Abstract
Antibody mimic proteins (AMPs) are poly-peptides that bind to their target analytes with high affinity and specificity, just like conventional antibodies, but are much smaller in size (2–5 nm, less than 10kDa). In this report, we describe the first application of AMP in the field of nanobiosensors. In2O3 nanowire based biosensors have been configured with an AMP (Fibronectin, Fn) to detect nucleocapsid (N) protein, a biomarker for severe acute respiratory syndrome (SARS). Using these devices, N protein was detected at sub-nanomolar concentration in the presence of 44 µM bovine serum albumin as a background. Furthermore, the binding constant of the AMP to Fn was determined from the concentration dependence of the response of our biosensors.
Keywords: Biosensor, nanowire, nucleocapsid (N) protein, antibody mimetic protein
Introduction
In less than a decade, biosensors based on nanowire/carbon nanotube transistors have successfully made the transition from proof of concept1 to highly selective, ultra sensitive devices capable of detecting specific proteins and DNA sequences.2–8 These devices utilize a capture agent on the sensor surface to selectively bind the target biomolecules. The captured biomolecules affect the electronic properties of the nanowires/nanotubes, resulting in an electronically readable signal. Capture agents commonly used in nanobiosensors include antibodies, oligonucleotides, and small ligands (e.g. biotin).1– 4, 8
Antibody mimic proteins (AMPs) are a class of affinity binding agents developed by in vitro selection techniques.9, 10 These AMPs can be evolved/engineered to improve recognition properties such as selectivity and binding affinity, with the potential to surpass antibodies and nucleotide aptamers. In contrast to typical antibodies, AMPs are stable to a wide range of pH and electrolyte concentrations, and are relatively small (usually 2–5 nm, less than 10 kDa). Moreover, it is expected that these peptide based affinity agents can be produced in large quantity, at relatively low cost. The combination of low cost, high binding affinity, chemical stability, and small size makes AMPs particularly attractive for use with nanowire/nanotube biosensors.
In this report, we introduce evolved AMPs as a new class of capture agents for nanowire/nanotube biosensors. These AMPs will allow us to build nanobiosensors for virtually any biomolecule with high sensitivity/selectivity, as demonstrated here for a protein related to severe acute respiratory syndrome (SARS), using devices based on In2O3 nanowires. Metal oxide nanowires, such as In2O3, ZnO, and SnO2, can be easily derivatized and their surface do not possess an insulating, native oxide layer (e.g. SiO2 on Si nanowires) that may decrease the nanowire sensitivity.8 Thus, it is worthwhile to investigate metal oxide nanowires as alternative nanomaterials to silicon nanowires for biosensing applications. We demonstrate that our technology platform, entailing In2O3 nanowire FETs combined with AMPs, can be used as a diagnostic tool with the potential to serve as a cost-effective, rapid, portable system. A fibronectin-based protein (Fn) was employed as an example of AMP capture agent to selectively recognize and bind the nucleocapsid (N) protein. The N protein is a biomarker associated with the SARS coronavirus.11 Our platform is capable of specifically detecting the N protein at sub-nanomolar concentrations, in the presence of 44 µM bovine serum albumin (BSA) as a background. This sensitivity, while comparable to current immunological detection methods, can be obtained in a relatively short time and without the aid of any signal amplifier, such as fluorescence labeled reagents. Ultimately, we show that our platform can also be used to accurately determine the dissociation constant of the N protein and Fn by applying a conventional Langumir model to the concentration-dependent sensing response.
