Abstract
Nanoparticles have shown tremendous potential for effective drug delivery due to their tiny size and cell membrane penetration capabilities. Cellular targeting with nanoparticles is often achieved by surface modifications followed by ligand conjugation. However, the efficiency of the nanoparticles reaching the target cells and getting internalized depends on the stability of targeting ligands and the chemical nature of the ligand nanoparticle binding. Recent advancements in nanobiomaterials research have proven the superoxide dismutase (SOD) mimetic activity of cerium oxide nanoparticles (CNPs) in protecting cells against oxidative stress. Due to their excellent biocompatibility, CNPs can be used as potential drug carrier that can transport and release drugs to the malignant sites. Here we combine single molecule force spectroscopy (SMFS) and density functional theory (DFT) simulations to understand the interaction between transferrin, a ligand protein over-expressed in cancer cells, and CNPs. SMFS studies demonstrate an increase in the transferrin adhesion to the nanoparticles surface with an increase in positive zeta potential of CNPs. Binding energy values obtained from DFT calculations predict an increase in bond strength between the transferrin and CNPs upon surface protonation and charge modification. Transferrin conjugated CNPs were tested for their binding stability and preferential cellular uptake efficiency by incubating them with human lung cancer cells (A549) and normal embryo lung cells (WI-38). The results demonstrate the importance of tuning the surface properties of nanoparticles for better ligand adsorption and cellular uptake.
Keywords: single molecule force spectroscopy, atomic force microscopy, transferrin, cerium oxide nanoparticles, density functional theory
With more than ten million new cases every year, cancer has become one of the most dreadful diseases worldwide. Current cancer treatments include surgical intervention, radiation therapy, and chemotherapeutic drugs, which often kill healthy cells and cause severe side effects.1 Site specific delivery of drugs and therapeutics can significantly reduce drug toxicity and increase the therapeutic effects.2 Recent advances in nanomedicine research have shown the capability of using nanoparticles (NPs) to treat intractable diseases (viral, genetic, cancerous) as they are the only microscopic objects that can gain access to the cells and function as therapeutic agents.3 NPs could be used as effective therapeutic agents for intracellular imaging, targeting, and in the delivery of drugs, genes and vaccines.1, 4-6 Cellular targeting is often achieved by bioconjugation of polymeric (drug carriers), metallic, or ceramic NPs with ligand proteins that can bind to the overexpressed receptors on the target cells.1, 7 NP-protein conjugation is often influenced by the surface potential differences, and understanding their specific interactions leading to their cellular uptake is crucial for targeted drug delivery.8
CNPs have been shown to be biocompatible, nontoxic and have excellent antioxidant properties in the prevention of reactive oxygen species (ROS) mediated cell damage in animals.9 CNPs have been found to prevent healthy cell damage during radiation therapy10 and provide neuroprotection to spinal cord neurons11 by acting as an antioxidant. Due to their smaller particle size, non-toxic nature and excellent biocompatibility, CNPs have the potential to be used as drug carrier and delivery agents.
Ligand-receptor mediated drug delivery and imaging systems have attracted primary focus in nanomedicine.12, 13 Transferrin (Tf) is a cellular targeting ligand protein (Supporting Information Figure S1) used for transportation and supply of iron to growing cells in the body. The iron uptake process involves binding of iron carrying Tf to the Tf receptors (TfRs) expressed on the cells followed by their internalization via receptor mediated endocytosis.14 With a motivation to develop a potential targeted drug delivery system, we have chosen CNPs as our model NP system and Holo-Tf (Tf carrying two iron ions) as the cellular targeting agent.15 Many studies have been carried out using Tf as a targeting ligand to deliver a wide range of therapeutic agents including nanoparticles into malignant sites that overexpress TfRs.15-19 Since surface potential differences play a major role in NP-protein adhesion,8 Tf could be attached to CNPs through electrostatic adsorption. During the transportation as well as cellular internalization process, changes in the local environmental pH could influence the surface charges and affect the NP-protein binding. Hence, a molecular level understanding of NP-protein binding is essential to develop effective cellular targets. No detailed studies have been reported so far on the molecular level interaction forces and adhesion behavior of Tf over nanoparticle surface.
Atomic force microscopy (AFM) based SMFS20-23 have proven to be the most versatile technique that induce molecular level interactions on surfaces using functionalized probes and monitor them in real time at sub-nanometer resolution. Here we have used SMFS technique to probe the interaction forces between Tf and CNPs with varying zeta potentials (ZPs), and an extensive study on the importance of Tf adhesion on nanoparticle surface for preferential cellular uptake is presented. Density functional theory (DFT) along with DFT+U technique is used to calculate the binding energy between the protein and the NP responsible for preferential cellular uptake. Both high resolution transmission electron microscopy (HRTEM) and inductively coupled plasma mass spectrometry (ICP-MS) measurements were performed on human lung adenocarcinoma epithelial cells and human embryo lung fibroblast cells to analyze and quantify the cellular internalization of transferrin conjugated CNPs (Tf:CNPs) and bare CNPs.
