Summary
Hfq is a conserved RNA-binding protein that regulates diverse cellular processes through post-transcriptional control of gene expression, often by functioning as a chaperone for regulatory sRNAs. Here, we explored the role of Hfq in enterohaemorrhagic E. coli (EHEC), a group of non-invasive intestinal pathogens. EHEC virulence is dependent on a Type III secretion system encoded in the LEE pathogenicity island. The abundance of transcripts for all 41 LEE genes and more than half of confirmed non-LEE-encoded T3 effectors were elevated in an EHEC hfq deletion mutant. Thus, Hfq promotes coordinated expression of the LEE-encoded T3S apparatus and both LEE- and non-LEE-encoded effectors. Increased transcript levels led to the formation of functional secretion complexes capable of secreting high quantities of effectors into the supernatant. The increase in LEE-derived transcripts and proteins was dependent on Ler, the LEE-encoded transcriptional activator, and the ler transcript appears to be a direct target of Hfq-mediated negative regulation. Finally, we found that Hfq contributes to the negative regulation of T3SSs in several other pathogens, suggesting that Hfq, potentially along with species-specific sRNAs, underlies a common means to prevent unfettered expression of T3SSs.
Keywords: Hfq, EHEC, Ler, Type III Secretion
Introduction
Hfq is a conserved protein that is an indispensable chaperone for a large class of small non-coding RNAs (sRNAs) in diverse bacterial species (Valentin-Hansen et al., 2004). These regulatory sRNAs typically function by base-pairing with target mRNAs, generally through regions of short, imperfect complementarity, and thereby alter the mRNAs’ stability or availability for translation (reviewed in (Storz et al., 2005)). Hfq stabilizes sRNAs and appears to promote formation of sRNA/mRNA complexes. In cells lacking Hfq, many such complexes do not form and the abundance of sRNAs is dramatically reduced. Together, sRNAs and Hfq have been shown to regulate a myriad of bacterial processes in a wide array of bacterial species (reviewed in (Waters and Storz, 2009)). In addition, Hfq has been shown to bind and directly regulate the translation of several mRNAs, not only leading to altered protein levels but influencing transcript abundance and stability as well (Hajnsdorf and Regnier, 2000; Vytvytska et al., 2000; Mohanty et al., 2004; Sittka et al., 2008; Urban and Vogel, 2008). Overall, RNA populations are significantly altered in cells lacking hfq. Although not all direct targets of Hfq show altered transcript levels (Updegrove et al., 2008), several studies have demonstrated that transcriptome analyses of hfq mutants can yield insight into Hfq and/or sRNA-mediated regulation (Ding et al., 2004; Guisbert et al., 2007; Sittka et al., 2008).
E. coli Hfq was originally identified as a host factor required for the replication of the RNA phage Qβ (Franze de Fernandez et al., 1968). Subsequent studies showed that an E. coli K12 hfq mutant exhibited highly pleiotropic phenotypes including reduced cell growth and increased sensitivity to a variety of stresses (Tsui et al., 1994), suggesting that Hfq is a global regulator. In part, the diverse effects of hfq disruption can be accounted for by the findings that Hfq/sRNAs influence the translation of the sigma factors RpoS and RpoE (Muffler et al., 1996; Thompson et al., 2007), data confirmed by microarray analyses of an E. coli K12 hfq mutant (Guisbert et al., 2007). Studies of additional bacterial species have revealed that Hfq can also have a critical impact on bacterial pathogenicity. For example, hfq mutants in Brucella abortus, Vibrio cholerae, Listeria monocytogenes, Neisseria meningitidis, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium are highly attenuated in animal models (Robertson and Roop, 1999; Sonnleitner et al., 2003; Christiansen et al., 2004; Ding et al., 2004; Sittka et al., 2007; Fantappie et al., 2009); however, these studies must be interpreted with some caution since hfq mutants often exhibit growth defects. In a few cases, Hfq/sRNAs have been shown to regulate processes directly linked to virulence. For example, Sittka et al. observed marked reductions in secretion of several type III secretion system (T3SS) effector proteins encoded by Salmonella pathogenicity island 1 in an S. Typhimurium hfq mutant (Sittka et al., 2007).
Despite the extensive knowledge of Hfq/sRNA-mediated regulation in E. coli K12, little is known about the contribution of Hfq or sRNAs to regulation in pathogenic E. coli such as enterohaemorrhagic E. coli (EHEC). EHEC are an emerging group of pathogenic E. coli that are a common cause of food-borne diarrhea in developed countries (reviewed in (Caprioli et al., 2005; Welinder-Olsson and Kaijser, 2005)). The typical clinical manifestations of infection with EHEC range from mild diarrhea to hemorrhagic colitis, and a subset of patients develop severe, life-threatening complications, including the hemolytic uremic syndrome. EHEC includes a number of serotypes, but the O157:H7 serotype accounts for most cases of EHEC-induced disease (Cohen and Giannella, 1992). While there is some variation in size and gene content of EHEC genomes, all are more than a megabase larger than E. coli K12. Much of the additional DNA found in EHEC strains (commonly referred to as “O-islands”) is found in mobile (or formerly mobile) genetic elements such as prophages (Hayashi et al., 2001; Perna et al., 2001). Currently, knowledge of Hfq/sRNA-mediated regulation of EHEC genes that lack homologs in E. coli K12 is lacking.
EHEC colonization of the host intestine is dependent on a T3SS, encoded in the Locus of Enterocyte Effacement (LEE) pathogenicity island (Elliott et al., 1998; Ritchie and Waldor, 2005). The T3SS enables EHEC to translocate effector proteins directly from the bacterial cytoplasm into the host cell. Some of the translocated effectors, including Tir and EspFu, subvert host cytoskeletal processes, enabling EHEC to ‘intimately adhere’ to the host intestinal epithelium and form ‘attaching/effacing’ (A/E) lesions (Kenny et al., 1997; Frankel et al., 1998; Hartland et al., 1999; Campellone et al., 2004). The LEE island consists of 41 genes, subdivided into 5 major operons (LEE1-LEE5), that encode the structural components and some of the regulators, effectors, and chaperones of the T3SS (Elliott et al., 1998; Mellies et al., 1999). Additional effector proteins are encoded in prophages outside of the LEE (Gruenheid et al., 2004; Tobe et al., 2006).