Results and Discussion
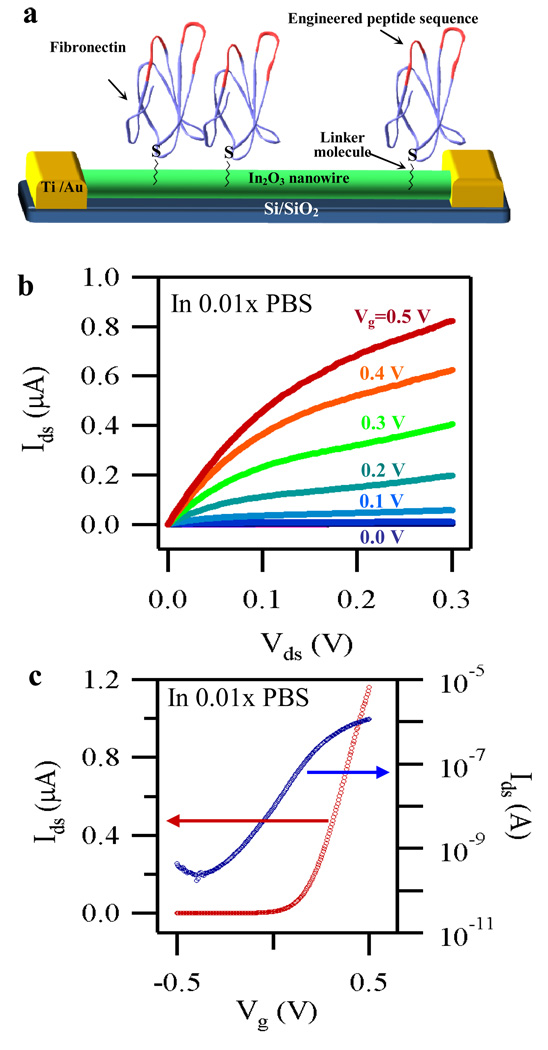
A schematic illustration of our fibronectin-based capture agent anchored to an In2O3 nanowire field-effect transistor is shown in Figure 1a Our Fn-based AMP was evolved using mRNA display from a large library of potential candidates and possesses a high binding affinity to the N protein (KD = 3.3 nM), as described in details elsewhere.12 The evolved portion of N-protein is highlighted in red in Figure 1a Our Fn probe was also engineered to have a single cystine residue near the C-terminus of the protein, remote from the binding site (Figure 1a and Figure S1). This unique thiol group allows the Fn anchoring to the nanowire to be carried out selectively, since the chosen linker molecule/chemistry (i.e. maleimide groups) gives a nanowire surface that is reactive only toward sulphydryl groups (see method and supporting information for details). This conjugation strategy allows every bound Fn to retain full activity, a clear advantage over antibodies, which are often bound to the nanowire surface via amine containing residues, randomly distribute on the antibody surface.2–7 Moreover, our Fn can also be easily configured with other functional groups, such as azides13, 14 or cyclopentadienes,15 that are useful in bioconjugation.
Figure 1.
(a) Schematic diagram showing Fn immobilized on the surface of an In2O3 nanowire FET device. The regions of Fn with the engineered peptide sequence are highlighted in red. Fn was attached to the NWs via the sulphydryl group of a cysteine near the C-terminus, remote from the binding site. (b) A family of Ids–Vds curves and (c) a typical Ids–Vg curve (plotted both in linear (red) and logarithmic (blue)) obtained from one of our devices operating with the liquid gate configuration.
Details of device fabrication, functionalization, and experimental conditions used here can be found in the method section. Briefly, In2O3 nanowires were grown via a laser ablation CVD method on a Si/SiO2 substrate, following a well established procedure in our laboratory.16–19 The nanowires were suspended in isopropanol and deposited onto another Si/SiO2 substrate. The position of the source and drain (S-D) electrodes was defined by photolithography, with channel length and width of 2.5 µm and 780 µm, respectively. Metal deposition on the pre-patterned surface followed by lift-off completed the device fabrication. Immediately after cleaning, the nanowire devices were submerged in a 0.1 mM aqueous solution of 6-phosphonohexanoic acid followed by baking under inert atmosphere, resulting in the binding of the phosphonic acid to the surface of the In2O3 NWs. The S-D contacts were then wired to a custom-made printed circuit board and a mixing cell was assembled on the device chip (supporting information, Figure S3). This mixing cell was used to deliver and handle all the chemical reagents necessary to complete the surface modification and all the buffer solutions during active measurements.
We have then measured the device characteristics utilizing a liquid gate electrode20 while using the above described set up. Our In2O3 NW FET devices exhibit excellent transistor behavior in 0.01x phosphate buffered saline (PBS) solution. The linear behavior of the source/drain current versus source/drain voltage (Ids–Vds) curves at Vds, ≤ 0.08 V (Figure 1b) suggests good contact between the nanowires and source/drain electrodes. Strong gate dependence was also observed for a typical device, as shown by the Ids versus liquid gate voltage (Ids–Vg) curves shown in linear (red curve) and logarithmic scale (blue) in Figure 1c. The on/off ratio and transconductance were ~4.6 × 103 and ~3.6 µS, respectively. These results confirm the stability of our devices under active measurement conditions.
Further surface modification of the nanowires conferred our devices with the desired biological recognition properties. The carboxylic acid functional groups on the NW surface were activated with EDC and the activated COOHs were allowed to react with BMPH, resulting in the formation of a NW surface reactive toward the unique thiol present on the Fibronectin probe molecule. The functionalized devices were stored submerged in 1× PBS at 4 °C.