Results and Discussion
Cerium oxide nanoparticles
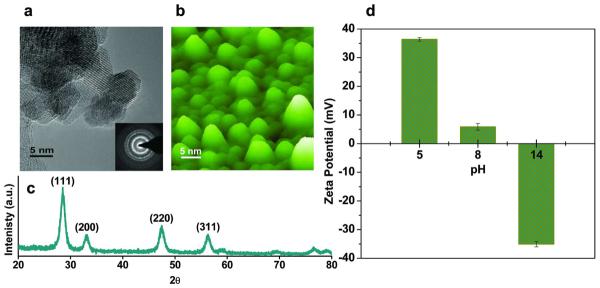
HRTEM image of base precipitated CNPs (mean size of 8.2 ± 1 nm) and AFM image of the same CNPs coated on a silicon substrate are shown in Figure 1a, and 1b, respectively. X-ray diffraction (XRD) pattern of CNPs shows a fluorite structure with dominant (111) planes (Figure 1c). CNPs in water showed a positive ZP of +39.6±0.631 mV in the synthesized condition, while Tf dissolved in deionized water exhibited a negative ZP of -19.9 ± 0.9 mV. In order to study the electrostatic interaction of CNPs with negatively charged Tf, CNPs with different surface charges were prepared by treating them with acidic and basic pH buffers.8 CNPs treated with pH buffers 5, 8 and 14 showing a ZP values of +36.4±0.7, +5.9±1.2, and -35.1±0.9 mV, respectively, were used for SMFS studies (Figure 1d).
Figure 1.
Particle size, crystal structure and surface potentials of CNPs. (a) HRTEM (b) AFM image and (c) XRD spectrum of fluorite structured CNPs; (d) ZP plot of CNPs treated with different pH buffers. AFM and HRTEM images show that the CNP particle size is between 7 - 10 nm. XRD pattern and HRTEM images shows the presence of dominant (1 1 1) planes of cerium oxide fluorite structure. Positively charged CNPs were obtained by acidic buffer treatment (more H+ ions on surface) while basic buffer treatment (more OH- ions on surface) resulted in negatively charged CNPs. The isoelectric point (pl) is observed at 8.5. Each sample was analyzed 30 times (n=30) and the average values of ZP with standard deviation (mean±sd) are plotted here.
Single molecule interaction of transferrin with cerium oxide nanoparticles
SMFS measurements were carried out using a Tf conjugated AFM tip (Figure 2a-2c) on CNPs (with zeta potentials of +36.4±0.7, +5.9±1.2, and -35.1±0.9 mV) coated on silicon substrate. To understand the interaction mode and conformational changes of Tf, SMFS measurements were also conducted on an atomically smooth silicon substrate in aqueous medium (Supporting Information Figure S2). Figure 2d-2f shows the force-extension profile of Tf coated AFM tip interaction with CNPs of varying surface potentials. The corresponding unbinding force and unbinding length histograms of rupture events are also shown in Figure 2g-2l. These results indicate that the adhesion between Tf and CNPs decreases on tuning CNP surface charges from positive to negative ZP values, with a maximum for +36.4±0.7 mV ZP. The force spectrum corresponding to CNPs of +36.4±0.7 mV ZP (Figure 2d), shows multiple interaction events due to the unbinding of proteins from the CNP sample at three different sites (strong interaction between Tf and CNPs could result in the adhesion of multiple proteins from the AFM tip at different sites on the sample). The highest unbinding force is observed at the first dip where the magnitude of pull-off force is ~2.835 nN. As the ZP is directly related to the particle surface potential, a strong attraction is observed between the Tf and positive CNPs due to their high surface charge difference. To confirm that the observed changes in the force signals were indeed originated from the single molecule rupture events, the force extension profiles were fitted using entropic elasticity models, which predict the relationship between the applied force and the stretching of a polymer molecule.24 The elastic properties of these stretching events can be described by the worm-like chain (WLC) model.23, 25 According to this model, the force needed to stretch a linear polymer molecule in a medium to a length x is given by
| (1) |
where Kb is the Boltzmann constant, T is the absolute temperature of the medium, b is the persistence length (length of a stiff segment of the protein chain), L is the contour length (length of the completely stretched chain) and x is the distance between the attachment points of the protein (extension or end-toend distance between the tip and sample). The stiffest element in a peptide chain is the single amino acid unit and it has a length of ~0.38 nm which corresponds to the persistence length b in the WLC model.26 WLC fit yielded values of b = (0.024±0.003) nm and L = (423.2±5.7) nm for the first interaction event (Figure 2d). Such a low persistence length (~0.024nm) is unphysical (one order less than the values typically quoted for pulling of single proteins25) and the contour length is also higher than the maximum possible length (~257 nm) of a fully extended single Tf. This could be due to the simultaneous unfolding of multiple interacting proteins