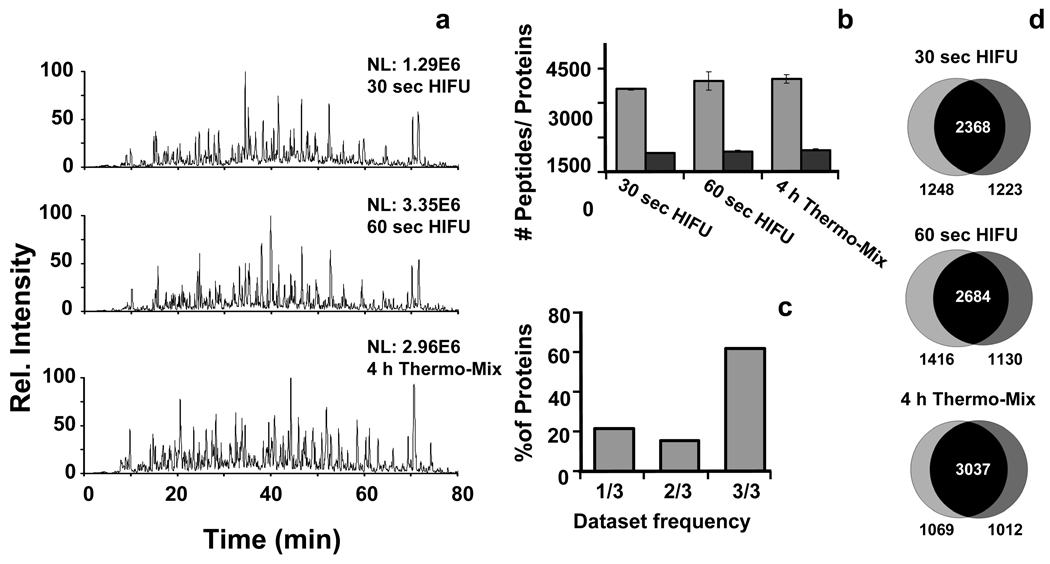
A S. oneidensis (an anaerobic soil bacteria) cell lysate was digested to evaluate the utility of the immobilized trypsin combined with ultrasound on a complex proteome sample. S. oneidensis cell lysates were digested in duplicate using HIFU and immobilized trypsin for 30 s, 60s, or a standard 4-h digestion (i.e., control) at 37 ºC with mixing at 1500 revolutions per minute (rpm). Representative chromatograms of each digestion and the amount of information generated in terms of unique peptide and protein identification was similar in all cases (Figure 2a and b) and the reproducibility of peptide identification among duplicates was >80% (Figure 2d). In addition to being comparable in terms of digestion efficiency and reproducibility to standard digestions, the combined use of immobilized trypsin with HIFU provides significant advantages over the traditional protocol in that the digestion procedure is complete within a few minutes rather than a few hours and the trypsin can be removed quickly. Furthermore, by rolling-up the identified peptides to the protein level, we show (Figure 2c) that more than 60% of the identified proteins are identified in three out of the three experiments, which represents an extraordinary achievement in terms of reproducibility in a proteomic experiment.
Figure 2.
Evaluation of the digestion process combining ultrasonic energy and immobilized trypsin using a Shewanella oneidensis cell extract. a) The representative chromatograms corresponding to different digestion times show a great similarity which is further demonstrated by b) the histograms obtained for each sample. Grey bars indicate unique peptide identifications and dark bars indicate the number of identified proteins. c) Histograms obtained by analyzing the percentage of proteins common to the different datasets. d) Venn diagrams showing the peptide overlap between technical duplicates.