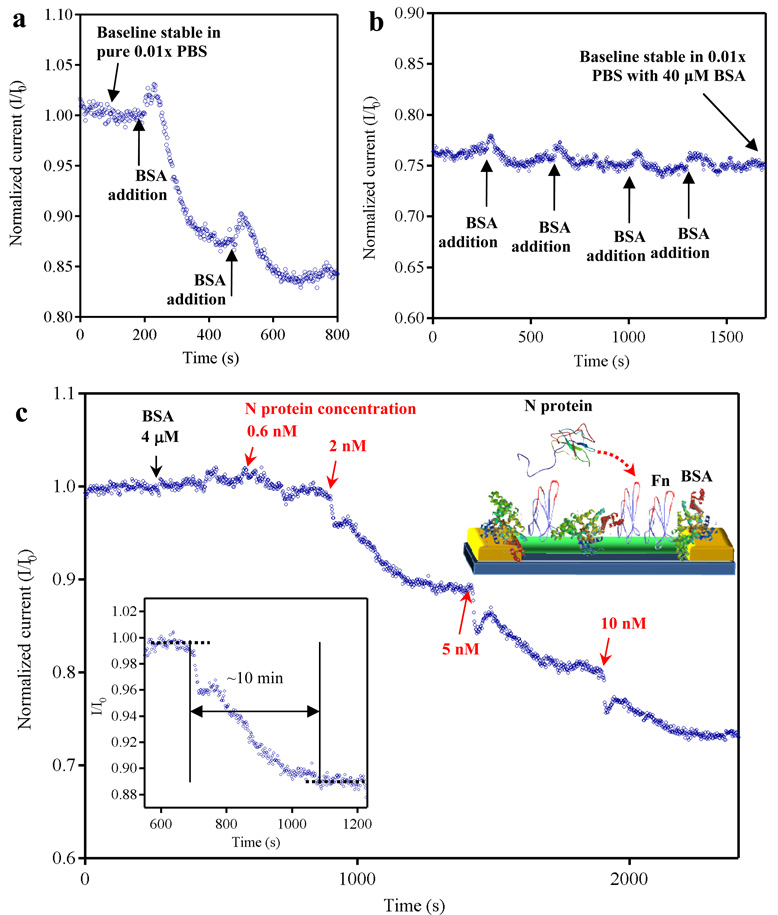
The normalized electrical response of an Fn-modified nanowire device is shown in Figure 2a–c, where we have plotted Ids divided by the Ids at t = 0 s, referred to as I/I0. The device was operated at Vds = −200 mV and Vg = −100 mV. Under such experimental conditions, a baseline signal was quickly established in pure 0.01× PBS buffer, as indicated in Figure 2a. We note that the leakage current between source and drain through the buffer is negligible compared to the conduction through nanowires. (see supporting information.) A shift in the baseline level is often observed when transitioning from pure buffer to protein-rich buffer, attributed to non-specific binding interactions of proteins with the nanowire device. These non-specific binding phenomena, if not adequately mitigated, may lead to false positive results. Passivating regions of the device that are subject to non-specific binding with a “blocking agent” (typically BSA or Tween 20), as traditionally used in bioanalytical assays such as ELISA,21, 22 is technique useful to minimize false positives during active measurements. We have employed BSA as “blocking agent” for our nanowire devices including the source-drain electrodes. Aliquots of a 10 mg/mL solution of BSA were used to increase the protein concentration of the buffer in contact with the nanowires. After each BSA addition, the baseline re-equilibrated to a lower value of S-D current, as shown in Figure 2a and 2b. Saturation of non-specific binding sites was achieved at 40µM concentration of BSA, as indicated in Figure 2b. We note that BSA passivation is widely employed in many standard detection techniques such as ELISA; however, it may be possible that the BSA passivation step can be eliminated by increasing the surface coverage of Fn. The baseline stability in a protein-rich medium was then tested by increasing the BSA concentration by 10% (4 µM) (shown with a black arrow in Figure 2c). The device showed no significant response, which confirmed that sites for non-specific binding were blocked. The conductance of the device rapidly decreased (4%) upon exposure the nanowire sensor to a solution containing 0.6 nM of N protein in 44 µM BSA. We further tested the response of our devices to higher N-protein concentrations. N-protein solutions were prepared by successive additions of small aliquots of a 100±30 nM stock solution of N-protein in 0.01× PBS containing 44 µM BSA. When the N protein concentration was progressively increased to 2 nM, 5 nM, and 10 nM, we observed a consistent decrease in device conductance of 12%, 22%, and 31%, respectively, relative to the baseline. The pI of In2O3 is ~8,23 and the pI of N protein is ~10,24 while the PBS buffer we used had pH = 7.4. The pI of the N protein indicates that our target analyte possesses a net positive charge under our measurement conditions. However, some regions of N proteins are locally negatively charged, such as the C-terminus of N protein, which is rich in serine groups (pI ~ 5).25 The pI of In2O3 indicates that the surface of our In2O3 nanowires is positively charged. Thus, it might be possible that the positive charges on In2O3 nanowires attract the negatively charged regions of N protein captured on the nanowire surface. These serine residues could induce a chemical gating effect on the nanowires, causing a decrease in the conductance of our device. Other non-negligible contributions to the sensing mechanism could come from an increase of the carrier scattering due to the binding of N protein. We note that the sensing mechanism for nanobiosensors is still a topic of intense debate in the literature26–30 and it is currently under intensive investigations in our research group.
Figure 2.