attached to the tip27 (since the Tf backbone is folded into multiple helical groups, pulling Tf itself involves stretching of many interacting protein chains). The second and third unbinding force dips correspond to a magnitude of ~748 pN and ~499 pN, respectively. These additional dips in the force-extension spectrum reveal multiple protein interactions with the NP surface. For these interactions we used a persistence length value of 0.5 nm and the WLC model yielded a counter length value of 203.1±3.1 nm and 83.5±6.7 nm for the second and third dip, respectively. These contour lengths were smaller than an estimated fully extended length of Tf (polypeptide chain of 678 amino acids corresponds to ~257 nm in length). Hence the second dip could be as a result of single molecule stretching of peptide chain involving unfolding of both the domains (N-lobe and C-lobe) and the third dip could be likely due to the unfolding of part of another peptide chain (most likely just one of the two domains of the protein is involved in stretching). This kind of multiple protein stretching event happens due to the strong interaction of Tf with high positive CNPs and most of the SMFS measurements carried out on these samples showed similar multiple stretching events. According to the unbinding force histogram analysis shown in Figure 2g, the majority of rupture force events observed are between 150 to 225 pN with a maximum force at ~9.0 nN. Even though they had a wide spectrum of force range, events with a force of magnitude higher than 1.5 nN were mostly non-repeating single events. Similarly majority of protein unbinding lengths fall between 25 to 50 nm (Figure 2h). Figure 2e shows the Tf interaction spectrum with CNPs having moderate ZP (+5.9±1.2 mV). The force-extension spectrum shows two successive single molecule interaction events and the corresponding unbinding forces are ~318 pN and ~596 pN, respectively. WLC fit with a persistence length of 0.5 nm yielded contour lengths of 215.4±2.5 nm and 130.9±2.0 nm. The lower unbinding force observed is due to the weak interaction between Tf and CNPs with lower positive surface potentials. Unbinding force histogram analysis shows that the maximum number of rupture force events is observed between 75 to 150 pN with a maximum force at ~1.1 nN (Figure 2i), and the maximum number of protein unbinding lengths falls between 0 to 25 nm (Figure 2j). Similarly, Figure 2f shows the force spectrum of Tf interacting with high negative ZP (-35.1±0.9 mV) CNPs, and the unbinding force observed is ~349 pN. WLC fit with persistence length of 0.5 nm yielded a contour length of 251.2±3.7 nm. Repeated SMFS measurements on these samples yielded a force of interaction much lower than 349 pN and within the range of 0 to 125 pN (mostly a flat force spectrum at 0 nN was obtained due to the absence of interaction events). Unbinding force histogram analysis showed almost negligible interaction force events with a maximum unbinding force at ~900 pN (Figure 2k). Here above 50 pN, most of the events were non-repeating single events and the unbinding length histogram exhibited a peak close to zero nm due to higher number of negligible interaction events (Figure 2l). The lower unbinding force and length observed is due to the strong repulsion between Tf and negatively charged CNPs. Characteristic domain unfolding of Tf was observed in some of the force extension profiles (Supporting Information Figure S3) and were very similar to the domain unfolding usually seen in SMFS of other proteins.25 SMFS studies conducted on NP samples revealed a strong binding affinity of Tf to protonated CNP surface. To understand the nature and type of Tf bonding over CNP surface, theoretical simulation studies have been carried out.
Figure 2.
Schematic diagram of AFM probe functionalized with Tf interacting with positively charged CNPs, the force-extension spectrum and the force, length histograms obtained on CNPs with different ZPs. For protein-CNP interaction studies, (a) succinimide functionalized AFM tip was bioconjugated with Tf to form (b) Tf terminated AFM probe. (c) Interaction of Tf terminated AFM tip with protonated CNPs. Force-extension plots of Tf interacting with CNPs of ZP (d) +36.4±0.7 mV; (e) +5.9±1.2 mV; and (f) -35.1±0.9 mV. The corresponding force (g, i, and k) and length histograms (h, j, and l) are also shown. Force and length histograms were obtained by conducting multiple SMFS measurements on each sample. The total number of force and length values analyzed were n=273 (g and h), 206 (i and j) and 210 (k and l).