The EHEC T3SS, like T3SSs found in other pathogens, is thought to be regulated by contact with host cells; however, the factors and mechanisms that mediate contact-dependence of the EHEC T3SS have not been determined (Roe et al., 2004; Brutinel and Yahr, 2008). The LEE genes are regulated through a complicated and only partially understood regulatory cascade. LEE1 is the first operon to be transcribed, and a number of transcriptional regulators, including QseA, Per, IHF, H-NS, GadX, hha, pch, EtrA and EivF, have been implicated in its activation in response to various environmental stimuli (Friedberg et al., 1999; Mellies et al., 1999; Bustamante et al., 2001; Shin et al., 2001; Yona-Nadler et al., 2003; Iyoda and Watanabe, 2004; Sharma and Zuerner, 2004; Zhang et al., 2004; Russell et al., 2007). LEE1 encodes the LEE-encoded regulator (Ler), the main transcriptional activator of the island, which activates transcription of LEE2-LEE5 by antagonizing the repressor H-NS (Elliott et al., 2000; Bustamante et al., 2001). In addition, Ler functions as a repressor at LEE1, creating a negative feedback loop (Berdichevsky et al., 2005). Ler also regulates grlRA, LEE-encoded genes that lie outside of the principal 5 LEE operons, which encode a repressor (GrlR) and activator (GrlA) respectively. GrlA binds the LEE1 promoter and activates transcription of LEE1, while GrlR antagonizes this process by sequestering GrlA and preventing its interaction with DNA (Creasey et al., 2003; Iyoda et al., 2006; Russell et al., 2007; Huang and Syu, 2008). Together, these regulators modulate LEE gene expression by controlling the levels of Ler. Hfq-independent post transcriptional mechanisms of regulation of LEE gene expression have also been reported (Roe et al., 2003; Tsai et al., 2006; Campellone et al., 2007; Lodato and Kaper, 2009).
In this study, we investigated the role of Hfq in the regulation of EHEC gene expression. Comparison of the transcriptomes of a wt and Δhfq EHEC strain revealed that this RNA chaperone negatively regulates the expression of all 41 LEE genes and of approximately half of confirmed non-LEE encoded T3 effectors. Moreover, we observed elevated secretion of LEE-encoded T3 effector proteins by the hfq mutant. The increases in LEE mRNA and in effector secretion result from increased transcription of LEE operons and are dependent on Ler. The increase in mRNA for non-LEE encoded effectors is partially dependent upon Ler; however, several effectors demonstrate Ler-independent regulation as well, hinting at a complex pathway likely involving multiple sRNAs and targets. Hfq and sRNA(s) likely act directly upon ler mRNA, as translation of this transcript is increased in the absence of Hfq. Finally, we present evidence that negative regulation of T3S by Hfq is not limited to EHEC, but is common to multiple strains of pathogenic E. coli as well as other bacterial species, including V. cholerae and P. aeruginosa.
Results
LEE transcript levels are increased in EDL933Δhfq
Pioneering studies in nonpathogenic E. coli K12 have revealed that Hfq/sRNA-mediated regulation influences diverse cellular processes. However, little is known about Hfq/sRNA-mediated regulation in pathogenic E. coli such as EHEC. To begin to assess the role of Hfq/sRNA-mediated gene regulation in EHEC O157:H7, we compared the transcriptomes of the EHEC clinical isolate EDL933 and an isogenic mutant with an in frame deletion of hfq, using Affymetrix microarrays. These analyses revealed that 316/5349 genes, or 5.9% of the genome, had altered transcript abundance in the hfq mutant, and hence are likely to be regulated either directly or indirectly by an sRNA. Of these genes, 173 showed increased expression (ratio ≥2.0) and 143 showed decreased expression (ratio ≤0.5). A similar percentage of genes were found to be regulated by Hfq/sRNAs in experiments conducted with hfq mutants of E. coli K12 and Vibrio cholerae (Ding et al., 2004; Guisbert et al., 2007; Sittka et al., 2008).
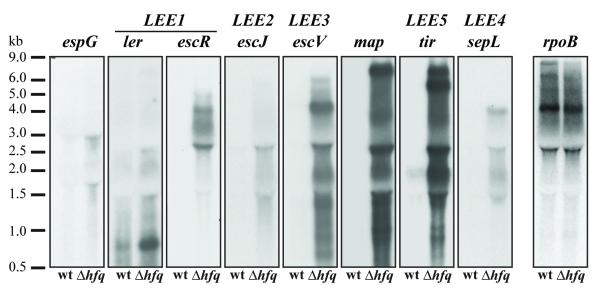
Notably, the genes with increased transcript abundance included all 41 genes encoded within the Locus of Enterocyte Effacement (LEE), a pathogenicity island critical for the virulence of EHEC O157:H7 (Table 1). Northern analyses confirmed that transcripts from each of the 5 major operons within LEE (which contain the majority of genes within this locus) were more abundant in the hfq mutant than in RNA from wt cells. As suggested by the microarray analyses, RNA from late log phase cultures of wt EDL933 grown in LB contained low, at times undetectable, levels of transcripts from ler and escR (LEE1), escJ (LEE2), escV (LEE3), sepL (LEE4), and tir (LEE5). These observations are consistent with previous ones indicating that LB media does not induce production of/secretion from the T3SS encoded within LEE. In contrast, transcripts from these genes were readily detectable in RNA from the hfq mutant (Figure 1). Additionally, transcripts for the LEE-encoded effectors espG and map, which are not transcribed within LEE1-5, were more abundant in RNA from the hfq mutant than in wt EDL933 RNA (Figure 1). There was no appreciable difference in the abundance of rpoB transcripts in the hfq mutant (Figure 1), demonstrating that the absence of hfq does not lead to indiscriminate elevation of mRNAs. Together, the Northern and microarray analyses suggest that Hfq, potentially with the aid of sRNAs, enables coordinated repression of the virulence genes encoded within the EHEC LEE.
Table 1.
