Abstract
A simple method to determine the relative stereochemistry of statine amino acids (γ-amino-β-hydroxyacids) using 1H NMR spectroscopy is described. Configurational assignment of statine units within complex natural products is possible without degradation or derivatization as the syn and anti diastereomers can be distinguished using a combination of chemical shift and coupling constant information derived from the α-methylene ABX system. Seventy-three examples are provided demonstrating the scope and limitations of the methodology. These examples range in complexity from simple statine units to cyclic depsipeptides, such as tamandarin B.
Introduction
Modern structure determination of small molecules remains a challenging problem. The number of structural reassignments reported each year is testimony to the myriad of potential pitfalls.1 While classical approaches to structure determination rely heavily on chemical degradation, most modern approaches use non-destructive techniques to provide the same connectivity information.2 Any structure determination encompasses three discrete assignments: planar, relative and absolute. Advances in NMR instrumentation and NMR pulse sequences have greatly simplified the assignment of the planar structure, i.e., the constitutional connectivites between the various nuclei.3 Conversely, relative4 and absolute stereochemical assignments5 are becoming more challenging as the isolation of submicromolar quantities of metabolites becomes increasingly more common.6
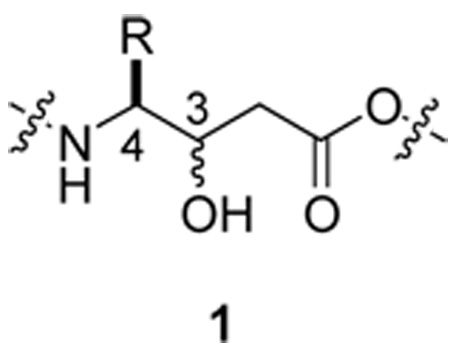
Statine-containing peptides exemplify these challenges. Statine amino acids (1), γ-amino-β-hydroxy acids, are present in several compounds including the anticancer agents didemnin B,7 tamandarin B,8 and hapalosin.9 This isostere is often associated with improved pharmacokinetic properties and increased potency.10,11 While the planar moiety is easily identified by conventional NMR spectroscopy, there is no simple method to determine the relative configuration of the vicinal chiral centers bearing the amine and the alcohol. Traditional solutions to this problem have involved either hydrolysis and subsequent HPLC/GC comparison with synthetic standards or conversion to the oxazolidine/oxazolidinone derivative.
Ideally, a simple NMR-based method could be used to determine the relative configuration of the statine unit within the intact peptide. This would be analogous to the Universal NMR databases developed by Kishi for configurational assignment of polyketides, which utilizes the distinctive 13C NMR chemical shift patterns of the diastereomers.12 Several years ago we noted that it might be possible to distinguish the two diastereomers of 4-amino-3-hydroxy-5-phenylpentanoic acid using the proton signals for H2-2,13 but little supporting evidence was available at that time for the generality of the method.
Results and Discussion
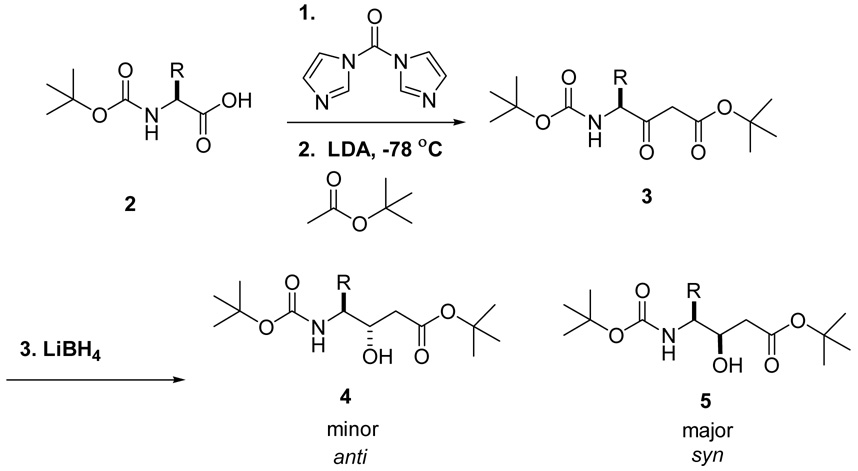
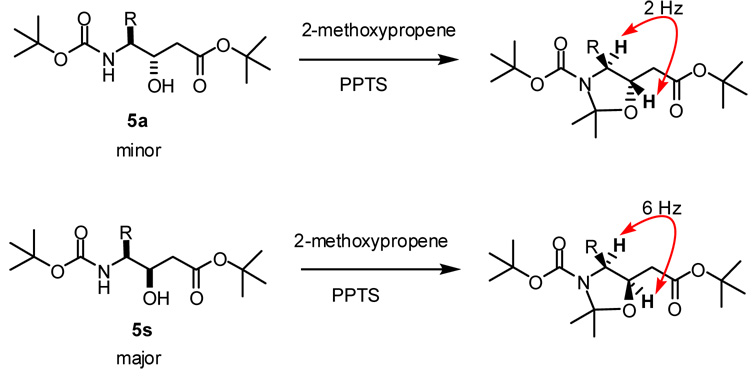
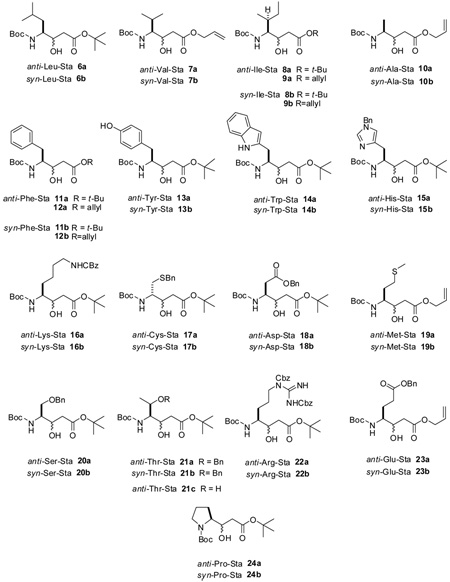
To explore these issues, we synthesized a variety of γ-amino-β-hydroxy acids derived from the proteogenic amino acids using a simple procedure (Scheme 1). The imidazole activated Boc-amino acid was coupled with the enolate derived from t-butyl acetate or allyl acetate. Subsequent reduction with lithium or sodium borohydride produced a mixture of C-3 diastereomers,14 which were separated by normal phase chromatography to yield the protected statine units 4 and 5. As expected, the syn diastereomer, formed via a chelation controlled reduction,14 was the major product in all cases.15 This stereochemical outcome was confirmed by converting representative compounds 11, 12, 16, 21, and 22 to the oxazolidine derivatives (Scheme 2)16 and analysis of the vicinal proton-proton coupling constant between the two chiral centers. The syn and anti derivatives displayed 3JH4,H3 values of approximately 6 and 2 Hz, respectively.17,18
Scheme 1.