DFT simulation of protein interaction with protonated cerium oxide nanoparticles
Under physiological pH conditions, Tf exhibits higher number of negatively charged sites on its surface. These sites contain carboxylate ions in the form of aspartate (Asp) and glutamate (Glu) amino acid residues 28 that can interact with protonated CNPs to form hydrogen bonds with the NP surface. Both these residues are terminated with carboxyl ions at each end. DFT simulation studies were conducted to understand the nature of bonding between the carboxyl ion of the protein and the protonated CNPs. For simplicity, we modeled the NP-protein interaction by considering one glycinate ion (the simplest amino acid structure with one carboxyl ion) interacting with one CNP. Figure 3a shows the atomic structural model of protonated CNP interacting with the carboxyl ion of the glycinate. This octahedral CNP was derived from a bulk fluorite lattice that exposes the most stable (111) facets. To simulate the surface properties of CNPs with respect to their surface charge conditions, different levels of protonation varying from one to three H+ ions on the surface of CNP were considered. This is achieved by terminating up to three neighboring oxygen atoms on the same NP facet with one H+ ion each. Relaxed configuration of triple protonated CNP interacting with the carboxyl ion of the glycinate is shown in Figure 3a. All of the H+ ions interacted strongly with the NP and formed strong hydrogen bonds (more covalent in nature) with its surface oxygen atoms. The length of the O-H bond obtained here is 0.991 Å. The projection of electron localization function (ELF)29 on different planes (Figure 3b and 3c) passing through oxygen atoms of the protonated CNP and the glycinate ion reveals the formation of weak hydrogen bonds (more ionic in nature) between one of the carboxyl oxygen atoms and multiple protons on the CNP surface (Figure 3b). Here, the equilibrium O-H distance obtained is 1.87 Å. Meanwhile, the second oxygen atom of the carboxyl ion, which is in an equilibrium position with respect to one of the NP surface protons, formed a strong hydrogen bond with that proton (Figure 3c). This hydrogen bonding forms the basis for strong interaction of proteins with CNPs. Here, the equilibrium O-H distance obtained is 1.36 Å. Figure 3d shows the energy profiles of protonated CNP interacting with the carboxyl ion of the glycinate as a function of the number of protons on the NP surface. The blue curve represents the total ground state energies of CNPs interacting with the carboxyl ion of the glycinate, while the red curve represents the sum of their individual energies when they are separated by infinite distance (no interaction with each other). Both the curves show a decrease in the ground state energies with the addition of protons on CNP surfaces. However, the ground state energies of the carboxyl ion of the glycinate interacting with doubly and triply protonated CNP are not much different. This is due to the bending of bonds as a result of relaxation of weak hydrogen bond forming oxygen atoms to their equilibrium positions (total energy increases as the bond angles are bent from their normal configuration). The decrease in ground state energies with increase in numbers of protons on NP surfaces showed that the triply protonated CNP's surface is at a more favorable energy state for carboxyl ion interaction than the singly protonated CNP's surface. The total energies of CNP glycinate complex at binding configuration and at a large separation are represented by the red and the blue curves in Figure 3d. The difference of these total energies amounts to the binding energy between the protonated CNP and the carboxyl ion of the glycinate and is represented by cyan curve as shown in Figure 3d. The binding energy values displayed a tendency to increase with an increase in the number of protons on the NP surface. The binding energies between the carboxyl ion of a glycinate and a CNP covered by 1 to 3 protons are 2.81 eV, 4.03 eV and 4.19 eV, respectively. Hence, it is clear that adding more protons to the CNP surface could enhance the NP-protein binding. Both SMFS and DFT simulation studies conducted on CNPs revealed that Tf forms strong hydrogen bonds with a protonated CNP surface. The ligand-receptor mediated internalization of CNPs depends on the stability of Tf adhesion on CNP surface in a physiological environment. To test the stability of Tf coating over protonated CNP surface and its cellular uptake efficiency, Tf:CNPs were incubated with human lung adenocarcinoma epithelial cells (A549) and human embryo lung fibroblast cells (WI-38).
Figure 3.
Atomic structural model and energy profile of carboxyl ion of a glycinate interacting with a CNP. (a) Side view of the relaxed configuration of a negatively charged glycinate ion on a triply protonated facet of CNP. (b and c) Representation of projection of electron localization function (ELF) on different planes passing through oxygen atoms of a protonated CNP and a glycinate ion, revealing the formation of weak and strong hydrogen bonds between them. (d) Representation of ground state energy profile. Blue curve represents the ground state energy levels of a protonated CNP interacting with the carboxyl ion of a glycinate and the red curve represents the sum of the ground state energies of a protonated CNP and a glycinate ion located at infinite distance. The binding energy (cyan curve) between the protonated CNP and carboxyl ion of the glycinate was evaluated as the difference between the red and blue energy curves. ELF plots also reveal that all the protons formed strong hydrogen bonds with oxygen atoms of cerium oxide. Color code blue (ELF=0) and red (ELF=1.0) represents full absence and full presence of electron pair in actual point of space.
Transferrin adsorption and cellular uptake of cerium oxide nanoparticles
CNPs with a high positive ZP of +39.6±0.631 mV were used for protein adsorption and cellular uptake studies. Due to a strong electrostatic attraction, Tf was strongly bonded to CNPs (the higher the surface charges of NPs, the higher the protein adsorption). The ZP obtained on CNPs after Tf coating was +24.0±2.1 mV (ZP differences between CNPs and Tf:CNPs indicate the presence of Tf on CNPs). The protein adsorption on CNPs was confirmed by conducting FTIR (Supporting Information Figure S4) and XPS (Supporting Information Figure S5) analyses on Tf:CNPs. The amount of Tf adsorption on CNPs was quantified by conducting UV absorbance spectroscopy measurements at 280 nm wavelength (Supporting Information Figure S6) on Tf:CNP solutions. Tf:CNPs showed a protein adsorption of approximately 500 mg/g of CNPs.