All LEE-encoded transcripts are increased in EDL933Δhfq.
| Gene ID | Gene Name | hfq/wt ratio | Gene ID | Gene Name | hfq/wt ratio |
|---|---|---|---|---|---|
| Z5100 e | espF | 11.5 | Z5122 b | sepZ | 3.5 |
| Z5102 e | 15.0 | Z5123 b | 6.2 | ||
| Z5103 e | escF | 15.1 | Z5124 b | escJ | 7.8 |
| Z5104 e | 8.1 | Z5125 b | sepD | 6.3 | |
| Z5105 e | espB | 4.2 | Z5126 b | escC | 6.7 |
| Z5106 e | espD | 5.5 | Z5127 b | cesD | 9.0 |
| Z5107 e | espA | 4.2 | Z5128 | grlA | 3.7 |
| Z5108 e | sepL | 7.8 | Z5129 | grlR | 3.1 |
| Z5109 | escD | 7.4 | Z5131 | 8.1 | |
| Z5110 d | eae | 11.5 | Z5132 a | escU | 5.3 |
| Z5111 d | cesT | 16.6 | Z5133 a | escT | 5.8 |
| Z5112 d | tir | 6.2 | Z5134 a | escS | 4.1 |
| Z5113 | map | 8.0 | Z5135 a | escR | 4.9 |
| Z5114 | cesF | 7.2 | Z5136 a | 4.2 | |
| Z5115 c | espH | 2.0 | Z5137 a | 2.8 | |
| Z5116 c | sepQ | 4.9 | Z5138 a | 2.4 | |
| Z5117 c | 4.9 | Z5139 a | 2.2 | ||
| Z5118 c | 4.9 | Z5140 a | ler | 2.2 | |
| Z5119 c | escN | 4.1 | Z5142 | espG | 6.8 |
| Z5120 c | escV | 8.4 | Z5143 | 16.3 | |
| Z5121 c | mpc | 16.5 |
LEE1
LEE2
LEE3
LEE5
LEE4
Figure 1. Northern blots of LEE transcripts in wt and Δhfq EDL933.
Transcript levels in wt and Δhfq EDL933 were assessed by Northern blot using in vitro transcribed probes to the genes listed above the blot. Multiple bands likely reflect processing and/or partial degradation of transcripts. The RNA ladder is marked on the left.
EDL933 hfq mutants acquire suppressor mutations
As has been reported for hfq mutants of other bacterial species (Tsui et al., 1994; Sonnleitner et al., 2003; Ding et al., 2004), deletion of hfq impaired the growth of EHEC O157:H7. When grown in rich media such as LB, EDL933Δhfq reached a lower density than wt EDL933, and exhibited a longer growth lag when emerging from stationary phase (Figure 2A and data not shown). This growth defect was exacerbated when the bacteria were cultured in minimal media (M9) or DMEM (Figure 2A). EDL933Δhfq also formed small colonies on LB agar; however, we observed that these small colonies frequently gave rise to larger, approximately wt-sized colonies, probably due to acquisition of suppressor mutations (Figure 2B). To determine whether all putative suppressors were equivalent, we performed Northern blot analyses on RNA isolated from small and large colonies of several independently-derived hfq mutants. These experiments revealed that transcript levels for map (Fig. 2C and data not shown) and other genes within the LEE (data not shown) were consistently elevated in RNA from small colonies of hfq mutants. In contrast, these transcripts were only more abundant than wt levels in RNA from a subset of the large hfq colonies. These data suggest that the increased LEE gene expression observed in the hfq mutant may impede the growth of this strain, and that processes which downregulate LEE transcripts can help to restore normal growth. However, they also suggest that LEE-independent suppressor mechanisms can arise, and thus that overexpression of LEE is not the only factor impairing growth of the hfq mutant. Both classes of suppressors shed light on the processes underlying LEE gene upregulation, as described below.
Figure 2. Influence of deletion of hfq and ler on growth and expression of map in EDL933.
A) Growth kinetics of wt (black), Δhfq (green), Δler (orange), and Δler Δhfq (blue) EDL933 in LB, M9, and DMEM. The growth rates of the indicated strains were measured in a plate reader. A representative experiment is shown and the curves are averages of quadruplicate wells. B) Colony sizes of wt EDL933 and the indicated deletion mutants on LB agar are shown. Black arrows denote “large” colonies and orange arrows denote “small” colonies. C) Transcript levels of map in left panel: EDL933, EDL933Δhfq large #1, EDL933Δhfq small #1, EDL933Δhfq large #2, EDL933Δhfq small #2 and in right panel: EDL933 wt, ES669, ES669/pUC19, ES669/pES153, ES450, ES450/pUC19, ES450/pES153 were detected by Northern blot using an in vitro transcribed probe. The RNA ladder is marked on the left.
As we wished to explore the mechanism by which deletion of hfq increased LEE transcripts, and were concerned that acquisition of diverse suppressor mutations could complicate our experiments, we explored the utility of one of the large colonies of EDL933Δhfq that maintained elevated LEE transcript levels as the basis for additional studies. This strain (ES450) is presumed to contain a suppressor mutation and hence is not fully isogenic (other than hfq) with EDL933. Consequently, we generated a new “wt” control strain (ES669) by replacing the mutant hfq in ES450 with the wt gene. Northern analyses indicated that LEE transcripts were equally abundant in ES669 and EDL933 (Figure 2C and data not shown), suggesting that the mutation within hfq, rather than the unknown suppressor mutation, accounts for elevation of LEE transcripts in ES450. Furthermore, complementation analyses confirmed that the absence of hfq was solely responsible for the increase in LEE transcripts in ES450. Introduction of a plasmid borne copy of hfq (pES153, a pUC19 derivative containing hfq with its native promoter) into ES450 reduced map transcripts to wild-type levels, while introduction of pUC19 into ES450 had no effect (Fig. 2C). Neither plasmid altered map transcript levels when introduced into ES669. Taken together, these results demonstrate that the isogenic pair of ES669 and ES450 behaves like wt and hfq EDL933 with respect to over-expression of LEE genes, and that this phenotype is due to the absence of hfq. In order to minimize selective pressure for the development of suppressor mutations in strains lacking hfq, which might mask the LEE-related phenotype of hfq mutants, ES450, ES699, and derivatives were used for the majority of additional experiments.
LEE expression is increased in EDL933Δhfq in a ler-dependent manner
Previous analyses of LEE gene expression have shown that transcription of LEE2-LEE5 is activated by the LEE-encoded regulator (Ler), whose transcript we have found to be elevated in hfq mutants. Hence, it seemed likely that the increase in LEE transcripts in the hfq mutant was due to overexpression of ler. To explore this possibility, we first confirmed that deletion of hfq resulted in increased amounts of Ler protein as well as RNA. We replaced the endogenous ler locus of ES699 with ler-his6 and then assessed levels of Ler-His6 in cell pellets from overnight cultures of this strain and an hfq derivative. Ler-His6 was undetectable in the wt background, but could readily be seen in the hfq mutant, indicating that the increased ler transcripts seen in hfq strains result in increased amounts of Ler protein (Figure 3A).