Synthesis of Statine Units.
Scheme 2.
Conversion to the Oxazolidine Derivatives.
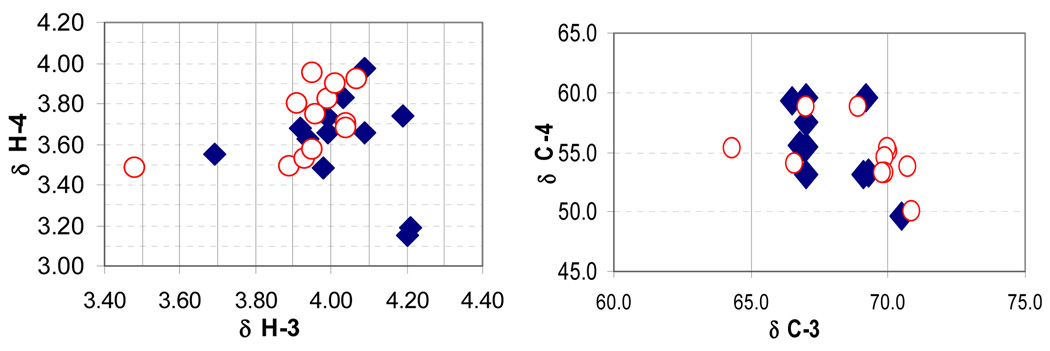
A comparison of the spectral data for the diastereomers revealed no predictive clustering of carbon or proton chemical shifts that could be used to assign the relative configuration of an unknown statine unit (Figure 1).19 Likewise, the magnitude of the 3J values between H-3 and H-4 did not provided a consistent means of distinguishing the respective diastereomers.20 These results are likely due to the conformational flexibility between the two chiral centers C-3 and C-4 and are consistent with the trends observed for 1,2-diols21 and for α,γ-dimethyl-β-hydroxycarbonyls.22
Figure 1.
a) Chemical shifts (ppm) for H-3 and H-4 of the syn (blue diamond) and anti (red circle) statine diastereomers. b) Chemical shifts for C-3 and C-4 of the syn (blue diamond) and anti (red circle) statine diastereomers.
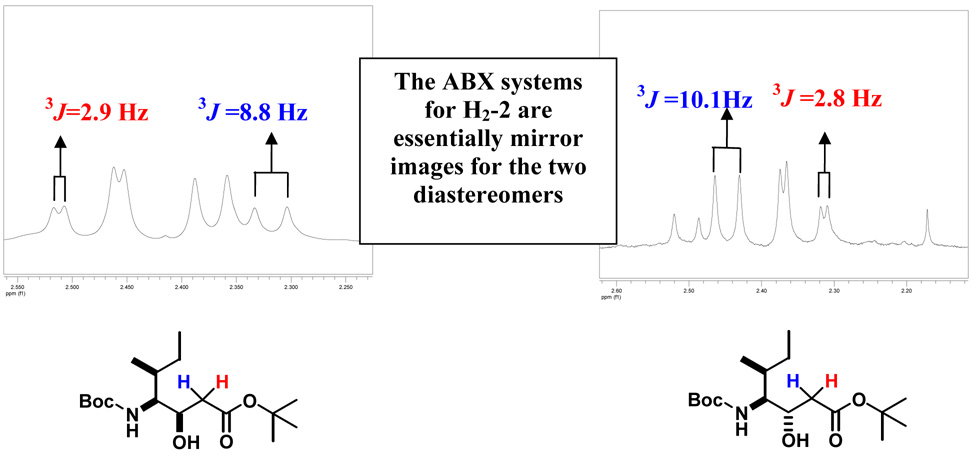
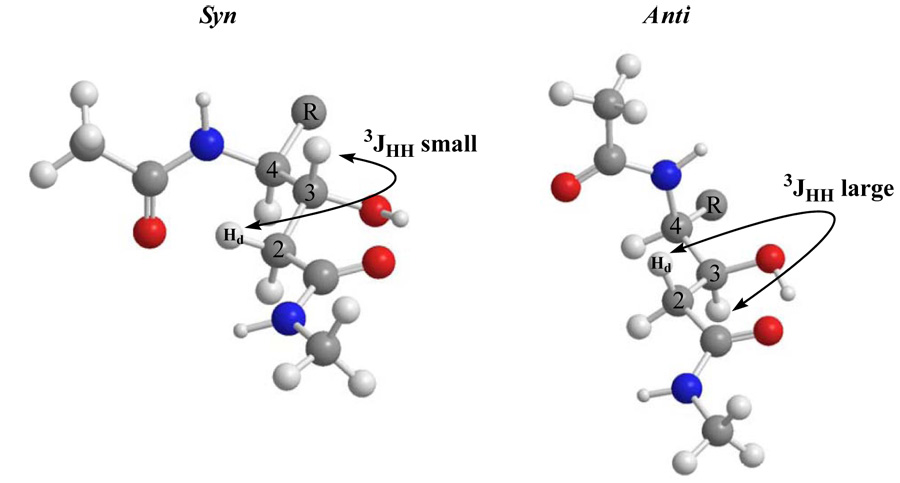
A more detailed analysis of the NMR spectroscopic data for the diastereomeric statine derivatives revealed a diagnostic pattern associated with the methylene protons (H2-2) though. When NMR spectra were recorded in CDCl3, the ABX patterns of the α-methylene protons of the two diastereomers were essentially mirror images. For the syn diastereomer, the downfield proton signal of the methylene pair was consistently a doublet of doublets with a small vicinal coupling to H-3. Conversely, for the anti diastereomer, the downfield methylene proton signal displayed a large vicinal coupling to H-3. The upfield protons of these pairs showed the opposite pattern, i.e., the syn and anti diastereomers had large and small vicinal couplings to H-3, respectively. Figure 2 shows the AB portion of the 1H NMR spectra of 8a and 8b where this trend is clearly observed. On average, the magnitude of the 3J values between the downfield resonance of H2-2 and H-3 was 10 Hz for the anti diastereomer and 2 Hz for the syn diastereomer (Figure 3).
Figure 2.
Expansions of the H2-2 ABX systems in the 1H NMR spectra of two diastereomers (syn & anti). These patterns are essentially mirror images allowing assignment of the relative configuration.