Normalized electrical output (I/I0) versus time of a single operating device. (a)–(b) Response curves to passivation upon addition of successive aliquots of BSA. Upon increasing the concentration of BSA (from pure 0.01× PBS), the baseline re-equilibrates at lower values of S-D current until stability is ultimately reached at 40 µM BSA, in 0.01× PBS. (c) Response for a nanowire device functionalized with Fn. The red arrows indicate the times when the solution was raised to a given concentration of N protein. The inset on the right side is the configuration of our device during active sensing measurements. BSA protein was used to block sites for non-specific binding. The Fn probe molecule was then used to specifically capture the target N protein. The inset on the left side is to show the plateau and the definition of response time.
We have defined the response time to be the time necessary to achieve equilibrium after a change in the concentration of the N protein. Under our active measurement conditions, the response time turned out to be in the order of ~10 minutes as shown in Fig. 2c inset for one measurement. This response time can be considered relatively short when compared to the time required to produce a signal using other diagnostic technologies such as ELISA (~hours),31 which requires multistep analysis. Thus, while detecting the N protein in the nanomolar range can also be achieved using current immunological clinical tests, our nanowire sensors offer additional advantages such as label free detection and a comparatively short response time.
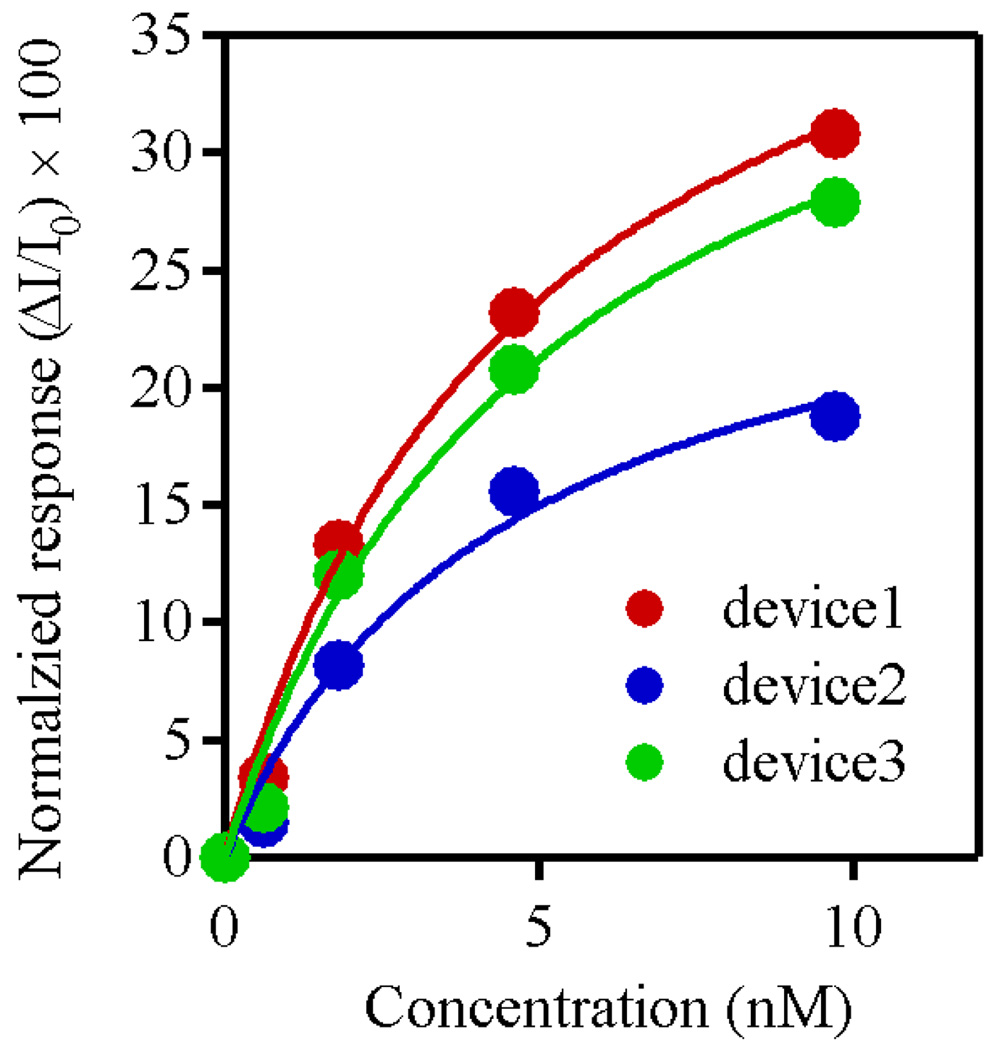
Three devices were tested in parallel, and all the devices showed a quantitatively similar concentration dependence for their response. Plots of sensor response versus N-protein concentration for these three devices are shown in Figure 3 (dots), confirming the reproducibility of the results. These plots were fitted using a conventional Langmuir isotherm model32,33 (solid line) and these fits were used to estimate the dissociation constant of Fn to the N protein. In applying this model, we assumed that the response of the sensor is proportional to the number of captured molecules on the sensor surface, such that I/I0 ∝ Fn surface coverage. The following equation was used to describe the concentration dependence of our sensor response:
where A is a coefficient that converts surface coverage into electrical response, α is a constant, and n is the concentration of N protein. The values of A and α were determined using the least-square method to give the best fit to the experimental values. The dissociation constant was estimated by calculating the solution of n in the following equation:
where θ is the percentage coverage of the surface, since the definition of dissociation constant is an analyte concentration when half of the sites are occupied. Application of this analytical model yields a dissociation constant of 4.9 ± 0.4 nM, which is close to the value of the dissociation constant (KD = 3.3 nM) obtained from measurements of surface plasmon resonance (SPR).12 The close match of the dissociation constant illustrates the validity of our assumption and the Langmuir isotherm model. The small difference may come from the fact that the measurements were done in buffers with different ionic strengths (0.01× PBS for nanobiosensor and 1× PBS for SPR).