Figure 3. Influence of Ler on expression of LEE operons in EDL933Δhfq.
A) Western blot analysis of pellet fractions of ES669 ler-his6 and ES669 ler-his6 Δhfq. Ler-His6 was detected using an α-His antibody, H-NS and RNAP were detected by using α-H-NS and α-RNAP antibodies, respectively. B) Transcript levels of map in ES669, ES669Δler, ES450, and E669Δler Δhfq were detected by Northern blot using an in vitro transcribed probe. The RNA ladder is marked on the left. C) Transcriptional fusions of the map and LEE5 promoters to lacZ were integrated into the chromosome of ES669, ES669Δler, ES450, and ES669Δler Δhfq and β-galactosidase activities were measured in triplicate overnight cultures. The data presented are averages + SD from 3 independent experiments.
To assess the extent to which overproduction of Ler accounts for upregulation of LEE-encoded genes, we compared expression of these genes in ES699 and isogenic ler, hfq, and ler hfq strains. Deletion of hfq had dramatically different consequences in the wt and ler backgrounds. Northern blot assays revealed that the marked increase in transcripts for map (Figure 3B) and representative genes of LEE2-LEE5 (data not shown) seen when hfq is deleted from a wt strain were mostly absent when hfq was deleted from a ler mutant, strongly suggesting that Hfq’s effect on LEE gene expression is largely dependent upon Ler upregulation.
Transcription reporter fusions were generated for several LEE-encoded genes to confirm that the Ler-dependent increase in transcripts was due to Ler’s previously described role as a transcription activator. Chromosomal lacZ fusions to the map and LEE5 promoters were generated in ES669 as well as its ler, hfq, and ler hfq derivatives, and their activity was monitored in overnight cultures grown in LB. Both fusions demonstrated increased activity in the hfq mutant as anticipated. This increase was dependent on the presence of ler, as transcription in the ler hfq double mutant was significantly lower than in the hfq mutant, and comparable to that seen in a strain lacking ler alone (Figure 3C). The variability in activity displayed by the fusions in all backgrounds is in line with previous reports of heterogeneous expression of LEE genes at the single cell level (Roe et al., 2003; Roe et al., 2004). Taken together, these results suggest that the absence of Hfq leads to increased levels of Ler, which subsequently leads to enhanced transcription from LEE promoters and increased levels of LEE gene transcripts.
Type III secretion is increased in EDL933Δhfq
Next, we explored whether deletion of hfq led to increased secretion of LEE-encoded T3 effector proteins as well as an increase in LEE transcripts. We first used SDS-PAGE followed by silver staining to examine supernatant fractions from cells cultured overnight in LB, a condition in which T3S has been reported to be off in EHEC. We observed a significant increase in the number of proteins in the supernatant fraction of the hfq mutant (ES450) as compared to the wt (ES669) sample (Figure 4A, lanes 4 and 1). While this paper was in preparation similar results were reported by Hirakawa, et al. (Hirakawa et al., 2009). Protein secretion by our hfq strain was reduced to wild-type levels by introduction of the Hfq-encoding plasmid pES153 and was unaffected by introduction of pUC19, the empty vector control. Neither plasmid had an effect on secretion by ES669. Disruption of ler and of escN (which encodes an essential structural component of the EHEC T3SS) prevented much of the increase in protein secretion observed for the hfq strain. Together, these data suggest that several of the proteins secreted at high levels by the hfq mutant are ler-regulated T3-secreted effectors (Figure 4B, lane 4, bands marked by black arrows). Supernatants from the hfq mutant also had increased levels of an as yet unidentified protein whose expression and/or secretion was dependent on ler but not dependent on escN (Figure 4B, lanes 4 and 6, band labeled with a white arrow). Thus, Hfq appears to influence protein secretion both via the LEE-encoded T3SS and by another secretion system.
Figure 4. Effect of deletion of hfq on Type III secretion.
Supernatant (sup) only (A,B) or supernatant and pellet (C) fractions of various derivatives of ES669 were analyzed by SDS-PAGE followed by silver stain (A, B) or western blot (C). The molecular weight marker is shown on the left. The black arrows denote the most prominent examples of ler/escN-dependent secreted proteins and the white arrow denotes a ler-dependent escN-independent secreted protein. C) Strains are derivatives of ES669 map-his6 (top 2 panels) or ES669 tir-his6 (lower 2 panels) with additional genetic changes as noted in the figure. The upper panel of each set was probed with α-His antisera and the lower panel was probed with α-RNAP antibody.
Since we could not identify the proteins observed by silver stain definitively, we examined secretion of two of the known LEE-encoded T3 effectors — Map and Tir - using Western blots. We generated chromosomal C-terminal His6 fusions for both genes and then compared production and secretion of these proteins in the wt strain and strains lacking escN, ler, hfq, or combinations of these genes. Map-His6 and Tir-His6 were found in the supernatant fractions of the hfq mutant only (Figure 4C, sample 4). As expected, no secretion was detected from the wt strain, and deletion of either ler or escN in the hfq mutant background abolished secretion of the effectors. Map-His6 and Tir-His6 were present in the pellet fraction of the hfq and escN hfq mutants, although Tir-His6 was partially degraded in the hfq escN mutant and migrated at a smaller size (data not shown). Neither protein was detected in the pellet fraction from the ler hfq mutant. Thus, in the absence of Hfq, formation of a functional T3S apparatus and secretion of T3 effectors into the surrounding milieu is no longer regulated by environmental stimuli such as contact with a eukaryotic cell and occurs under in vitro “off” conditions. Furthermore, production of the effectors by the hfq mutant is dependent on Ler, while their secretion is dependent on a functional secretion apparatus.
Over-expression of T3SS is detrimental to EHEC growth
Additional support for Ler’s role in activation was provided by analysis of some of the large hfq colonies described above that did not maintain elevated expression of map and other LEE genes. Sequencing of the ler promoter and coding sequences in these strains revealed that one mutant contained a 24 bp duplication in the N-terminal portion of ler. Identification of this mutation suggested that disruption of ler might also function as a suppressor of the “small colony” phenotype of the hfq mutant. This hypothesis was confirmed by creating an in frame deletion within ler in wt EDL933 prior to introduction of the hfq deletion. EDL933ΔlerΔhfq grew more rapidly than the hfq mutant, although not as rapidly as wt, and formed large colonies only (Figure 2A and 2B). Together, these data suggest that overproduction of the T3SS encoded within LEE imposes a significant burden upon EHEC that is detrimental to growth, providing a biological rationale for the repression of these genes observed within wt EHEC under standard laboratory growth conditions.