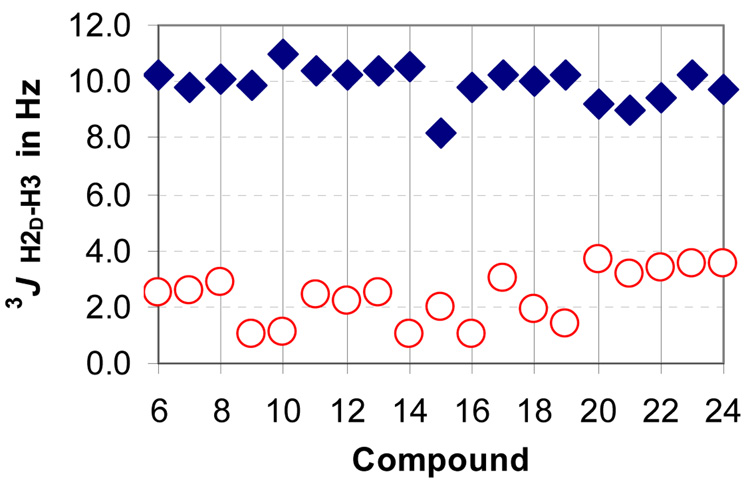
Figure 3.
Comparison of the vicinal proton-proton coupling constants between H-2D and H-3 for the anti (blue diamond) and syn (red circle) diastereomers for compounds 6–24
Technically in a second-order ABX system, such as shown in Figure 2, only JAB and the sum JAX + JBX can be determined directly from the spectrum. In other words, exact values for JAX and JBX, specifically in this case 3JH3,H2d and 3JH3,H2u, cannot be determined by simple subtraction of the line frequencies.2 In practice outside a small community, this issue is largely ignored and coupling constants in second-order ABX systems are extracted as if these are first-order. We have chosen to analyze all the ABX systems in this manner as it provides a simple way of designating “large” and “small” for this technique.
Table 1 shows the complete data set for 48 synthetic statine derivatives in CDCl3. In all cases, the proton signals for H2-2 were unambiguously identified by either simple inspection of the 1HNMR spectra or through a combination of 1D TOCSY and selective decoupling experiments. The anti derivatives clearly showed the diagnostic pattern, with the downfield methylene resonance displaying a large vicinal coupling to H-3, while the upfield proton had a small vicinal coupling. These could be easily distinguished from the corresponding syn derivatives based on the pattern shown in Figure 2.
Table 1.
1H NMR data for H2-2 ABX system of Statine Derivatives
| Anti Compound | Parent AA | δHa (JAX) | δHb (JBX) | Syn Compound | δHa (JAX) | δHb (JBX) |
|---|---|---|---|---|---|---|
| 6ad | Leucine | 2.47 (8.2) | 2.45 (0.4) | 6bd | 2.29 (3.7) | 2.28 (10.0) |
| 7a | Valine | 2.66 (9.8) | 2.54 (3.0) | 7b | 2.63 (2.6) | 2.50 (8.9) |
| 8a | Isoleucine | 2.48 (10.1) | 2.39 (2.8) | 8b | 2.48 (2.9) | 2.35 (8.8) |
| 9a | Isoleucine | 2.60 (9.9) | 2.31 (2.2) | 9b | 2.43 (1)a | 2.31 (8.7) |
| 10a | Alanine | 2.56 (11.0)d | 2.53 (0.7)d | 10bb | 2.54 (2.2) | 2.51 (9.4) |
| 11a | Phenylalanine | 2.51 (10.4) | 2.27 (2.4) | 11b | 2.51 (2.4) | 2.41 (8.9) |
| 12a | Phenylalanine | 2.61 (10.2) | 2.41 (2.5) | 12b | 2.61 (2.2) | 2.52 (8.2) |
| 13a | Tyrosine | 2.51 (10.4) | 2.28 (2.6) | 13b | 2.51 (2.5) | 2.41 (9.0) |
| 14a | Tryptophan | 2.52 (10.5) | 2.25 (2.6) | 14b | 2.54 (1)a | 2.46 (8.3) |
| 15a | Histidine | 2.58 (8.2) | 2.43 (4.7) | 15b | 2.55 (2.0) | 2.51 (8.6) |
| 16a | Lysine | 2.48 (9.8) | 2.36 (3.1) | 16bb | 2.45 (0.7) | 2.41 (9.3) |
| 17a | Cysteine | 2.47(10.2) | 2.26 (1)a | 17b | 2.48 (3.0) | 2.35 (9.0) |
| 18ac | Aspartate | 2.47 (10.0) | 2.40 (2.6) | 18b | 2.53 (1.9) | 2.46 (9.0) |
| 19a | Methionine | 2.60 (10.2) | 2.53 (3.0) | 19b | 2.58 (1.4) | 2.54 (9.4) |
| 20a | Serine | 2.43 (9.2) | 2.31 (4.0) | 20b | 2.46 (3.7) | 2.40 (8.5) |
| 21a | Threonine | 2.39 (9.0) | 2.28 (1.6) | 21b | 2.50 (3.2) | 2.33 (8.7) |
| 21c | Threonine | 2.53 (9.8) | 2.43 (3.3) | ------ | ------ | ------ |
| 22a | Arginine | 2.40 (9.4) | 2.30 (3.3) | 22b | 2.37 (3.4) | 2.28 (8.9) |
| 23a | Glutamate | 2.60 (10.2) | 2.53 (2.2) | 23bd | 2.56 (3.5) | 2.53 (8.9) |
| 24ad | Proline | 2.36 (9.7) | 2.33 (2.5) | 24b | 2.54 (3.5) | 2.44 (8.5) |
broad proton signal with no discernable coupling to Hx that for illustrative purposes is considered approximately a 1 Hz coupling.
Data recorded at 800 MHz.
Coupling constants determined though a combination of 1D-TOCSY and selective homonuclear decoupling at 500 MHz.
Coupling constants and chemical shifts determined by comparison of simulated and experimental NMR spectra.