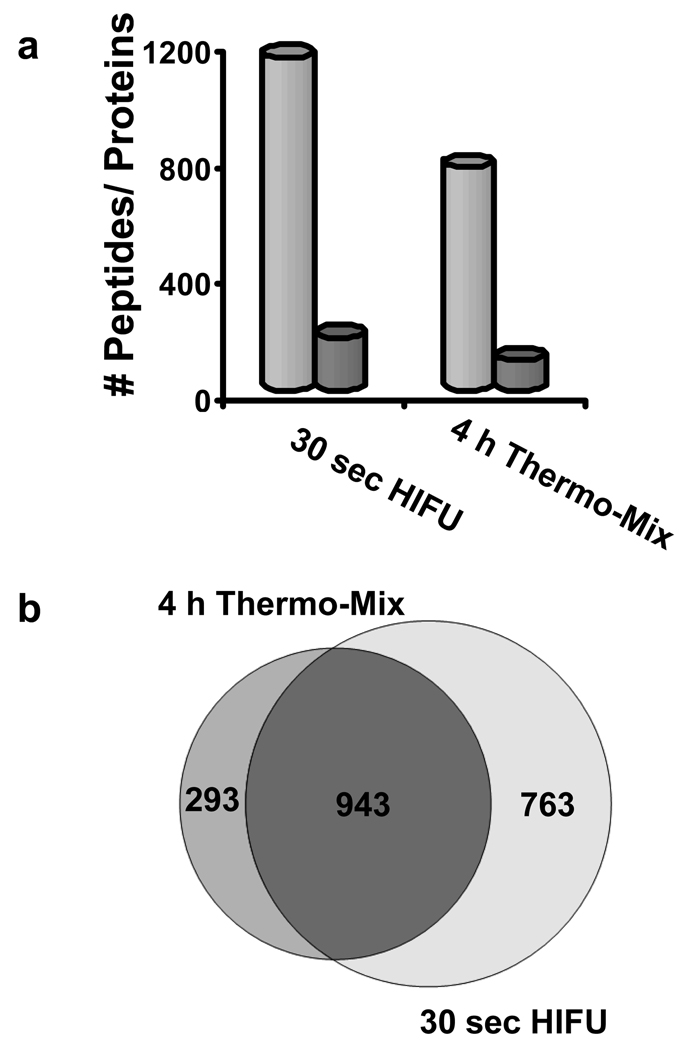
Having demonstrated that the method worked well on prokaryotic samples, the strategy was next evaluated using a more complex proteome represented by a non-depleted mouse plasma sample. The plasma was diluted, reduced, and then alkylated during a 3 min ultrasonic irradiation. Trypsin beads were added, and the plasma-bead mixture was sonicated for 30 s. For comparison, we employed a standard 4-h digestion using a thermomixer set at 37 ºC with shaking at 1500 rpm. The histogram in Figure 3a shows the number of unique peptides identified with a false discovery rate (FDR) of < 1% along with the corresponding number of proteins identified. A 76% overlap of shared peptides was identified between the 4 h thermo-mix digestion and the 30 sec HIFU digestion relative to all peptides identified in the 4 h thermo-mix digestion and a 55% overlap of shared peptides was identified in the 30 sec HIFU digestion relative to all peptides identified in the 30 sec HIFU digestion (Figure 3b). Overall, the HIFU digestion identified 470 more peptides (∼ 40%) than the 4 h thermo-mix digestion. The overlap between the two digestion methods was above the levels expected even between technical replicates of the same sample in a typical shotgun proteomics experiment21.
Figure 3.
a) Histogram showing the number of identified peptides (light-grey bars) and proteins (dark-grey bars) comparing a 30 s HIFU digestion versus a 4 h digestion using the thermomixer. b) Venn diagram showing the overlap between the samples in terms of peptide identifications.
Evaluation of HIFU and immobilized trypsin for the digestion and 18O labeling of complex protein mixtures
18O labeling strategies are useful for quantitative protein comparisons among samples 4, 22. However, C-terminal oxygens can back exchange over time in the presence of 4, 21residual active trypsin and non-labeled aqueous solutions (i.e., when reconstituted in solution for LC-MS analysis), and this can lead to inaccurate quantitation. HIFU coupled with immobilized trypsin beads can reduce such back exchange by enabling fast removal of the trypsin-coated beads. To determine whether this method allowed for complete and reproducible labeling, we applied the labeling method to a mouse serum proteome for 30, 60 and 90 s. Afterwards, the samples were quickly acidified to quench any remaining trypsin activity, the trypsin was removed by filtering the beads, and the 16O and 18O labeled samples were mixed in a 1-to-1 ratio and dried.