Transcripts of non-LEE dependent effectors are increased in EDL933Δhfq
As described in several other pathogens encoding T3SSs, EHEC strains contain loci encoding T3 effector proteins that are not genetically linked to the locus encoding the secretion apparatus (i.e. to the LEE island) (Garmendia et al., 2005; Tobe et al., 2006; Dean and Kenny, 2009). Many of these non-LEE-encoded effectors are encoded within prophages and different strains appear to encode varying numbers and sets of effectors. Using the list of known and predicted effectors derived by Tobe et al. (Tobe et al., 2006), we examined our microarray data to determine whether transcripts for any non-LEE—encoded effectors were increased. Among the 29 confirmed non-LEE encoded effectors, transcripts of 16 (55%) were increased at least 2-fold (Table 2). In addition, homologs of 3 predicted but unconfirmed effector genes also had significantly increased transcript levels. Northern blot assays of a subset of the 16 apparently upregulated genes confirmed that all had increased transcript abundance in the hfq mutant. Increased transcript levels were detected for espFu, espJ, espK, nleA, and nleG2-2 (Figure 5A). Together, these data suggest that Hfq regulation of EHEC T3S extends outside of the LEE pathogenicity island to encompass many other effectors encoded throughout the genome.
Table 2.
Transcripts of many non-LEE encoded effectors are increased in EDL933Δhfq
| EDL933 Gene ID | Gene Name | hfq/wt ratio | SAKAI Gene ID |
|---|---|---|---|
|
| |||
| Z0065 | 0.8 | ECs0061 | |
|
| |||
| Z0986 | nleC | 1.1 | ECs0847 |
| Z0989 | 1.2 | ECs0848 | |
| Z0990 | nleD | 0.9 | ECs0850 |
|
| |||
| Z1019 | 0.8 | ECs0876 | |
|
| |||
| Z1822 | 3.2 | ECs1560 | |
| Z1823/1824 b | 5.1/3.8 | ECs1561 | |
| Z1828 c | 2.2 | ECs1567 | |
| Z1829 | espK | 1.6 | ECs1568 |
|
| |||
| Z2077 | 1.0 | ECs2226 | |
|
| |||
| Z2339/Z2149 d | nleG2-2 | 2.3 | ECs1994/2156 |
| Z2338/Z2150 d | 3.3 | ECs1995/2155 | |
| Z2337/Z2151 d | 2.7 | ECs1996/2154 | |
|
| |||
| Z2565 | 1.0 | ECs1825 | |
|
| |||
| Z3071 | espJ | 2.4 | ECs2714 |
| Z3072 | espFu/tccP a | 2.4 | ECs2715 |
|
| |||
| Z3918 | 1.0 | ECs3485 | |
| Z3919 | 1.2 | ECs3486 | |
| Z3920 | espW | 1.4 | ECs3487 |
|
| |||
| Z4326 | 2.3 | ECs3855 | |
| Z4328 | nleB | 2.0 | ECs3857 |
| Z4329 | nleE | 2.0 | ECs3858 |
|
| |||
| Z5211 | 1.3 | ECs4653 | |
|
| |||
| Z6010 | nleG | 1.1 | ECs1824 |
|
| |||
| Z6025 | 3.2 | ECs1810/1811 | |
| Z6024 | nleA | 12.0 | ECs1812 |
| Z6021 | 2.4 | ECs1814 | |
| Z6020 | nleF | 2.3 | ECs1815 |
|
| |||
|
| |||
| Z2076 e | 3.1 | ECs2228 | |
|
| |||
| Z2075 e | 3.1 | ECs2229 | |
|
| |||
| Z5636 e | 2.4 | ECs5021 | |
|
| |||
Two accepted gene names
The sequence encoding ECs1561 in SAKAI corresponds to sequence in EDL933 annotated to contain two genes: Z1823 and Z1824. A single basepair insertion within the codon for amino acid 190 creates a frameshift in EDL933 creating a stop codon after amino acid 194.
There is no annotated gene in the intergenic region between Z1827 and Z1829, however the intergenic sequence is identical to the SAKAI sequence encoding ECs1567.
Both SAKAI and EDL933 genomes contain two nearly identical (>95% identity) copies of these 3 genes. The microarray contains one set of probes for these genes. Transcripts from either operon would anneal to the microarray probes.
Hypothetical effectors not confirmed experimentally.
Figure 5. Northern blots of several non-LEE encoded T3S effectors.
Transcript levels in wt and Δhfq EDL933 in A) and in ES669, ES669Δler, ES450, and ES669ΔhfqΔler in B). In vitro transcribed probes to the genes listed above the blots were used in each case. An RNA ladder is marked on the left of each panel.
Next, we examined whether the increased abundance of non-LEE effector transcripts is dependent on ler. Similar to what we observed for LEE-encoded genes, the increased abundance of nleA transcripts in the hfq mutant required ler. Northern analyses revealed that ES699 lacking both ler and hfq contained the same level of nleA transcripts as did ES699 lacking ler alone (Figure 5B). This result is consistent with published reports that nleA is regulated by Ler (Roe et al., 2007). However, it appears that increased Ler is not the only factor that promotes expression in the hfq mutant of several non-LEE effector loci including espFu, espJ, espK, and nleG2-2. ES699 lacking both ler and hfq contained fewer transcripts for these genes than did ES699Δhfq; however, the abundance of these transcripts was still markedly higher than in ES699 Δler (Figure 5B and data not shown). Together these data suggest that the hfq mutant’s increased expression of some non-LEE encoded effectors, such as nleA, can be attributed solely to increased production of Ler, while regulation of many other non-LEE-encoded effectors is mediated both by Ler and by additional factors. These results hint at a complex regulatory network that may involve multiple Hfq targets and/or sRNAs.
Translation and transcription of ler are increased in EDL933Δhfq
Production of Ler is clearly enhanced in EHEC strains lacking hfq, with dramatic consequences for expression of the genes that it regulates; however, it cannot be determined from the analyses presented thus far whether ler is a direct target of Hfq and Hfq-dependent sRNAs. Hfq/sRNA-mediated regulation of gene expression is typically exerted at the post-transcriptional level, relying on mechanisms such as altered transcript stability or availability of ribosome binding sites. Altered transcription in hfq mutants generally reflects indirect regulation by Hfq, mediated by a direct target that lies upstream. However, in analyses of ler expression, the complex autoregulatory scheme that controls this gene’s transcription must also be taken into account.