Figure 3.
Normalized response from three devices versus concentration of N protein (dots). These plots can be fitted using a Langmuir isotherm model (solid line).
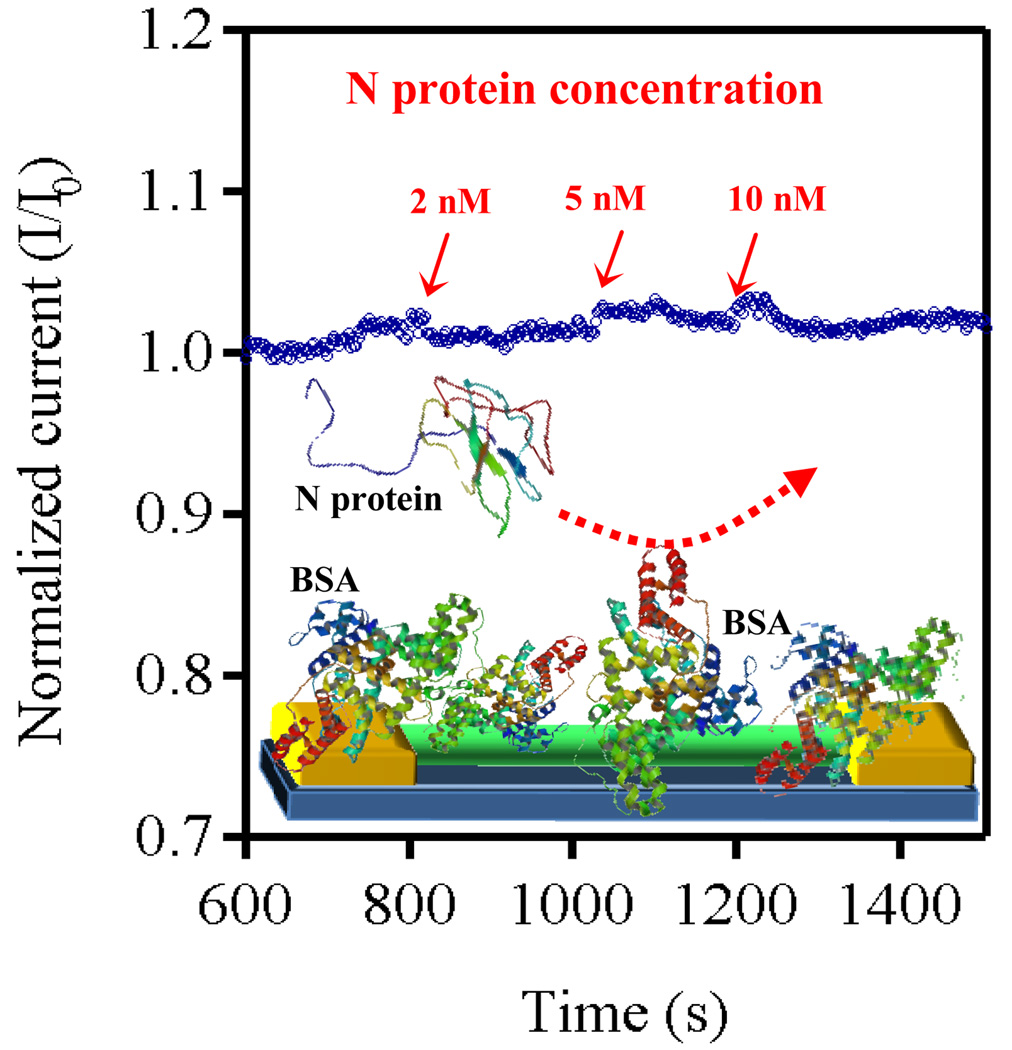
We conducted further experiments to confirm the role of Fn as a selective capture probe. A nanowire surface, previously activated for bioconjugation, was treated with 2-mercaptoethanol, prior to Fn. The Fn capture probe is not expected to bind to the nanowire surface coated with 2-mercaptoethanol moieties, and thus this device should not specifically recognize the N protein. A baseline was established for this device after saturation of any site for non-specific binding with a 40 µM solution of BSA as for the device with Fn (Fig. S5). This device was then sequentially exposed to a 2, 5, and 10 nM solution of N protein, while still in the presence of 40 µM BSA. (Fig. 4) We did not observe any significant responses, in sharp contrast to the response observed when we used a device functionalized with Fn, confirming that our Fn based capture probe can selectively capture the N protein.