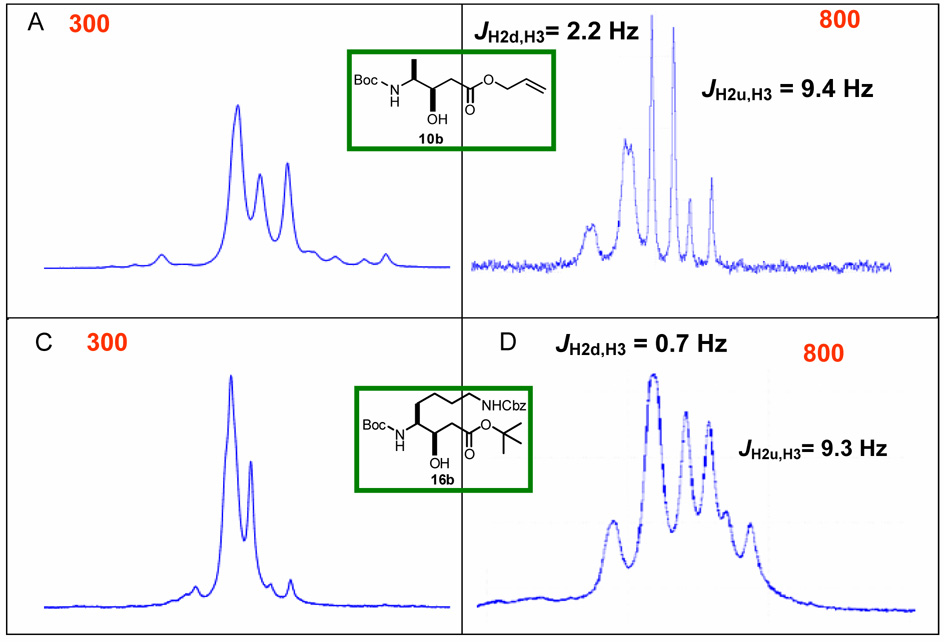
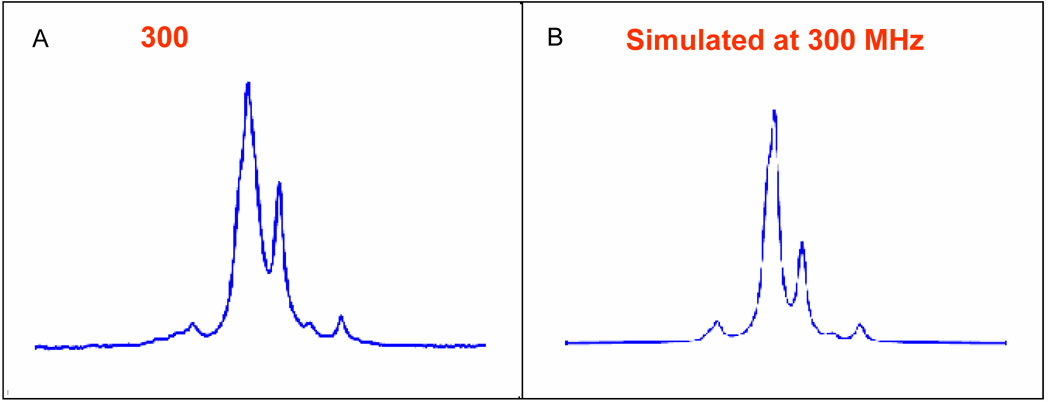
It is worth noting that in some cases, the proton resonances for the ABX system were strongly second order at 300 MHz. The statine units derived from Ala (10a–b) displayed such a large distortion that magnitude of the coupling constants could not be directly extracted at 300 MHz. In general, this effect was more prevalent with the syn diastereomers as lysine (16b), glutamate (17b), methionine (19b), and proline (24b) derivatives all had strongly distorted ABX spin systems for H2-2. Since second order systems occur when ΔJ/(Δδ in Hz) < 6, increasing the spectrometer frequency provided one simple solution. Figure 4 shows expansions of the ABX systems for the two derivatives that displayed the largest second order distortions at 300 MHz and the corresponding spectra at 800 MHz.23 In both cases, the higher field strength resolves the signals into a pattern consistent with the rest of the data set. Coupling constants in these second order systems could also be obtained by comparison of the AB systems of experimental and simulated NMR spectrum at 300 MHz (Figure 5).24 Coupling constants and chemical shifts in Table 1 for the anti-Ala (10a), syn-Pro (24b), and syn-Glu-Sta (23b) were determined in this manner.
Figure 4.
Expansion of the AB pattern for H2-2 in the 1H NMR spectra a) 10b at 300 MHz b) 10b at 800 MHz c) 16b at 300 MHz d) 16b at 800 MHz
Figure 5.
a) 300 MHz Spectrum of 16b in CDCl3 at 300 MHz b) Simulated Spectrum at 300 MHz (δHa 2.426, δHb 2.385, 2JAB= −15.84, 3JAX= 0.7 Hz, 3JBX= 9.3 Hz)
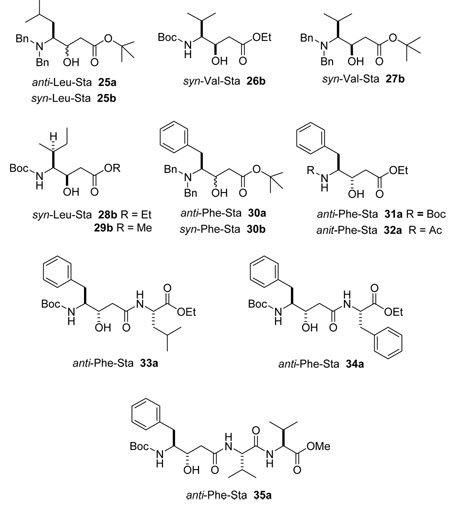
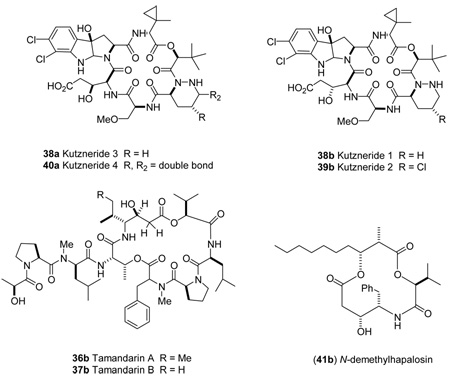
A number of statine containing compounds have been described.25 Table 2 summarizes some additional reports consistent with our observations. The diagnostic trend was observed with statine derivatives when the amine was protected as the tert-butoxycarbonyl, acetyl, and dibenzyl derivatives and with a variety of different protecting groups on the C-1 ester. These latter examples (25a, 25b, 30a, 30b) suggest the configuration of statine units containing 3° amines can be assigned using this technique. The generality of the trend is further exemplified by the linear di-and tripeptides 33a, 34a, 35a, which contain additional chiral centers. Natural products of significant structurally complexity can also be assigned using this approach. For example, the configuration of the statine unit within the didemnin26/tamandarin and hapalosin series of cyclic natural products (36b, 37b, 41b) can be correctly predicted as syn based on analysis of the reported NMR data. Likewise, the epimeric derivatives belonging to the Kutzneride class of compounds (38–40) can also be correctly assigned with the syn derivatives displaying the smaller three-bond proton-proton coupling to the downfield signal of H2-2.27
Table 2.