Cellular uptake of NPs involves a two step process. The first step is the binding of NPs to the cell membrane and the second is cellular internalization.30 After the adsorption of the NPs on the cell membrane, the uptake occurs via several possible mechanisms: pinocytosis, receptor-mediated endocytosis, or phagocytosis.31 Since cancer cells have a higher growth rate, they over-express TfRs on their surface and will absorb iron at a higher rate. Hence NPs linked with Tf would be taken up efficiently. To understand the mechanism of CNP uptake in cancer and healthy cells, we have chosen A549 and WI-38 cells, respectively, as model cell systems. The cells were incubated with varying concentrations (from 100 nM to 100 μM) of Tf:CNPs and bare CNPs. Figures 4a and 4b show the percent of CNP cellular uptake by A549 and WI-38 cells obtained from ICP-MS measurements (ICPMS measurements were made after diluting the NPs incubated cell samples to ppb level concentrations and the % uptake are reported with respect to the molar concentrations at which the cells were incubated). A549 cells incubated with 100 nM concentration of CNPs, exhibited an enhanced uptake (~9.7 ng/ml) of Tf:CNPs compared to bare CNPs (~5.2 ng/ml). However at 1 μM concentration, A549 cells exhibited a reduced uptake (~51.8 ng/ml) of Tf:CNPs than that of bare CNPs (~61.1 ng/ml). Both 10 μM and 100 μM CNP incubated cells showed a higher uptake (~0.9 μg/ml and ~11.3 μg/ml, respectively) of Tf:CNPs and a lower uptake (~0.7 μg/ml and ~6.8 μg/ml, respectively) of bare CNPs similar to cells incubated with 100 nM concentrations of CNPs. Among all the concentrations, A549 cells incubated with concentrations of 100 nM, 10 μM, and 100 μM exhibited preferential uptake of Tf:CNPs compared to bare CNPs (Figure 4a). An enhanced uptake of Tf:CNPs by A549 cells indicates that the uptake in cancer cells is most likely by receptor-mediated endocytosis.32 Figure 4c shows the proposed model of TfR mediated endocytosis of CNPs by A549 cancer cells. The first step is the binding of Holo-Tf carrying CNPs to the TfRs on the cell membrane (SMFS studies reported earlier have shown an unbinding force of ~56 pN and an unbinding length of ~44 nm between holo-Tf coated tip and and TfR functionalized mica substrate at pH 7.433). The complex is then taken up inside the cell and transferred to the endosomal compartment where it is acidified (pH changes from ~7.4 to ~5.5). At a pH close to 5.4 (pl of Tf), Tf becomes neutrally charged and triggers the release of CNPs and iron as there is no more interaction with Tf. Recent studies have shown that Tf conjugated quantum dots (Tf:Qdots), after internalization through receptor mediated endocytosis remained in the endocytic structures and were not efficiently exocytosed.32 This could be due to the strong interaction of Tf with the quantum dots. Similar to Tf:Qdots, Tf:CNPs can also get trapped inside the endocytic structures. To understand the exocytosis mechanism of Tf:CNPs, a detailed investigation has to be carried out. Due to their high growth rate, cancer cells need more iron and thus will eventually internalize more iron carrying Tf:CNPs. The highest percent uptake (~66%, corresponds to ~11.3 μg/ml) of Tf:CNPs was seen in cells incubated with 100 μM concentration of CNPs (Figure 4a). A549 cells incubated with a concentration of 100nM exhibited the highest difference in uptake (~87%, from ~5.2 ng/ml to ~9.7 ng/ml) upon coating with Tf. Figure 4d shows the TEM images of A549 cells incubated with a concentration of 100 nM of Tf:CNPs. The core-shell like particles seen in TEM images, indicate the presence of a thin layer of Tf coating on CNPs. Large core size (~500nm) indicates the formation of NP agglomerates during the uptake process. Although the percentage of uptake was lower compared to Tf:CNPs, A549 cells showed considerable uptake of bare CNPs (Figure 4a). In the case of bare CNPs, the initial binding process can be visualized as a strong attraction between the positively charged CNPs and the negatively charged cell, facilitating their adsorption on the cell membrane and subsequent internalization via a nonspecific phagocytosis or pinocytosis processes.8 The relatively lower uptake of bare CNPs by A549 cells is also evident from the lack of particles seen in the TEM images of cells incubated with a concentration of 100nM of bare CNPs (Supporting Information Figure S7a).
Figure 4.
Cellular uptake of CNPs. (a and b) Cellular uptake of the CNPs by A549 cancer cells and WI-38 healthy cells at different concentrations of CNPs. (c) The cycle of TfR mediated cellular uptake of Tf:CNPs. (d and e) TEM images of A549 cancer cells incubated with Tf:CNPs and WI-38 healthy cells incubated with bare CNPs. (a) A549 Cells treated with 100 nM, 10 μM and 100 μM concentrations showed preferential cellular uptake of Tf:CNPs while (b) WI-38 cells treated with 100 nM, 1 μM, and 10 μM concentrations exhibit an increased uptake of bare CNPs. The bar chart is obtained by conducting ICP-MS analysis on 24 sets of A549 and WI-38 cells. The data is expressed as mean±sd. (c) Holo-Tf carrying CNP binds to TfR molecules expressed on the cell surface. After endocytosis via clathrin coated pits formed on the plasma membrane, the complex is taken up inside the plasma membrane by forming an endocytic vesicle. (d) The core shell like particles seen in A549 cells indicates the presence of Tf:CNPs (e) Solid spherical particles seen in WI-38 indicate the presence of bare CNPs. Agglomerated CNPs of size ~500 nm is observed in both the cases. Inset shows the magnified view of particles. White doted circles show the position of the CNPs inside the core shell structure.