Figure 4.
A control device without the Fn capture probe does not respond to the presence of N protein.
In conclusion, we have demonstrated that AMPs can be employed as capture agents in nanobiosensors. Sensors based on In2O3 nanowires were modified with a fibronectin-based binding agent that can selectively detect the SARS biomarker N protein. The N protein was detected at a sensitivity comparable to current immunological detection methods (subnanomolar concentration), but obtained within shorter time and without the aid of labeled reagents. We believe that nanowire biosensor devices functionalized with engineered proteins (evolved to possess elevated affinity toward target molecules) can have important potential applications ranging from disease diagnosis to homeland security. This report also demonstrates the potential for nanobiosensors to be used as an accurate, convenient, and rapid tool to measure the dissociation constants for biological complex systems, such as antibody-antigen, protein-ligand, oligonucleotides, etc..
Method
Materials
Phosphate buffer saline (PBS, pH = 7.40, 10 mM phosphates, 137 mM NaCl, 2 mM KCl) was purchased from Mediatech Inc. Water was of high purity (HPLC grade). [(2-N-morpholino) ethanesulfonic acid] buffer (MES buffer, 100 mM, pH =5.2) was purchased from Fluka. (1-Ethyl-3-[ 3-Dimethylaminopropyl]carbodiimide hydrochloride) (EDC) was purchased from Aldrich. (N-[β-Maleimidopropionic acid] hydrazide, trifluoroacetic acid salt) (BMPH) was purchased from Pierce. 6-Phosphohexanoic acid was purchased from Aldrich. Bovine serum albumin (BSA) was purchased from Sigma. Aqueous solution of 0.01% Au nanoparticles (mean diameter: 9.6 nm) were purchased from BBInternational. Indium arsenide was purchased from Alfa Aesar. Stock solutions of Fibronectin (1 µM in 1× PBS buffer) and N protein (200 nM in 1× PBS buffer) were provided by Prof. Ren Sun’s research group. The N protein solution was buffer exchanged to 0.01× PBS before use with a NAP-5 column (Amersham Scientific). We note that during buffer exchange the concentration of the N protein is lowered due to about two fold dilution. We estimate the concentration of N protein in 0.01× PBS to be 100 nM with an error bar of ± 30%.
Device Fabrication
In2O3 nanowires were grown via a laser ablation CVD method on a Si/SiO2 substrate, following a well established procedure in our laboratory.4, 17 Briefly, a stock aqueous solution of Au nanoparticles, with a diameter of about 10 nm, was obtained by a 1:10 dilution of the purchase solution in isopropanol. A veil of this diluted Au nanoparticle solution was allowed to dry on a Si/SiO2 surface (typical size: 1–2 cm2), resulting in a uniform surface coating of Au nanoparticles. These nanoparticles were then employed as a catalyst for the laser ablation promoted growth of indium oxide nanowires, using InAs as a source of In. Details of the nanowire growth are described elsewhere.16, 18, 19 The as-grown nanowires were suspended in isopropanol by sonication and this suspension was deposited onto another Si/SiO2 substrate (typically on a 3” Si wafer with a surface coverage of 1 nanowire per 100 µm2). The position of the source and drain (S-D) electrodes was defined by photolithography. The S-D electrodes were designed to have an interdigitated interface at the semiconductor channel, leading to FETs with channel length and width of 2.5 µm and 780 µm, respectively. Metal deposition (5 nm of Ti and 50 nm of Au) on the pre-patterned surface followed by lift-off completed the device fabrication.
Surface Functionalization
In2O3 NW device cleaning procedure
In2O3 NW devices were submerged for 5 minutes each in boiling tetrachloroethylene, acetone, ethanol, and water. The NW devices were rinsed with ethanol, dried with nitrogen, placed in a ozone/UV chamber for 8–10 minutes, and then submerged in hot water (80–90 °C) for 5 minutes.