Relevant Literature Examples
| Anti Compound | Parent AA | δHa (JAX) | δHb (JBX) | Syn Compound | δHa (JAX) | δHb (JBX) | Ref |
|---|---|---|---|---|---|---|---|
| 25a | Leucine | 2.34 (7.9) | 2.27 (3.4) | 25b | 2.48 (2.9) | 2.29 (10.0) | 28 |
| 26a | Valine | NA | NA | 26b | 2.58 (2.9) | 2.47 (9.1) | 29 |
| 27a | Valine | NA | NA | 27b | 2.64 (2.1) | 2.32 (10.6) | 28 |
| 28a | Isoleucine | NA | NA | 27b | 2.60 (2.7) | 2.48 (9.1) | 30 |
| 29a | Isoleucine | NA | NA | 29b | 2.60 (2.7) | 2.42 (8.7) | 30 |
| 30a | Phenylalanine | 2.40 (9.5) | 2.04 (2.4) | 30b | 2.63 (2.5) | 2.22 (10.1) | 28 |
| 31a | Phenylalanine | 2.59 (9.9) | 2.37 (2.6) | 31b | NA | NA | 31 |
| 32a | Phenylalanine | 2.57 (10.3) | 2.38 (3.0) | 32b | NA | NA | 31 |
| 33a | Phenylalanine | 2.54 (9.8) | 2.28 (3.1) | 33b | NA | NA | 32 |
| 34a | Phenylalanine | 2.38 (9.0) | 2.19 (4.0) | 34b | NA | NA | 32 |
| 35a | Phenylalanine | 2.55 (9.5) | 2.32 (3.0) | 35b | NA | NA | 32 |
| 36a | Isoleucine | NA | NA | 36b | 3.25 (1)a | 2.44 (8.1) | 8 |
| 37a | Isoleucine | NA | NA | 37b | 3.25 (2.7) | 2.48 (7.5) | 8 |
| 38a | Hydroxyglutamic acid | 2.62 (8.8) | 2.54 (5.0) | 38b | 2.74 (3.7) | 2.69 (10.5) | 33 |
| 39a | Hydroxyglutamic acid | 2.65 (9.2) | 2.55 (4.6) | 39b | NA | NA | 33 |
| 40a | Hydroxyglutamic acid | NA | NA | 40b | 2.62, m | 33 | |
| 41a | Phenylalanine | NA | NA | 41b | 2.65 (3.6) | 2.57 (5.6) | 34 |
| 42a (DMSO) | Tryptophan | NA | NA | 42b | 2.35 (3.7) | 2.28 (9.1) | 35 |
| 43a (DMSO) | Tyrosine | NA | NA | 41b | 2.38 (3.7) | 2.29 (9.5) | 36 |
| 44a (MeOH) | Leucine | 2.36 (7.2) | 2.27 (6.7) | NA | NA | NA | 37 |
broad proton signal with no discernable coupling to Hx that for illustrative purposes is considered approximately a 1 Hz coupling.
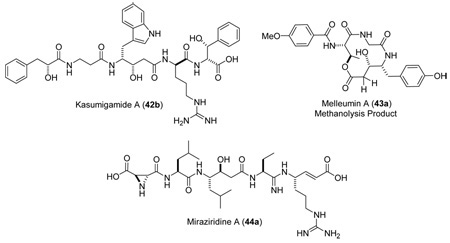
Natural products (42–44) in solvents other than CDCl3 show the same diagnostic patterns, and while it would be tempting to apply this methodology, our data indicates that caution must be exercised. As illustrated in Table 3 there is a clear solvent effect, which is more pronounced for the anti derivatives than the syn. In some cases, such as 6a and 7a in DMSO and MeOH-d4 the methylene protons become magnetically equivalent.38 In other cases, the diastereomers of the pairs 10a/10b and 11a/11b are indistinguishable in acetone-d6 and DMSO-d6, respectively, as in both cases, the downfield methylene proton displays the small vicinal proton coupling. Application of this methodology in these instances would lead to an erroneous assignment of the anti diastereomer as syn. Thus, CDCl3 insoluble compounds should be converted to a soluble derivative prior to analysis.39
Table 3.
Solvent Effect
| Anti Compound | Solvent | δHa (JAX) | δHb (JBX) | Syn Compound | δHa (JAX) | δHb (JBX) |
|---|---|---|---|---|---|---|
| 6a | CDCl3 | 2.47 (10.2) | 2.34 (2.4) | 6b | 2.47 (2.5) | 2.34 (8.9) |
| 7a | DMSO-d6 | 2nd order | ------ | 7b | 2.36 (2.5) | 2.16 (9.6) |
| 6a | DMSO-d6 | 2.20 d (2H, 7.0) | ------ | 6b | 2.39 (3.2) | 2.15 (9.3) |
| 11a | DMSO-d6 | 2.33 (4.2) | 2.16 (8.8) | 11b | 2.39 (3.9) | 2.16 (9.3) |
| 6a | CD3OD | 2.27 d (2H, 6.8) | ------ | 6b | 2.39 (3.2) | 2.15 (9.3) |
| 7a | Acetone-d6 | 2.47 (2 H, 6.6) | 7b | 2.60 (2.6) | 2.35 (9.1) | |
| 10a | Acetone-d6 | 2.50 (4.2) | 2.43 (8.8) | 10b | 2.57 (3.5) | 2.39 (9.3) |
This approach is not without precedent as a similar technique was proposed by Roush et al. to assign the relative stereochemistry of β-hydroxy ketones with alkyl substituents in the γ-position.40 Two differences should be noted. First in Roush’s method, varying the solvent had no noticeable effect on the pattern in the 1H NMR spectra for the two compounds examined. Second, with those substrates C-5 must be branched, e.g., analogous to 7a, for the two diastereomers to be distinguishable.40a The latter is not the case for statine moieties as is illustrated by examples 6, 10–20 and 22–23 in Table 1. These compounds are not branched C-5, yet are distinguishable.