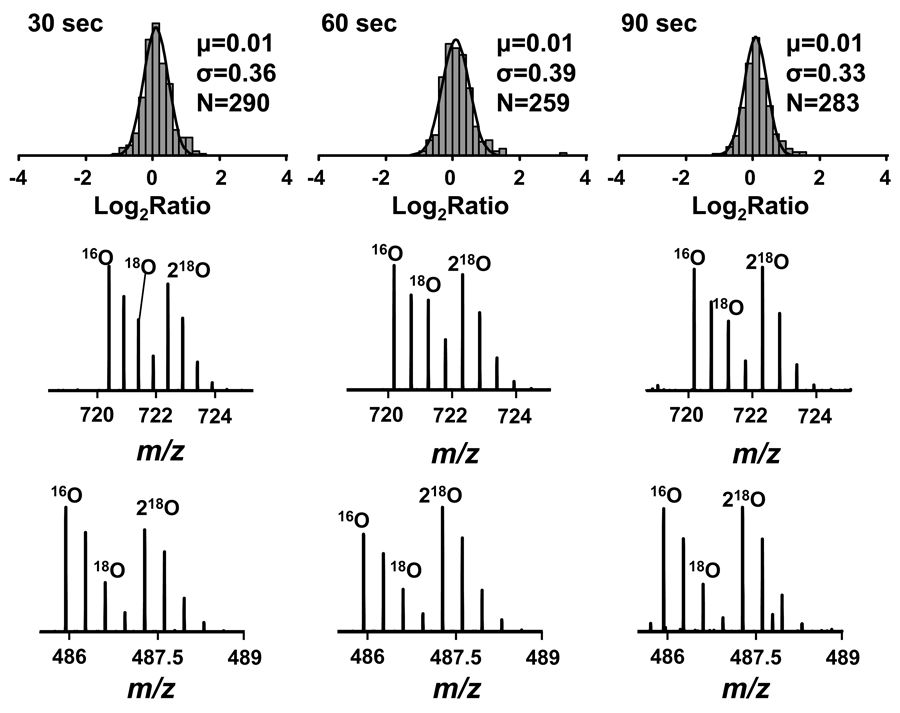
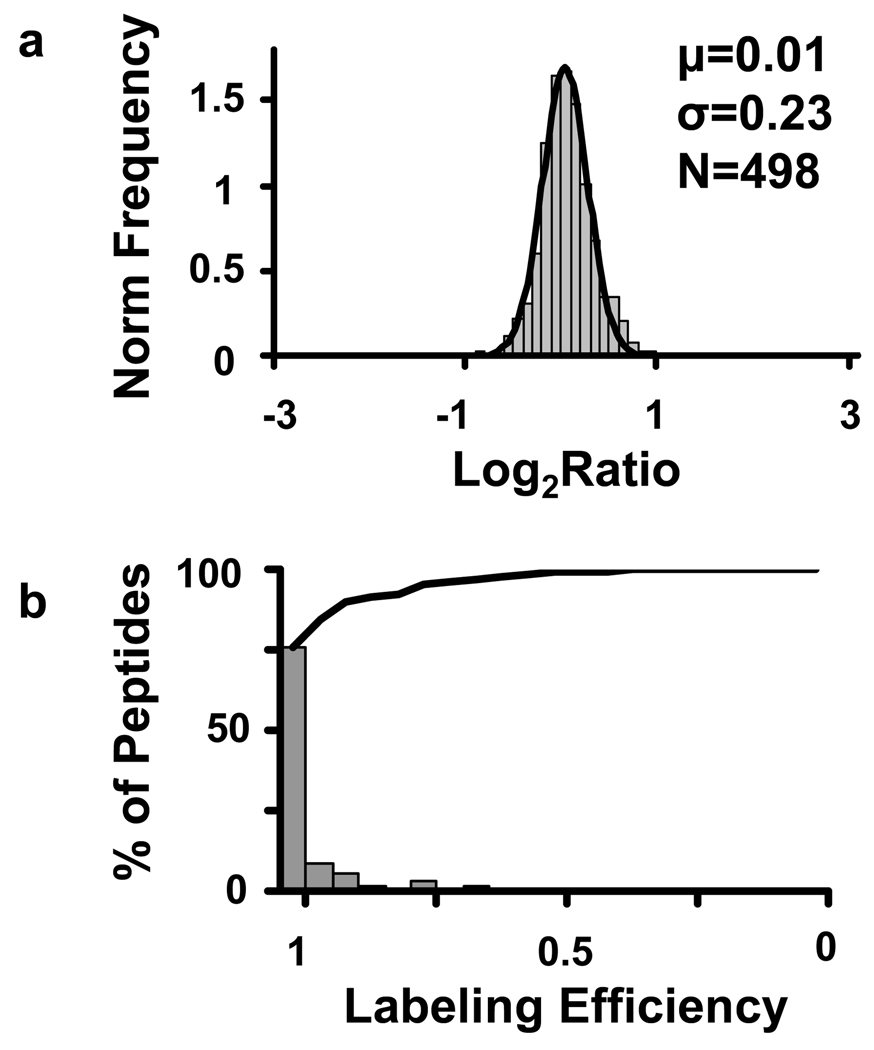
Statistical analysis shows that all quantified peptides followed a normal distribution with statistically significant changes (i.e., those with an FDR < 10%) occurring in < 1% of the identified peptides (Figure 4). This level of false identifications remained consistent through multiple replicate analyses (data not shown), which highlights the reproducibility and accuracy of the method for quantitative experiments. As a control experiment to evaluate the complete proteomic workflow, two identical aliquots of Shewanella oniedensis were subjected to reduction and alkylation, digestion, and labeling by using immobilized enzymes and HIFU. As shown in Figure 5 more than 90% of the quantified pairs had a labeling efficiency of >90%, which allowed for accurate peptide quantitation, even with an irradiation time of only 30 s.