WI-38 cells incubated with three different concentrations of bare CNPs under similar conditions exhibited enhanced uptake compared to Tf:CNPs (Figure 4b). Here the mechanism of uptake is similar to that seen in the case of bare CNPs in A549 cells. Figure 4e shows TEM images of WI-38 cells incubated with a concentration of 100 nM of bare CNPs. The solid spherical particles seen in TEM images indicate the presence of CNP agglomerates. It is clear from the ICP-MS data (Figure 4b) that the Tf:CNPs had a reduced uptake in these cells. At 100 nM, 1 μM and 10 μM concentrations, WI-38 cells exhibited a lower uptake (~5.5 ng/ml, ~94.4 ng/ml and ~0.9μg/ml, respectively) of Tf:CNPs compared to bare CNPs (~12.8 ng/ml, ~122.7 ng/ml and ~1.2 μg/ml, respectively). However, 100 μM CNP treated cells showed a higher uptake (~7.2 μg/ml) of Tf:CNPs that that of bare CNPs (~4.5 μg/ml). The TEM images of WI-38 cells incubated with 100 nM of Tf:CNPs also displayed less numbers of particles inside the cells and most of them were trapped near the cell wall (Supporting Information Figure S7b). This indicates that Tf:CNPs interact with the WI-38 cell membrane to a lesser extent as compared to bare CNPs. Besides reducing the CNPs effective surface potentials (from +39.6±0.631 mV to +24±2.1 mV), Tf coating can also induce steric hindrance between Tf and the negatively charged domains present on the cell membrane.30, 34 This could be the reason for the reduced cellular uptake of Tf:CNPs. WI-38 cells incubated with 100 nM of bare CNPs (Figure 4b) showed the highest percent of uptake (~75%, corresponds to ~12.8 ng/ml) and the uptake was reduced by ~57% (from ~12.8 ng/ml to ~5.5 ng/ml) upon coating with Tf. 100 μM treated WI-38 cells exhibited reduced uptake of bare CNPs as compared Tf:CNPs. At higher concentrations CNPs have a tendency to agglomerate,35 which could be the reason for the reduced uptake of bare CNPs seen at 100μM concentration (in the case of Tf:CNPs, larger agglomeration may not take place due to the steric hindrance between the Tf coated particles). The cellular uptake studies conducted on both A549 and WI-38 cells of Tf:CNPs and bare CNPs have shown that the interaction mechanism of nanoparticles with cells can be tuned by changing the NP surface conditions.
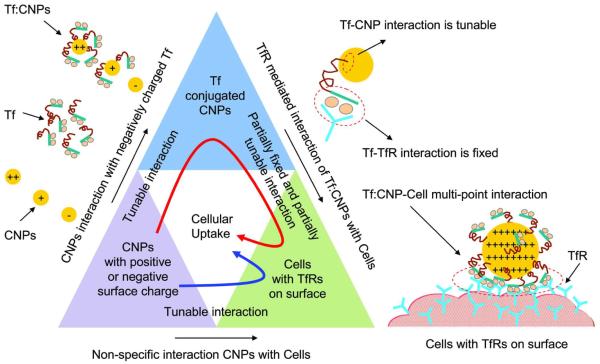
Tuning the surface charges of NPs (for enhanced ligand coverage) is a key requirement to achieve efficient cellular targeting and preferential drug delivery to malignant sites in the body. The interaction forces acting between NPs and biological systems (proteins and cells) at different stages starting from ligand conjugation to cellular uptake plays a major role in the transportation and delivery of drugs. Figure 5 shows a schematic diagram of interaction forces acting at various stages of the Tf:CNP cellular uptake process. The specific interaction between a Tf (on NP surface) and a TfR (at cell surface) is fixed (unbinding force of ~56 pN and unbinding length of ~44 nm) and can not be tuned. However, through protonation, the interaction between Tf and CNPs can be enhanced (unbinding force of ~150 to 225 pN and unbinding length of ~25 to 50 nm for +36.4±0.7 mV ZP CNPs) to a greater extent. Since Tf loading on a NP surface depends mainly on the surface charge difference (electrostatic adhesion), it is possible to add more numbers of Tf to a highly positive CNP surface. A higher positive charge and more numbers of Tf on the CNP surface will lead to better cell adhesion through a large number of ligand receptor mediated interactions (tunable multi-point interaction events, Figure 5). Hence, tuning the NP surface with more protons can not only improve the adhesion of ligands with the NP surface (main focus of our study), but it can also enhance the adhesion of a ligand conjugated NP with the cell surface thereby enhancing the receptor mediated cellular uptake. Similarly the non-specific interaction can also be tuned by tuning the CNPs surface potentials.
Figure 5.
Schematic diagram of interaction forces acting at different stages of cellular uptake. The triangle blocks show the interaction pathways of cellular uptake of CNPs. CNPs with strong positive charge show better adsorption of Tf. The interaction between Tf and CNPs can be tuned by protonation; however, the interaction of Tf with TfR is fixed. CNPs with a strong positive charge lead to enhanced Tf adsorption and multiple interactions with the TfRs on the cell surface. Red curved arrow inside the triangle blocks indicates the receptor mediated cellular internalization pathway of positively charged CNPs and blue curved arrow indicates the non-specific cellular internalization pathway of both positively and negatively charged CNPs. The red dashed circle represents the domain of multi-point interaction between Tf:CNP and TfRs on cell surface.