To determine the mechanism(s) underlying ler upregulation in the hfq mutant, we first set out to examine whether translation of the ler transcript is increased in the hfq mutant. In particular, we explored the effect on translation of sequences extending from ler’s transcriptional start site, which we mapped by 5′RACE (data not shown) to the distal promoter previously identified by Sperandio et al., (Sperandio et al., 2002), to the translational start (Russell et al., 2007), since Hfq-dependent sRNAs often interact with sequences in the 5′ untranslated region (UTR) of target transcripts (Masse et al., 2003; Morita et al., 2006; Darfeuille et al., 2007). We adapted an approach first described by Urban and Vogel (Urban and Vogel, 2007), and fused this 169 bp sequence to a reporter gene (lacZ) and a non-native promoter (PLtetO) to generate pES154. A control plasmid, pES147, which contains a 77 bp multiple cloning site and a heterologous ribosome binding site instead of the 5′UTR sequences, was also generated. β-galactosidase activity generated from these plasmids was then assayed in strains ES669 and ES450. We detected 2.5-fold more β-galactosidase from pES154 in the hfq mutant than in the wt strain (Figure 6B). In contrast, β-galactosidase from pES147 was not elevated in the hfq mutant relative to the wt strain, suggesting that a non-specific increase in activity from PLtetO or lacZ does not account for the result with pES154. These data suggest that Hfq dampens translation of ler, either directly or with the help of an sRNA, and thus that at least some of the increased Ler observed in the hfq mutant is due to the absence of this inhibition. Since our results were obtained using a plasmid-borne reporter and a very potent exogenous promoter, which may yield more transcripts than the wt cell can typically repress, we suspect that the magnitude of Hfq’s repression may be greater on native ler transcript levels.
Figure 6. Effect of deletion of hfq on translation and transcription of ler.
A) Maps of pES147 and pES154. ‘Random sequence’ in pES147 refers to 77 bp of vector sequence containing a ribosomal binding site properly spaced in front of lacZ. pES154 is a translational fusion of the 5′UTR of ler to lacZ. B) Translation of lacZ from pES154 or pES147 was assessed in ES669 and ES450 by β-galactosidase assays of triplicate overnight cultures. The data is expressed as % of wt activity for each plasmid + SD. C) Transcriptional activity of Pler as measured by β-galactosidase activity. pES85 (Pler::lacZ in pVIK112) was integrated into ES669 and ES450 and β-galactosidase activity was measured in overnight cultures. The data presented are averages of 3 experiments + SD. D) Transcript levels of grlA in ES669, ES669Δler, ES450, and E669Δler Δhfq were detected by Northern blot using an in vitro transcribed probe. The RNA ladder is marked on the left.
We also explored whether transcriptional activity of a chromosomal ler::lacZ fusion is altered in the hfq mutant. This reporter was integrated at the endogenous ler locus of ES669 and ES450 but nonetheless preserved a wt copy of ler and its promoter. We found that β-galactosidase activity from the reporter was increased 6-fold in the hfq mutant relative to the wt strain (Figure 6C), suggesting that Hfq influences transcription as well as translation of ler. Such an effect could be due to increased production of the ler transcriptional activator GrlA, whose expression is activated by Ler and for which we had detected elevated transcript abundance in the hfq mutant (Table 1). Unlike results for transcripts for other genes in LEE, northern blot assays revealed that the increase in grlA transcripts in the hfq mutant is due largely, but not entirely, to increased Ler production; deletion of hfq from a ler mutant still resulted in a slight increase in grlA transcripts (Figure 6D). Additionally, while this paper was in preparation, Hansen and Kaper reported that the grlRA transcript is itself a target for Hfq and presumably a yet to be determined sRNA (Hansen and Kaper, 2009). Collectively, these data suggest that the abundant ler transcripts detected in the hfq mutant are a consequence both of indirect Ler autoactivation (via GrlA) and of a second, ler-independent pathway, Thus, there appear to be multiple processes, both transcriptional and translational, through which Hfq acts to regulate expression of Ler and ultimately of T3S in EDL933.
Since many transcriptional regulators in addition to GrlA have been reported to influence ler expression, we examined whether expression of these factors is also altered in the hfq mutant. We examined protein levels of the LEE repressor H-NS in pellet samples made from wt and hfq mutant EHEC, reasoning that the increase in LEE and/or ler transcripts could be mediated by loss of H-NS. However, equal amounts of H-NS were observed by Western blot in wt and hfq mutant samples, suggesting that reduction of H-NS does not contribute to elevated ler expression in the Δhfq mutant (Figure 3A). Additionally, our microarray data suggests that altered expression of QseA, IHF, hha, EtrA, EivF, and pch does not account for the changes in ler expression, since the abundance of their transcripts was not altered by deletion of hfq (data not shown). These data suggests ler and grlA may be the primary targets upon which Hfq acts to regulate expression of EDL933′s T3SS.
Hfq negatively regulates T3SS in multiple pathogens
Since there is heterogeneity among O157:H7 EHEC strains (Ogura et al., 2007) , we assessed whether negative regulation of T3 secretion by Hfq is restricted to EDL933. We made in-frame hfq deletions in two additional O157:H7 EHEC strains, 905 and 86-24, and in 5244/00, an O145:H- EHEC strain. Northern assays revealed that these 3 wt/mutant pairs showed patterns of gene expression comparable to those seen in the EDL933 background (Figure 7). Expression of LEE-encoded genes was low in the wt strains and markedly elevated in the hfq mutants, suggesting that Hfq has similar regulatory effects in most or all EHEC strains. Since EPEC, like EHEC also contains a T3SS encoded by the LEE pathogenicity island (Elliott et al., 1998), we compared expression of LEE genes in wt and hfq 2348/69, an O127:H6 serotype EPEC strain. The 2348/69Δhfq mutant had much higher levels of LEE transcripts than the wt strain (Figure 7), suggesting that Hfq negatively regulate T3S in EPEC as well as EHEC.
Figure 7. Northern blots of transcripts of genes encoded in T3SSs in multiple pathogens.
Transcript levels in wt and Δhfq derivatives of the strains listed were assessed by Northern blot using in vitro transcribed probes to the genes listed above the blots. An RNA ladder is marked on the left.