Attachment of the Fibronectin probe molecule
The attachment of the Fibronectin probe molecules is schematically illustrated in Figure S2. Immediately after cleaning, the nanowire devices were submerged in a 0.1 mM aqueous solution of 6-phosphonohexanoic acid for 16 hours at room temperature, resulting in the binding of the phosphonic acid residue to the surface of the In2O3 NWs (i). These devices were baked at 125 °C under nitrogen for 2 hr. These devices were then wired on a custom-made printed circuit board and a mixing cell was assembled on the device chip (Figure S3). This mixing cell was used to deliver and handle all the chemical reagents necessary for surface modification and all the buffer solutions during active measurements. The carboxylic acid functional groups on the NW surface were activated by placing in the mixing cell a 50 mM solution of EDC in MES buffer and allowed the activated COOHs to react with BMPH (present in the same solution at 5 mM concentration) for 60–90 minutes (ii). Unreacted, EDC-activated carboxyl groups were quenched with a 50 mM solution of ethanolamine (ii). This resulted in the formation of a NW surface reactive toward the unique thiol present on the Fibronectin probe molecule. This EDC/BMPH solution was removed from the mixing cell by progressive dilution with PBS buffer. A solution containing about 200 nM of Fn in 1× PBS (along with 1 equivalent of TCPE) was allowed to react with the NW surface, overnight, at 4 °C (iii). The unreacted maleimide groups were quenched with the addition of 10 uL of a 50 mM solution of 2-mercaptoethanol and allowed to react for 10–15 minutes (iii). This Fn solution was replaced with 1×PBS by progressive dilutions and the functionalized devices were stored submerged in 1× PBS at 4 °C.
Supplementary Material
Engineered Fibronectin, surface functionalization, experimental setup, measurement of leakage current, absolute response, and a control experiment. This material is available free of charge via the internet at http://pubs.acs.org.
ACKNOWLEDGMENTs
We acknowledge financial support from the Whittier Foundation and the National Institute of Health.
REFERENCES
- 1.Cui Y, Wei QQ, Park HK, Lieber CM. Nanowire Nanosensors for Highly Sensitive and Selective Detection of Biological and Chemical Species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 2.Patolsky F, Zheng G, Lieber CM. Nanowire Sensors for Medicine and the Life Sciences. Nanomedicine. 2006;1:51–65. doi: 10.2217/17435889.1.1.51. [DOI] [PubMed] [Google Scholar]
- 3.Stern E, Klemic JF, Routenberg DA, Wyrembak PN, Turner-Evans DB, Hamilton AD, LaVan DA, Fahmy TM, Reed MA. Label-Free Immunodetection with Cmos-Compatible Semiconducting Nanowires. Nature. 2007;445:519–522. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Curreli M, Lin H, Lei B, Ishikawa FN, Datar R, Cote RJ, Thompson ME, Zhou CW. Complementary Detection of Prostate-Specific Antigen Using In2O3 Nanowires and Carbon Nanotubes. J. Am. Chem. Soc. 2005;127:12484–12485. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 5.Curreli M, Zhang R, Ishikawa FN, Chang H-K, Cote RJ, Zhou C, Thompson ME. Real-Time, Label-Free Detection of Biological Entities Using Nanowire-Based Fets. IEEE Transactions on Nanotechnology. 2008;7:651–667. [Google Scholar]
- 6.Gruner G. Carbon Nanotube Transistors for Biosensing Applications. Anal. Bioanal. Chem. 2006;384:322–335. doi: 10.1007/s00216-005-3400-4. [DOI] [PubMed] [Google Scholar]
- 7.Allen BL, Kichambare PD, Star A. Carbon Nanotube Field-Effect-Transistor-Based Biosensors. Adv. Mater. 2007;19:1439–1451. [Google Scholar]
- 8.Bunimovich YL, Shin YS, Yeo WS, Amori M, Kwong G, Heath JR. Quantitative Real-Time Measurements of DNA Hybridization with Alkylated Nonoxidized Silicon Nanowires in Electrolyte Solution. J. Am. Chem. Soc. 2006;128:16323–16331. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binz HK, Amstutz P, Pluckthun A. Engineering Novel Binding Proteins from Nonimmunoglobulin Domains. Nat. Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 10.Binz HK, Pluckthun A. Engineered Proteins as Specific Binding Reagents. Curr. Opin. Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Zakhartchouk AN, Viswanathan S, Mahony JB, Gauldie J, Babiuk LA. Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein Expressed by an Adenovirus Vector Is Phosphorylated and Immunogenic in Mice. J. Gen. Virol. 2005;86:211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]