The experiments presented here not only show that the Tf can be effectively attached to the NP surface but also can be used as a targeting ligand in receptor mediated internalization of metal oxide NPs. Acidic buffer treatment of CNPs results in highly protonated surface and forms stable coating of Tf over CNPs. According to DFT simulation studies, the bonding between Tf and CNPs is mostly determined by the nature of hydrogen bonds forming between the NP surface protons and the carboxylate ions of the protein, and the binding energy between Tf and CNP increases with the number of surface protons present on the NPs. ICP-MS and TEM analysis conducted on both healthy and cancer cells incubated with CNPs shows that Tf:CNPs can be preferentially internalized by cancer cells. These results revealed the efficiency and physiological stability of Tf:CNP in receptor mediated cellular internalization process. They also demonstrate that the preferential cellular uptake of NPs can be achieved by binding with targeting ligands and it is essential to tune the surface charges of nanoparticles to achieve a better NP-ligand binding. Physiochemical modifications of nanoscale surfaces play a vital role in designing nanosize drug carriers which can preferentially target and deliver therapeutic drugs to malignant cells.
Materials and Methods
Cerium oxide nanoparticles
CNPs were synthesized by precipitation technique from cerium nitrate precursors.36 The particles were treated with acidic or basic pH buffers to induce different surface charges (see supporting information Material and Methods for more details).
Single molecule AFM experiments
Succinimide functionalized gold coated silicon nitride cantilevers (purchased from Novascan technologies) were treated with Tf solution to obtain Tf conjugated AFM probes. SMFM experiments were conducted on CNPs coated silicon surface placed in a liquid cell filled with deionized water. The forces of interaction between Tf and CNPs were measured by lowering the tip close to the NP surface and retracting it until the NP-protein bond broke (see supporting information Material and Methods for more details).
Supplementary Material
Acknowledgements
We thank the National Science Foundation (NIRT CBET 0708172) and National Institute of Health (R01: AG031529-01) for funding this work. Computational work was performed using (1) Stokes HPCC facility at UCF Institute for Simulation and Training (IST), (2) Bethe SMP server at UCF Nanoscience Technology Center (NSTC). The generous donation of the computer time is gratefully acknowledged.
Footnotes
Supporting Information Available. The structure and sequence of a human Tf (Figure S1); Cerium oxide nanoparticle synthesis; Tuning the surface charges of cerium oxide nanoparticles; Transferrin coating on cerium oxide nanoparticles; Primary culture of human lung adenocarcinoma epithelial cells and human embryo lung fibroblast cells; TEM biological cell sample preparations; Single molecule force spectroscopy (SMFS); Transferrin interaction with silicon (Figure S2); Characteristic domain unfolding of transferrin (Figure S3); Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (ATR-FTIR) analysis of transferrin coated ceria nanoparticles (Figure S4); X-ray Photoelectron Spectral (XPS) studies of transferrin coated ceria nanoparticles (Figure S5); UV-Vis analysis of transferrin coated ceria nanoparticles (Figure S6); TEM images of cells incubated with ceria nanoparticles (Figure S7); Computational details. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nano. 2007;2(12):751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Anabousi S, Laue M, Lehr C-M, Bakowsky U, Ehrhardt C. Assessing transferrin modification of liposomes by atomic force microscopy and transmission electron microscopy. Eur J Pharm Biopharm. 2005;60(2):295–303. doi: 10.1016/j.ejpb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv Colloid Interface Sci. 2007:134–135. doi: 10.1016/j.cis.2007.04.021. 167-174. [DOI] [PubMed] [Google Scholar]
- 4.Sun C, Lee JSH, Zhang M. Magnetic nanoparticles in MR imaging and drug delivery. Adv Drug Del Rev. 2008;60(11):1252–1265. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Z, Mumper RJ. Microparticles and nanoparticles as delivery systems for DNA vaccines. Crit Rev Ther Drug Carr Sys. 2003;20(23):103–137. doi: 10.1615/critrevtherdrugcarriersyst.v20.i23.10. [DOI] [PubMed] [Google Scholar]
- 6.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Del Rev. 2003;55(3):329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 7.Roy I, Ohulchanskyy TY, Pudavar HE, Bergey EJ, Oseroff AR, Morgan J, Dougherty TJ, Prasad PN. Ceramic-based nanoparticles entrapping water-insoluble photosensitizing anticancer drugs: a novel drug-carrier system for photodynamic therapy. J Am Chem Soc. 2003;125(26):7860–7865. doi: 10.1021/ja0343095. [DOI] [PubMed] [Google Scholar]
- 8.Patil S, Sandberg A, Heckert E, Self W, Seal S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials. 2007;28(31):4600–4607. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Patil S, Seal S, McGinnis JF. Rare earth nanoparticles prevent retinal degeneration induced by intracellular peroxides. Nat Nano. 2006;1(2):142–150. doi: 10.1038/nnano.2006.91. [DOI] [PubMed] [Google Scholar]