Finally, negative regulation of T3SS by Hfq is not limited to pathogenic E. coli that harbor the LEE pathogenicity island. A previously published report showed that an hfq mutant of Vibrio parahaemolyticus expresses higher levels of several T3S genes compared to wt (Nakano et al., 2008). We tested whether this theme extends to two additional gram negative pathogens, AM19226, a non-O1/non-O139 Vibrio cholerae strain, (Dziejman et al., 2005) and Pseudomonas aeruginosa strain PAO1, which both harbor pathogenicity islands that encode T3SSs. Northern blots of RNA from wt and hfq mutants of AM19226 and PAO1 revealed increased transcript abundance for both T3 effector and structural genes in the respective hfq mutants (Figure 7). Together, these observations suggest that negative regulation of T3S by Hfq is a conserved mechanism among diverse gram-negative pathogens.
Discussion
We have found that Hfq acts a global regulator of gene expression in EHEC, directly or indirectly influencing transcript abundance of at least 5.9% of EHEC genes. The altered expression profile that results from hfq deletion severely compromised EHEC growth and prompted emergence of diverse suppressor mutations. Hfq regulates numerous genes that are shared by E. coli K12 and EHEC; however, the absence of this RNA chaperone also altered the expression of many EHEC-specific genes. In particular, we found that levels of all transcripts for the EHEC T3SS encoded in the LEE pathogenicity island and many transcripts for prophage-encoded T3 effector proteins were elevated in the hfq mutant. The increase in transcript levels was correlated with release of high quantities of T3SS effectors into the supernatant in a T3S-dependent manner, under conditions where T3S is not ordinarily observed. Elevated LEE transcripts in the hfq mutant were a consequence of increased Ler production, whereas the abundance of many non-LEE-encoded effectors was only partially dependent on this LEE-encoded transcriptional activator. Translation as well as transcription of ler was increased in the hfq mutant, suggesting that Ler may be a direct target of Hfq and sRNA-mediated regulation. Finally, we found that Hfq contributes to the negative regulation of T3SSs in several other pathogens, suggesting that Hfq, potentially along with species-specific sRNAs, underlies a common means to prevent unfettered expression of T3SS’s.
Our EHEC hfq deletion mutant had a severe growth defect, reflected in its formation of small colonies. These frequently gave rise to larger colonies, prompting us to explore the genetics of mutations that enhance the growth of an hfq mutant. Our studies suggest that there are two classes of suppressors, one that reduces LEE transcript levels and one that does not. Thus, restoration of wt expression of the numerous LEE-encoded factors typically overproduced by the hfq mutant is one of several means for relieving the significant burden imposed by the multifaceted disruption of gene regulation in this genetic background. We utilized a member of the other class of suppressor mutants for our studies, although the identity of the suppressor is not yet known. Future studies will explore the nature of such apparently LEE-independent suppressors, which should yield greater understanding of the physiology of hfq mutants. Recently, deep sequencing technology has enabled identification of analogous suppressor mutations that relieve the growth defect of a strain lacking rpoE (Davis and Waldor, 2009).
The phenotype of an hfq mutant with a suppressor mutation in ler strongly suggested that Ler is a key mediator of LEE overexpression by the hfq mutant. Additional genetic studies, which took advantage of the non-ler-linked suppressor mutation, confirmed that the hfq mutant’s marked elevation in LEE transcripts depends on ler, which is also overexpressed. At least two mechanisms underlie this increase in Ler. First, there is increased translation of the ler message in the hfq mutant, suggesting that this message is a direct target of Hfq, either acting alone or in conjunction with an sRNA. Interestingly, Hfq controls Salmonella pathogenicity island 1 (SPI-1) gene expression in a similar fashion - by targeting the hilD mRNA, which encodes a key SPI-1 encoded transcription factor (Sittka et al., 2008). However, while the effect of Hfq on ler expression is negative, the effect on hilD is positive. We also found that transcription of ler is increased in the hfq mutant. This may reflect Ler’s activation of one of its own activators, GrlA, since our Northern analyses indicate that grlA transcripts increase in the hfq mutant in a largely Ler-dependent manner. Thus, GrlA may contribute to a positive feedback loop that enhances transcription of ler. In addition, while our paper was in preparation, Hansen and Kaper reported that grlA transcripts are more stable in cells lacking Hfq, and therefore proposed that grlA is a direct target of Hfq (Hansen and Kaper, 2009). Such direct regulation of grlA by Hfq is consistent with our observation that some of the increase in grlA transcripts in the hfq mutant is independent of ler. Thus, it appears that there are multiple mechanisms by which Hfq regulates expression of Ler and other LEE-encoded genes. Ler translation is enhanced, suggesting a direct role for Hfq in ler regulation; however, ler transcription is also elevated, potentially due to increased production of grlA, which may itself be subject to both direct and indirect consequences of Hfq’s absence. Future studies will focus on elucidating the mechanistic details of these regulatory processes.
Our microarray analyses revealed that Hfq coordinates expression of the LEE-encoded T3S apparatus with expression of both LEE- and non-LEE- encoded effectors. In the EHECΔhfq strain, transcripts for more than half of non-LEE-encoded effectors were elevated along with all LEE transcripts. Culture conditions which induce expression of some LEE operons were previously reported to also induce expression of several non-LEE encoded effector genes (Deng et al., 2005; Roe et al., 2007), but the factors/mechanisms accounting for this observation have not been revealed. Coordinate expression of certain non-LEE encoded effectors, such as NleA (Figure 5B and (Roe et al., 2007)) with LEE-encoded effectors appears to be largely explained by their common dependence on Ler for transcription. However, the elevation of many non-LEE effector transcripts in EHECΔhfq was at least partially independent of ler (Figure 5B and data not shown), indicating that another factor besides Ler is involved in synchronizing effector expression. Identification of this Hfq-dependent factor is the subject of on-going studies.
A common feature among T3SSs encoded by bacterial pathogens is that they are silent under many growth conditions, yet effector translocation and gene expression can be activated by bacterial contact with eukaryotic cells. For several systems, including P. aeruginosa and Yersinia sp., intricate pathways explaining contact dependence have been proposed (reviewed in (Brutinel and Yahr, 2008)); for other pathogens, these pathways remain to be elucidated. Our finding that Hfq negatively regulates T3SSs in a variety of pathogens suggests that similar regulatory processes may have developed in these organisms, despite the diversity of the T3SS effectors, chaperones, and regulators that they produce. Hfq/sRNA-mediated regulation of T3S could provide an effective mechanism to tightly control contact dependence, since sRNA regulation is thought to enable rapid responses to changing environmental conditions yet can also suppress fluctuations in transcription to prevent untimely activation of a given pathway (reviewed in (Levine and Hwa, 2008)). Given the deleterious effects on growth that ensued from inappropriate expression of EHEC’s T3SS, as well as the likely benefits for pathogens in reacting quickly to an altered host milieu, the apparent widespread coupling of Hfq/sRNA control mechanisms to the regulation of T3S appears reasonable.