- 12.Liao HI, Olson CA, Ishikawa FN, Curreli M, Chang H-K, Thompson ME, Zhou C, Roberts RW, Sun R. 2009 Manuscript in Preparation. [Google Scholar]
- 13.Bunimovich YL, Ge G, Beverly KC, Ries RS, Hood L, Heath JR. Electrochemically Programmed, Spatially Selective Biofunctionalization of Silicon Wires. Langmuir. 2004;20:10630–10638. doi: 10.1021/la047913h. [DOI] [PubMed] [Google Scholar]
- 14.Rohde RD, Agnew HD, Yeo WS, Bailey RC, Heath JR. A Non-Oxidative Approach toward Chemically and Electrochemically Functionalizing Si(111) J. Am. Chem. Soc. 2006;128:9518–9525. doi: 10.1021/ja062012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yousaf MN, Houseman BT, Mrksich M. Using Electroactive Substrates to Pattern the Attachment of Two Different Cell Populations. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5992–5996. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Bao M, Wang KL, Li C, Lei B, Zhou C. One-Dimensional Transport of In2O3 Nanowires. Appl. Phys. Lett. 2005;86:213101–213103. [Google Scholar]
- 17.Curreli M, Li C, Sun YH, Lei B, Gundersen MA, Thompson ME, Zhou CW. Selective Functionalization of In2O3 Nanowire Mat Devices for Biosensing Applications. J. Am.Chem. Soc. 2005;127:6922–6923. doi: 10.1021/ja0503478. [DOI] [PubMed] [Google Scholar]
- 18.Lei B, Li C, Zhang D, Tang T, Zhou C. Tuning Electronic Properties of In2O3 Nanowires by Doping Control. Appl. Phys. A: Mater. Sci. Process. 2004;79:439–442. [Google Scholar]
- 19.Li C, Zhang D, Han S, Liu X, Tang T, Lei B, Liu Z, Zhou C. Synthesis, Electronic Properties, and Applications of Indium Oxide Nanowires. In Molecular Electronics Iii. 2003;Vol.1006:104–121. doi: 10.1196/annals.1292.007. [DOI] [PubMed] [Google Scholar]
- 20.Rosenblatt S, Yaish Y, Park J, Gore J, Sazonova V, McEuen PL. High Performance Electrolyte Gated Carbon Nanotube Transistors. Nano Lett. 2002;2:869–872. [Google Scholar]
- 21.Stadtherr K, Wolf H, Lindner P. An Aptamer-Based Protein Biochip. Anal. Chem. (77) 2005:3437–3443. doi: 10.1021/ac0483421. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Curr. Protoc. Mol. Biol. Hoboken: John Wiley and Sons Inc.; 2007. [Google Scholar]
- 23.Kosmulski M. Pristine Points of Zero Charge of Gallium and Indium Oxides. J. Colloid Interface Sci. 2001;238:225–227. doi: 10.1006/jcis.2001.7484. [DOI] [PubMed] [Google Scholar]
- 24.Mark J, Li XG, Cyr T, Fournier S, Jaentschke L, Hefford MA. Sars Coronavirus:Unusual Lability of the Nucleocapsid Protein. Biochem. Biophys. Res. Commun. 2008;377:429–433. doi: 10.1016/j.bbrc.2008.09.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai MMC, Cavanagh D. The Molecular Biology of Coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair PR, Alam MA. Screening-Limited Response of Nanobiosensors. Nano Lett. 2008;8:1281–1285. doi: 10.1021/nl072593i. [DOI] [PubMed] [Google Scholar]
- 27.Heller I, Janssens AM, Mannik J, Minot ED, Lemay SG, Dekker C. Identifying the Mechanism of Biosensing with Carbon Nanotube Transistors. Nano Lett. 2008;8:591–595. doi: 10.1021/nl072996i. [DOI] [PubMed] [Google Scholar]
- 28.Stern E, Wagner R, Sigworth FJ, Breaker R, Fahmy TM, Reed MA. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007;7:3405–3409. doi: 10.1021/nl071792z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang XW, Bansaruntip S, Nakayama N, Yenilmez E, Chang YL, Wang Q. Carbon Nanotube DNA Sensor and Sensing Mechanism. Nano Lett. 2006;6:1632–1636. doi: 10.1021/nl060613v. [DOI] [PubMed] [Google Scholar]
- 30.Chen RJ, Choi HC, Bangsaruntip S, Yenilmez E, Tang XW, Wang Q, Chang YL, Dai HJ. An Investigation of the Mechanisms of Electronic Sensing of Protein Adsorption on Carbon Nanotube Devices. J. Am. Chem. Soc. 2004;126:1563–1568. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- 31.Crowther JR. Elisa: Theory and Practice. New York: Humana Press; 1995. [DOI] [PubMed] [Google Scholar]
- 32.Langmuir I. The Constitution and Fundamental Properties of Solids and Liquids Part I Solids. J.Am. Chem. Soc. 1916;38:2221–2295. [Google Scholar]
- 33.Halperin A, Buhot A, Zhulina EB. On the Hybridization Isotherms of DNA Microarrays: The Langmuir Model and Its Extensions. J. Phys.-Condens. Matter. 2006;18:S463–S490. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Engineered Fibronectin, surface functionalization, experimental setup, measurement of leakage current, absolute response, and a control experiment. This material is available free of charge via the internet at http://pubs.acs.org.