Figure 4.
Results obtained in a negative control experiment using HIFU and immobilized trypsin for post-digestion labeling. Upper panel shows the results obtained by fitting the histogram of the log2(16O/18O) ratios to a Gaussian function for different irradiation times. The bottom panel shows representative MS spectra for two peptides from the experiment: VVIEDGVGDAVLTR, PPTVTITSR.
Figure 5.
Summary of the results obtained following HIFU assisted digestion and labeling with immobilized trypsin applied to the Shewanella oneidensis proteome. Panel a) Histogram of the log2(16O/18O) ratios fitted to a Gaussian function. b) Histogram of the labeling efficiency calculated from the quantified peptides. The solid line represents the cumulative frequency of the labeling efficiency.
The work presented here demonstrates that the use of immobilized trypsin in the presence of ultrasonic energy produces a faster and more complete digestion and/or 18O exchange compared to classic thermo-mixing techniques. Several components in this method contribute to the acceleration of the process. First, in the immobilized form, trypsin maintains an active conformation in the presence of ultrasonic energy and other harsh conditions, such as the presence of organic solvents, and elevated temperatures. Second, the increased load capacity of immobilized trypsin increases the reaction rate due to the higher enzyme-to-protein ratio. Third, with our method, we create microstreaming events in the solution, which are propagated by the immobilized trypsin beads. Microstreaming is a phenomenon whereby the acoustic waves are reflected back and forth in a medium due to the presence of micro particles or air bubbles22–24, and has been applied to accelerate DNA hybridization reactions in biochips25, 26. The reflected energy creates supplementary disturbances that further enhance reactant transport and mixing. This phenomenon is often incorporated into ultrasound processes to avoid a potential drawback of sonication, which is that acoustic waves in the liquid can lose energy due to resistance as they propagate through the solution. These loses are considerable and especially prevalent when the waves encounter the plastic microfuge tube walls that absorb most of the energy22.
CONCLUSIONS
We successfully demonstrated a method for coupling HIFU with immobilized trypsin to perform ultra-rapid digestion and/or 18O labeling for proteome applications. The method proved successful for rapidly digesting the globular protein bovine serum albumin and the complex proteome mixtures of S. oneidensis lysates and mouse plasma. Although the denaturing effects of ultrasonic irradiation enhanced the digestion of the plasma proteins, no appreciable differences were observed between the novel and traditional protocols, even though the digestion time was reduced from 4 h to less than a few minutes. When the technique was applied to post-digestion 18O labeling experiments, more than 90% of the quantified pairs had a labeling efficiency of >90%, which allowed for accurate peptide quantitation, even with an irradiation time of only 30 s. In addition, the digestions proved to be extremely robust and reproducible, as illustrated by the extremely tight Gaussian distributions. When added together, the labeled and unlabeled samples had equal ratios, which further highlight the reproducibility of this method for both digestion and labeling.
We anticipate the ultra-rapid labeling method presented here will become the mainstay for quantitative proteome analysis for not only two sample comparisons but for large-scale multiple sample analyses as well. The combined use of highly stable enzymes and ultrasound provides a means of dramatically reducing both sample preparation times and digestion and labeling costs. The method is compatible with many downstream workflows in proteome analysis such as HPLC fractionation and it is amenable to automation making faster, more reproducible high-throughput proteomics achievable. Overall, the combination of highly stable enzymes and HIFU cavitation microstreaming in microfluidic devices holds the potential for making the ultimate lab-on-a-chip a reality for proteomic experiments.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Tyler Heibeck, Dr. Weijun Qian, Dr. Matthew Monroe, Angela Norbeck, and Penny Colton for their helpful assistance and suggestions. Moreover, the EMSL High-Throughput Proteomics group for technical assistance in the mass spectrometry part. Portions of this work were supported by the NIH National Center for Research Resources (RR018522), NIH National Cancer Institute (R21 CA12619-01), and the Pacific Northwest National Laboratory’s (PNNL) Laboratory Directed Research and Development Program. PNNL is operated for the U.S. Department of Energy by Battelle under contract DE-AC05-76RLO1830.