- 10.Tarnuzzer RW, Colon J, Patil S, Seal S. Vacancy engineered ceria nanostructures for protection from radiation-induced cellular damage. Nano Lett. 2005;5(12):2573–2577. doi: 10.1021/nl052024f. [DOI] [PubMed] [Google Scholar]
- 11.Das M, Patil S, Bhargava N, Kang JF, Riedel LM, Seal S, Hickman JJ. Auto-catalytic ceria nanoparticles offer neuroprotection to adult rat spinal cord neurons. Biomaterials. 2007;28(10):1918–1925. doi: 10.1016/j.biomaterials.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vyas SP, Singh A, Sihorkar V. Ligand-receptor-mediated drug delivery: an emerging paradigm in cellular drug targeting. Crit Rev Ther Drug Carr Sys. 2001;18(1):1–76. [PubMed] [Google Scholar]
- 13.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotech. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 14.Gomme PT, McCann KB, Bertolini J. Transferrin: structure, function and potential therapeutic actions. Drug Discov Today. 2005;10(4):267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- 15.Qian ZM, Li H, Sun H, Ho K. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol Rev. 2002;54(4):561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 16.Iinuma H, Maruyama K, Okinaga K, Sasaki K, Sekine T, Ishida O, Ogiwara N, Johkura K, Yonemura Y. Intracellular targeting therapy of cisplatin-encapsulated transferrinpolyethylene glycol liposome on peritoneal dissemination of gastric cancer. Int J Cancer. 2002;99(1):130–137. doi: 10.1002/ijc.10242. [DOI] [PubMed] [Google Scholar]
- 17.Ishida O, Maruyama K, Tanahashi H, Iwatsuru M, Sasaki K, Eriguchi M, Yanagie H. Liposomes bearing polyethyleneglycol-coupled transferrin with intracellular targeting property to the solid tumors in vivo. Pharm Res. 2001;18(7):1042–1048. doi: 10.1023/a:1010960900254. [DOI] [PubMed] [Google Scholar]
- 18.Yang PH, Sun X, Chiu JF, Sun H, He QY. Transferrin-mediated gold nanoparticle cellular uptake. Bioconjugate Chem. 2005;16(3):494–496. doi: 10.1021/bc049775d. [DOI] [PubMed] [Google Scholar]
- 19.Chithrani BD, Chan WCW. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007;7(6):1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 20.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, uuml, ller DJ. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288(5463):143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 21.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264(5157):415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 22.Smith BL, et al. Molecular mechanistic origin of the toughness of natural adhesives, fibres and composites. Nature. 1999;399(6738):761–763. [Google Scholar]
- 23.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393(6681):181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 24.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265(5178):1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 25.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by afm. Science. 1997;276(5315):1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 26.Mueller H, Butt H-J, Bamberg E. Force measurements on myelin basic protein adsorbed to mica and lipid bilayer surfaces done with the atomic force microscope. Biophys J. 1999;76(2):1072–1079. doi: 10.1016/S0006-3495(99)77272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JB, Kindt JH, Drake B, Hansma HG, Morse DE, Hansma PK. Bone indentation recovery time correlates with bond reforming time. Nature. 2001;414(6865):773–776. doi: 10.1038/414773a. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, Liu R, Zak O, Aisen P, Chance MR. Structural allostery and binding of the transferrin.receptor complex. Mol Cell Proteomics. 2005;4(12):1959–1967. doi: 10.1074/mcp.M500095-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Silvi B, Savin A. Classification of chemical-bonds based on topological analysis of electron localization functions. Nature. 1994;371(6499):683–686. [Google Scholar]
- 30.Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Intracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coating. Biomaterials. 2003;24(6):1001–1011. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]
- 31.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 32.Tekle C, Deurs B. v., Sandvig K, Iversen T-G. Cellular Trafficking of Quantum Dot-Ligand Bioconjugates and Their Induction of Changes in Normal Routing of Unconjugated Ligands. Nano Lett. 2008;8(7):1858–1865. doi: 10.1021/nl0803848. [DOI] [PubMed] [Google Scholar]
- 33.Yersin A, Osada T, Ikai A. Exploring transferrin-receptor interactions at the single-molecule level. Biophys J. 2008;94(1):230–240. doi: 10.1529/biophysj.107.114637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakkar H, Lowe PA, Price CP, Newman DJ. Measurement of the kinetics of protein uptake by proximal tubular cells using an optical biosensor. Kidney Int. 1998;54(4):1197–1205. doi: 10.1046/j.1523-1755.1998.00098.x. [DOI] [PubMed] [Google Scholar]
- 35.Babu S, Velez A, Wozniak K, Szydlowska J, Seal S. Electron paramagnetic study on radical scavenging properties of ceria nanoparticles. Chem Phys Lett. 2007;442(46):405–408. [Google Scholar]
- 36.Babu S, Schulte A, Seal S. Defects and symmetry influence on visible emission of Eu doped nanoceria. Appl Phys Lett. 2008;92(12):123112–3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.