Conformational preferences for 6a and 6b are known based on conformational computations.41 Analysis of these data suggests one possible explanation for the observed NMR patterns as shown in Figure 6. In both diastereomers, the carbonyls are in an eclipsed conformation with the C-2/C-3 bond coplanar to the carbonyl, but with the hydroxy C-O oxygen bond (+)-gauche and (−)-gauche to the carbonyl in the anti and syn diastereomers, respectively. Stabilization of these conformations by hydrogen bonding, analogous to that proposed in the Stiles-House model for α-alkyl-β-hydroxycarbonyls42 or Roush’s method for β-hydroxy ketones with alkyl substituents in the γ-position40 is suggested based on the work of Rich41 and Nisato,43 although not required.44 If these conformers predominate in CDCl3 then the pro-S methylene proton is deshielded in both diastereomers relative to the pro-R. This deshielding of the pro-S methylene proton may be due to the amide attached to C-4, in a fashion analogous to that reported by Whelton et al. for substituted cyclohexamines.45 It should be noted that for simplicities sake, we have chosen to depict only a single rotatmer between the vicinal chiral centers C-3/C-4, although at least two rotatmers exist in solution based on coupling constant analysis. A reversal of the relative population of the rotamers around the C-3/C-4 bond may ultimately be responsible for the solvent dependency. A more detailed conformational analysis of the factors responsible for the observed NMR patterns is underway.
Figure 6.
Proposed Conformational Model
The trend noted here provides the simplest methodology for the relative configurational assignment of CDCl3 soluble statine containing peptides. The methodology requires no degradation or derivatization, and is amenable to micromolar concentrations of analytes given the sensitivity limits of modern NMR spectrometers. Several examples above suggest this methodology is applicable to statine units within cyclic peptides/depsipeptides, although until a more complete understanding of the underlying factors responsible is available, we would urge caution in these constrained systems. While we have not found any examples, conformational constraints in cyclic systems could induce a reversal of the 1H NMR trend similar to that illustrated using solvents other than chloroform. For cyclic depsipeptides, conversion to a linear derivative would be advisable. Another fundamental limitation of the technique is obtaining sufficient dispersion in the ABX NMR spin system. Potential solutions to this issue include increasing the NMR field strength, although most of our data was acquired at 300 MHz, or simulation of the observed spin system to extract coupling constants through comparison with experimental spectra. As such, this methodology should be broadly applicable to the assignment of statine units within both natural products and synthetic derivatives using 1H NMR spectroscopy.
Experimental Section
General Procedure for Synthesis of β-keto esters.15
One gram of the protected amino acid (1 eq.) was dissolved in 20 mL of dry THF. To this solution was added 1.1 eq. of 1,1′-carbonyldiimidazole (recrystallized from THF) with stirring under dry nitrogen at room temperature. Butyl lithium (2.2M hexane solution, 3.3 equivalents) under nitrogen was diluted with 20 mL of THF and cooled to 0°C with an ice bath. To this was added dropwise diisopropylamine (3.6 equivalents). After stirring for ten minutes at 0°C, the solution was diluted with 70 mL of THF and cooled to −78°C. To the LDA solution was added either t-butyl acetate or allyl acetate (3.5 equivalents). After ten minutes, the Boc-amino acid/imidazole solution was cooled to −78°C and cannulated into the enolate solution under nitrogen. The reaction was allowed to stir for 30 minutes at −78°C then quenched with 50 mL of 10% citric acid and allowed to warm to room temperature. The aqueous solution was extracted with 200 mL EtOAc (4x 50 mL) and washed with 100 mL saturated NaHCO3 (2x50 mL), 50 mL brine, and dried over MgSO4. The solvent was evaporated and the crude material purified on silica eluting with mixtures of hexane/EtOAc).
Allyl 4(S)-[(tert-butoxycarbonyl)-amino]-5-methyl-3-oxohexanoate (4b)
amorphous powder: [α]27D = +8.5 (c 5.4, CHCl3); UV (i-PrOH) λmax (log ε) 248 (8.10) nm; IR (film) νmax : 3369, 1749, 1246, 1168 cm−1; 1H NMR (CDCl3 at room temperature) δ 5.90 (ddt, J =17.2, 10.4, 5.6 Hz, 1H), δ 5.33 (dd, J =17.2, 1.5 Hz, 1H), δ 5.28 (dd, J =10.4, 1.5 Hz, 1H) δ 5.06 (d, J =8.3 Hz, 1H), δ 4.63 (d, J =5.6 Hz, 2H), δ 4.32 (dd, J =8.7, 4.1 Hz, 1H), δ 3.57 (s, 2H), δ 2.24 (m, 1H), δ 1.48 (s, 9H), δ 1.01 (d, J =6.8 Hz, 3H), δ 0.82 (d, J =6.8 Hz, 3H); 13C NMR: δ 202.0, 166.2, 155.7, 131.5, 80.0, 66.0, 64.3, 47.0, 29.5, 28.2, 19.8, 16.7; HRESI-MS m/z 322.1636 [M+Na]+ [Calc. for C15H25NO5Na+, 322.1631, −3.8 ppm error].
General procedure for Reduction.15
To a flame dried flask was added 0.5 g (1 eq.) of BOC-β-keto-γ-amino t-butyl ester dissolved in 20 mL dry THF and cooled to −78°C under nitrogen. To this solution was added 1.3 eq. of LiBH4 (recrystallized from Et2O) which was stirred at −78°C until the reaction reached completion by as determined by TLC. The reaction was quenched by the addition of 1 N HCl and allowed to warm to room temperature over 15 min. This aqueous solution was extracted with 60 mL Et2O (3×20 mL) and then dried over MgSO4. Purification over silica eluting with hexane/EtOAc provided a mixture of diastereomers. HPLC purification on silica provided pure diastereomers.
Allyl 4(S)-[(tert-butoxycarbonyl)-amino]-5-methyl-3(S)-hydroxyhexanoate (7a)
amorphous powder: [α]27D = −1.7 (c 1.0, CHCl3); UV (i-PrOH) λmax (log ε) 248 (8.10) nm; IR (film) νmax 3375, 1715, 1506, 1261, 1172 cm−1; 1H NMR (CDCl3 at room temperature) δ 5.90 (ddt, J =17.2, 10.4 Hz, 1H), δ 5.33 (dq, J =17.2, 1.5 Hz, 1H), δ 5.26 (dd, J =10.4, 1.5 Hz, 1H), δ 4.83 (d, J =9.8 Hz, 1H), δ 4.60 (dt, J =5.8, 1.5 Hz, 2H), δ 4.20 (br. d, J =9.4 Hz, 1H), δ 3.20 (d, J =2.6 Hz, 1H), δ 3.15 (td, J =10.5, 1.7 Hz, 1H), δ 2.60 (dd, J =16.9, 9.8 Hz, 1H), δ 2.51 (dd, J =16.9, 3.2 Hz, 1H), δ 1.86 (dm,1H), δ 1.43 (m, 9H), δ 0.99 (d, J =6.7 Hz, 3H), δ 0.96 (d, J =6.8 Hz, 3H); 13C NMR: δ 173.3, 156.4, 131.7, 118.7, 79.2, 67.0, 65.5, 59.6, 39.0, 30.3, 29.7, 28.4, 19.8, 19.5; HRESI-TOF m/z 324.1767 [M + Na]+ [Calc. for C15H27NO5Na+, 324.1787, −4.5 ppm error].