Abbreviations
- ACN
Acetonitrile
- AMT
Accurate Mass and Time
- BSA
Bovine Serum Albumin
- ESI
Electrospray Ionization
- FDR
False Discovery Rate
- HIFU
High Intensity Focused Ultrasound
- HPLC
High Pressure Liquid Chromotography
- IAA
Iodoacetamide
- ICAT
Isotope-Coded Affinity Tags
- LC-MS/MS
Liquid Chromotography Tandem Mass Spectrometry
- PCR
Polymerase Chain Reaction
- SPE
Solid Phase Extraction
- TCEP
Tris[2-carboxyethyl]phosphine
- TFA
Trifluoroacetic acid
REFERENCES
- 1.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nature Biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Molecular and Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Analytical Chemistry. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Ferrer D, Ramos-Fernandez A, Martinez-Bartolome S, Garcia-Ruiz P, Vazquez J. Proteomics. 2006;6 Suppl 1:S4–S11. doi: 10.1002/pmic.200500375. [DOI] [PubMed] [Google Scholar]
- 5.Qian WJ, Liu T, Petyuk VA, Gritsenko MA, Petritis BO, Polpitiya AD, Kaushal A, Xiao W, Finnerty CC, Jeschke MG, Jaitly N, Monroe ME, Moore RJ, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, Smith RD. J Proteome Res. 2009;8:290–299. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freije JR, Mulder PP, Werkman W, Rieux L, Niederlander HA, Verpoorte E, Bischoff R. J Proteome Res. 2005;4:1805–1813. doi: 10.1021/pr050142y. [DOI] [PubMed] [Google Scholar]
- 7.Sevinsky JR, Brown KJ, Cargile BJ, Bundy JL, Stephenson JL., Jr Anal Chem. 2007;79:2158–2162. doi: 10.1021/ac0620819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason TJ, Lormer JP. Sonochemistry, theory, applications and uses of ultrasound in chemistry. 1st ed. Chischesteer, England: Ellis Horwood Limited; 1988. [Google Scholar]
- 9.López-Ferrer D, Cañas B, Vázquez J, Lodeiro C, Rial-Otero R, Moura I, Capelo JL. Trends in Analytical Chemistry. 2006;25:996–1005. [Google Scholar]
- 10.Lopez-Ferrer D, Capelo JL, Vazquez J. J Proteome Res. 2005;4:1569–1574. doi: 10.1021/pr050112v. [DOI] [PubMed] [Google Scholar]
- 11.Capelo JL, Ximenez-Embun P, Madrid-Albarran Y, Camara C. Anal Chem. 2004;76:233–237. doi: 10.1021/ac034871d. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Smith RD. Analytical Chemistry. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 13.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp Ii DG, Khan AH, Smith DJ, Smith RD. J Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, Yates IIIJR. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Ferrer D, Martinez-Bartolome S, Villar M, Campillos M, Martin-Maroto F, Vazquez J. Anal Chem. 2004;76:6853–6860. doi: 10.1021/ac049305c. [DOI] [PubMed] [Google Scholar]
- 17.López-Ferrer Daniel, Heibeck Tyloer H, Petritis Konstantinos, Hixson Kim K, Qian Weijun, Monroe Matthew E, Mayampurath Anoop, Moore Ronald J, Belov Mikhail E, Camp II David G, D SR. Journal of Proteome Research. 2008 doi: 10.1021/pr800161x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos-Fernandez A, Lopez-Ferrer D, Vazquez J. Mol Cell Proteomics. 2007;6:1274–1286. doi: 10.1074/mcp.T600029-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Anal Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 20.Lin S, Yao G, Qi D, Li Y, Deng C, Yang P, Zhang X. Anal Chem. 2008 doi: 10.1021/ac800023r. [DOI] [PubMed] [Google Scholar]
- 21.Resing KA, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, Old WM, Cheung HT, Russell S, Wattawa JL, Goehle GR, Knight RD, Ahn NG. Analytical Chemistry. 2004;76:3556–3568. doi: 10.1021/ac035229m. [DOI] [PubMed] [Google Scholar]
- 22.Shoh A. Ieee Transactions on Sonics and Ultrasonics. 1975;Su22:60–71. [Google Scholar]
- 23.Liu RH, Lenigk R, Druyor-Sanchez RL, Yang J, Grodzinski P. Anal Chem. 2003;75:1911–1917. doi: 10.1021/ac026267t. [DOI] [PubMed] [Google Scholar]
- 24.Liu RH, Yang J, Pindera MZ, Athavale M, Grodzinski P. Lab Chip. 2002;2:151–157. doi: 10.1039/b201952c. [DOI] [PubMed] [Google Scholar]
- 25.Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 26.Paxton WF, O'Hara MJ, Peper SM, Petersen SL, Grate JW. Anal Chem. 2008 doi: 10.1021/ac800160n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.