Allyl 4(S)-[(tert-butoxycarbonyl)-amino]-5-methyl-3(R)-hydroxyhexanoate (7b)
[α]24D = +7.9 (c 1.0, CHCl3); UV (i-PrOH) λmax (log ε) 203 nm (7.56), 205 nm (5.54); IR (Film) νmax : 3456, 3369, 1696 cm−1; 1H NMR (CDCl3 at room temperature) δ 5.98 (ddt, J =17.2, 10.4, 5.8 Hz, 1H), δ 5.33 (ddt, J =17.2, 2.7, 1.5 Hz, 1H), δ 5.24 (dd, J =10.4, 1.1 Hz, 1H), δ 4.57 (dt, J =5.8, 1.3 Hz, 2H), δ 4.50 (d, J =9.6 Hz, 1H), δ 3.89 (td, J =8.4, 2.9 Hz, 1H), δ 3.49 (m, 1H), δ 2.58 (dd, J =16.5, 2.9 Hz, 1H), δ 2.46 (dd, J =16.5, 8.8 Hz, 1H), δ 2.09 (m, 1H), δ 1.43 (m, 9H), δ 0.94 (d, J =6.9 Hz, 3H), δ 0.88 (d, J =6.8 Hz, 3H); 13C NMR: δ 172.9, 156.4, 131.8, 118.6, 79.6, 69.2, 65.5, 58.8, 38.3, 28.3, 27.5, 20.1, 16.3; HRESI-TOF m/z 302.1955 [M + H]+ [Calc. for C15H28NO5 +, 302.1962, 2.4 ppm error].
Supplementary Material
General experimental procedures for the preparation of and spectral data for compounds 6–24 are available free of charge via the internet at http://pubs.acs.org.
Acknowledgement
This work was funded by grants from the Victoria S. and Bradley L. Geist Foundation (20070461), the National Science Foundation (OCE04-32479), and the National Institute of Environmental Health Sciences (P50 ES012740). Funds for the upgrades of the NMR instrumentation were provided by the CRIF program of the National Science Foundation (CH E9974921) and the Elsa Pardee Foundation. The purchase of the Agilent LC-MS was funded by grant W911NF-04-1-0344 from the Department of Defense. We thank P. Moeller, NOAA, and W. Yoshida, UH Manoa, for the 800 and 500 MHz NMR data, respectively. B. Wong and S. Yuen aided in the collection of the physical data.
References and Notes
- 1.Nicolaou KC, Snyder SA. Angew. Chem., Int. Ed. 2005;44:1012–1044. doi: 10.1002/anie.200460864. [DOI] [PubMed] [Google Scholar]
- 2.Crews P, Rodriquez J, Jaspars M. Organic Structure Analysis. 1998 [Google Scholar]
- 3.Reynolds WF, Enriquez RG. J. Nat. Prod. 2002;65:221–244. doi: 10.1021/np010444o. [DOI] [PubMed] [Google Scholar]
- 4.Matsumori N, Kaneno D, Murata M, Nakamura H, Tachibana K. J. Org. Chem. 1999;64:866–876. doi: 10.1021/jo981810k. [DOI] [PubMed] [Google Scholar]
- 5.For a review on determining absolute configurations by NMR see: Seco JM, Quiñoá E, Riguera R. Chem. Rev. 2004;1:17–117. doi: 10.1021/cr2003344..
- 6.(a) Wolfender J-L, Queiroz EF, Hostettmann K. Mag. Res. Chem. 2005;43:697–709. doi: 10.1002/mrc.1631. [DOI] [PubMed] [Google Scholar]; (b) Bugni TS, Richards B, Bhoite L, Cimbora D, Harper MK, Ireland CM. J. Nat. Prod. 2008;71:1095–1098. doi: 10.1021/np800184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Rinehart KL, Jr, Gloer JB, Hughes RG, Jr, Renis HE, McGovren JP, Swynenberg EB, Stringfellow DA, Kuentzel SL, Li LH. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]; (b) Rinehart KL, Jr, Gloer JB, Cook JC, Jr, Mizsak SA, Scahill TA. J. Am. Chem. Soc. 1981;103:1857–1859. [Google Scholar]
- 8.Vervoort H, Fenical W, de Epifanio R. J. Org. Chem. 2000;65:782–792. doi: 10.1021/jo991425a. [DOI] [PubMed] [Google Scholar]
- 9.Stratmann K, Burgoyne DL, Moore RE, Patterson GML, Smith CD. J. Org. Chem. 1994;59:7219–7226. [Google Scholar]
- 10.Zuo Z, Luo X, Zhu W, Shen J, Shen X, Jiang H, Chen K. Bioorg. Med. Chem. 2005;13:2121–2131. doi: 10.1016/j.bmc.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Fehrentz JA, Chromier B, Bignon E, Venaud S, Chemann JC, Nisato D. Biochem. Biophys. Res. Commun. 1992;188:865–872. doi: 10.1016/0006-291x(92)91136-e. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Kobayashi Y, Tezuka K, Kishi Y. Org. Lett. 1999;1:2181–2184. doi: 10.1021/ol990379y. [DOI] [PubMed] [Google Scholar]
- 13.Williams PG, Yoshida WY, Moore RE, Paul VJ. J. Nat. Prod. 2003;66:1006–1009. doi: 10.1021/np030114z. [DOI] [PubMed] [Google Scholar]
- 14.Harris BD, Bhat KL, Joullie MM. Tetrahedron Lett. 1987;28:2837–2840. [Google Scholar]
- 15.In general, the anti diastereomer eluted first using normal phase HPLC, except for the threonine- and proline-derived statine units. While the order of elution was reversed for these two pairs of derivatives, the syn reduction product depicted dominates.
- 16.Ori M, Toda N, Takami K, Tago K, Kogen H. Tetrahedron. 2005;61:2075–2104. [Google Scholar]
- 17.Harris BD, Joullie MM. Tetrahedron. 1988;44:3489–3500. [Google Scholar]
- 18.The stereochemistry of 24a was determined by conversion to the oxazolidinone derivative through treatment with triphosgene. The stereochemistry was then assigned via analysis of the JH3,H4 coupling constant. See Wang M, Chen Y, Lou L, Tang W, Wang X, Shen J. Tetrahedron Lett. 2005;46:5309–5312..
- 19.A comparison of carbon and proton chemical shifts with the nature of the side chain (polar, nonpolar, aromatic, branched, etc) provided no predictive trend either.
- 20.There are reports of using 3JH3,H4 values for the relative stereochemical assignment of an Ala-Sta derivative (4-methylcarnitine; Comber RN, Brouillette WJ. J. Org. Chem. 1988;53:1121–1122.), but based on our data (see supporting) this is not a general trend.
- 21.Higashibayashi S, Kishi Y. Tetrahedron. 2004;60:11977–11982. [Google Scholar]
- 22.Heathcock CH, Pirrung MC, Sohn JE. J. Org. Chem. 1979;44:4294–4299. [Google Scholar]
- 23.At 500 MHz, the 1H NMR spectra for these compounds were second order.
- 24.Spin simulation were done using MestreC 4.9.8.0 in an iterative fashion comparing with the experimental spectra until identical patterns were obtained. Experimental data was used as a constraint in these simulations when possible, i.e. dH3, 2JH2u,H2d, 3JH3,H2d + 3JH3,H2u, ca. dH2.
- 25.A Scifinder Scholar search for the basic carbon skeleton yielded approximately 10 000 compounds, but the utility of this literature NMR data varied greatly. For example, H2-2 was frequently reported as a multiplet spanning a considerable chemical shift range. This indicated the methylene protons were probably diastereotopic (otherwise H2-2 would be a simple doublet), but provided no coupling constant information for comparison.
- 26.Sakai R, Stroh JG, Sullins DW, Rinehart KL. J. Am. Chem. Soc. 1995;117:3734–3748. [Google Scholar]
- 27.Zhang et al. have recently reported a new cyclic thiazolyl peptide Phillipmycin containing a statine unit, whose configuration can be correctly predicted using this method. See Zhang C, et al. J. Am. Chem. Soc. ASAP.
- 28.Andrés JM, Pedrosa R, Pérez A, Pérez-Encabo A. Tetrahedron. 2001;57:8521–8530. This reference also includes two Pro-Stat derivatives in which the ABX pattern appears to be reversed. We have synthesized one of these compounds, the Pro-Stat protected as a t-butyl ester and find the NMR data to be consistent with the rest of our data set.
- 29.Liang B, Richard DJ, Portonovo PS, Joullie MM. J. Am. Chem. Soc. 2001;123:4469–4474. doi: 10.1021/ja010222c. [DOI] [PubMed] [Google Scholar]
- 30.Rinehart KL, Sakai R, Kishore V, Sullins DW, Li KM. J. Org. Chem. 1992;57:3007–3013. [Google Scholar]
- 31.Doi T, Kokubo M, Yamamoto K, Takahashi T. J. Org. Chem. 1998;63:428–429. doi: 10.1021/jo9720400. [DOI] [PubMed] [Google Scholar]
- 32.Palomo C, Miranda JI, Linden A. J. Org. Chem. 1996;61:9196–9201. [Google Scholar]
- 33.Broberg A, Menkis A, Vasiliauskas R. J. Nat. Prod. 2006;69:97–102. doi: 10.1021/np050378g. [DOI] [PubMed] [Google Scholar]
- 34.Okuno T, Ohmori K, Nishiyama S, Yamamura S, Nakamura K, Houk KN, Okamoto K. Tetrahedron. 1996;52:14723–14734. [Google Scholar]
- 35.Ishida K, Murakami M. J. Org. Chem. 2000;65:5898–5900. doi: 10.1021/jo991918f.. The vicinal coupling constants for the methylene proton signals (dH 2.35 & 2.28) in Table 1 of this reference are transposed. We thank Drs. Ishida, Murakami, and Okada for providing copies of the original 1H NMR spectrum so that we could check this data. The NMR spectra is included in our supporting information.
- 36.Isolation: Nakatani S, Kamata K, Sato M, Onuki H, Hirota H, Matsumoto J, Ishibashi M. Tetrahedron Lett. 2005;46:267–271.. Synthesis: Hanazawa S, Arai M, Li X, Ishibashi M. Bioorg. Med. Chem. Lett. 2008;18:95–98. doi: 10.1016/j.bmcl.2007.11.005..
- 37.Nakao Y, Fujita M, Warabi K, Matsunaga S, Fusetani N. J. Am. Chem. Soc. 2000;122:10462–10463.. Synthesis: Konno H, Kubo K, Makabe H, Toshiro E, Hinoda N, Nosaka K, Akaji K. Tetrahedron. 2007;63:9502–9513..
- 38.Occasionally, H2-2 appeared as a simple doublet in the initial proton spectra of 6–24 immediately after silica chromatography. After 24h in CDCl3, these proton signals were generally resolved.
- 39.Because of this requirement of CDCl3 solubility, data for the deprotected compounds 6–24 was generally not included.
- 40.a) Roush WR, Bannister TD, Wendt MD, Van Nieuwenhze MS, Gustin DJ, Dilley GJ, Lane GC, Scheidt KA, Smith WJ., III J. Org. Chem. 2002;67:4284–4289. doi: 10.1021/jo0164148. [DOI] [PubMed] [Google Scholar]; b) Dias LC, Aguilar AM, Salles AG, Jr, Steil LJ, Roush WR. J. Org. Chem. 2005;70:10461–10465. doi: 10.1021/jo051773e. [DOI] [PubMed] [Google Scholar]
- 41.Rich DH, Terada Y, Kawai M. Int. J. Pept. Prot. Res. 1983;22:325–332. doi: 10.1111/j.1399-3011.1983.tb02098.x. [DOI] [PubMed] [Google Scholar]
- 42.a) Stiles M, Winkler RR, Chang Y-L, Traynor L. J. Am. Chem. Soc. 1964;86:3337–3342. [Google Scholar]; (b) House HO, Crumrine DS, Teranishi AY, Olmstead HD. J. Am. Chem. Soc. 1973;95:3310–3324. [Google Scholar]
- 43.Toniolo C, Valle G, Bonora GM, Lelj F, Barone V, Fraternali F, Callet G, Wagnon J, Nisato D. Int. J. Pept. Prot. Res. 1987;30:583–595. doi: 10.1111/j.1399-3011.1987.tb03369.x. [DOI] [PubMed] [Google Scholar]
- 44.Kitamura M, Nakano K, Miki T, Okada M, Noyori R. J. Am. Chem. Soc. 2001;123:8939–8950. doi: 10.1021/ja0110978. [DOI] [PubMed] [Google Scholar]
- 45.Whelton BD, Lowry BR, Carr JB, Huitric AC. J. Pharm. Sci. 1973;62:728–737. doi: 10.1002/jps.2600620505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
General experimental procedures for the preparation of and spectral data for compounds 6–24 are available free of charge via the internet at http://pubs.acs.org.