Experimental Procedures
Bacterial strains and culture conditions
The strains used in this study are listed in Table 3. All E. coli O157:H7 mutants generated in this study are derivatives of a streptomycin-resistant isolate of EDL933 (Perna et al., 2001). Bacteria were routinely cultured in LB at 37°C shaking at 250 r.p.m. unless otherwise stated. A SynergyHT microplate reader (BioTek, Winooski, VT) was used to determine the growth kinetics shown in Figure 2A. In these experiments, bacteria grown overnight on LB agar plates were resuspended in LB to an OD600=0.05, and diluted 1:20 into LB, M9, or DMEM containing appropriate antibiotics. Growth was monitored for 12 hours. Growth curve shown is the average of quadruplicate wells. Antibiotics were used at the following concentrations: streptomycin, 200 or 500 (for conjugations with BW20767)μg/mL; kanamycin, 50 μg/mL; carbenicillin, 50 μg/mL; gentamicin 30 μg/mL (P. aeruginosa) and 10 μg/mL buffered with 1M phosphate buffer (E. coli); irgasan, 10 μg/mL.
Table 3.
Strains used in this study
| Strain | Genotype | Derivation a | Referenceb |
|---|---|---|---|
| EDL933 | E. coli O157:H7 clinical isolate | (Perna et al., 2001) | |
| ES98 | EDL933 SmR | EDL933 | |
| ES138 | EDL933 SmR ΔlacZ | ES98 | |
| ES178 | EDL933 SmR ΔlacZ Δhfq | ES138 | |
| ES180 | EDL933 SmR ΔlacZ Δler | ES138 | |
| ES184 | EDL933 SmR ΔlacZ Δler Δhfq | ES180 | |
| ES450 | EDL933 SmR ΔlacZ Δhfq | ES138 | |
| ES669 | EDL933 SmR ΔlacZ * c | ES450 | |
| ES791 | EDL933 SmR ΔlacZ * ler-his6 | ES669 | |
| ES802 | EDL933 SmR ΔlacZ * ler-his6 Δhfq | ES791 | |
| ES726 | EDL933 SmR ΔlacZ * Δler | ES669 | |
| ES736 | EDL933 SmR ΔlacZ * Δler Δhfq | ES726 | |
| ES795 | EDL933 SmR ΔlacZ * ΔescN | ES669 | |
| ES798 | EDL933 SmR ΔlacZ * ΔescN Δhfq | ES795 | |
| ES681 | EDL933 SmR ΔlacZ * map-his6 | ES669 | |
| ES694 | EDL933 SmR ΔlacZ * map-his6 Δler | ES681 | |
| ES792 | EDL933 SmR ΔlacZ * map-his6 ΔescN | ES681 | |
| ES684 | EDL933 SmR ΔlacZ * map-his6 Δhfq | ES681 | |
| ES715 | EDL933 SmR ΔlacZ * map-his6 Δler Δhfq | ES694 | |
| ES799 | EDL933 SmR ΔlacZ * map-his6 ΔescN Δhfq | ES792 | |
| ES691 | EDL933 SmR ΔlacZ * tir-his6 | ES669 | |
| ES695 | EDL933 SmR ΔlacZ * tir-his6 Δler | ES691 | |
| ES794 | EDL933 SmR ΔlacZ * tir-his6 ΔescN | ES691 | |
| ES723 | EDL933 SmR ΔlacZ * tir-his6 Δhfq | ES691 | |
| ES724 | EDL933 SmR ΔlacZ * tir-his6 Δler Δhfq | ES695 | |
| ES801 | EDL933 SmR ΔlacZ * tir-his6 ΔescN Δhfq | ES794 | |
| 905 | E. coli O157:H7 clinical isolate | (Ritchie et al., 2003) | |
| ES379 | 905 Δhfq | 905 | |
| 86-24 | O157:H7 clinical isolate SmR | (Griffin et al., 1988) | |
| ES380 | 86-24 SmR Δhfq | 86-24 | |
| 5244/00 | O145:H- clinical isolate | (Karch et al., 1999) | |
| ES580 | O145:H- Δhfq | 5244/00 | |
| 2348/69 | SmR | (Levine et al., 1978) | |
| ES596 | O127:H6 2348/69 SmR Δhfq | 2348/69 | |
| MD992 | V. cholerae AM19226 SmR Δendo | (Tam et al., 2007) | |
| ES436 | AM19226 SmR Δendo Δhfq | MD992 | |
| PA01 | P. aeruginosa | (Stover et al., 2000) | |
| ES595 | PA01 Δhfq | PA01 |
Strain listed in the derivation column is the parent strain.
No reference denotes strain was created for this study.
* refers to the unknown suppressor mutation in this strain. This strain was made by replacing the hfq deletion of a large colony of ES450 with a wt copy of hfq by allelic exchange
Strain and plasmid construction
Laboratory stocks of DH5αλpir, SM10λpir, and BW20767 were used for cloning and for introduction of DNA into EDL933 by conjugation. Descriptions of the plasmids generated for this study are listed in Table 4. All inserts in plasmids which were generated by PCR were sequenced prior to use. Derivatives of the allele exchange vector pCVD442-lac (Butler and Camilli, 2004) were used to construct deletion mutants and to introduce His6-tags onto chromosomal genes in EHEC, EPEC, and V. cholerae as described previously (Skorupski and Taylor, 1996). Introduction of mutations into Δhfq strains proved difficult, thus all other mutations were introduced first, followed by deletion of hfq. Chromosomal transcriptional fusions to lacZ were made using pVIK112 derivatives (Kalogeraki and Winans, 1997). Allelic exchange in Pseudomonas aeruginosa was performed used pEXG2 (Rietsch et al., 2005).
Table 4.
Plasmids used in this study
| Plasmid | Description | Referencea |
|---|---|---|
| pCVD442-lac | allele exchange vector | (Butler and Camilli, 2004) |
| pES10 | pCVD442-lac ΔlacZ (deletes codons 359-637) | |
| pES30 | pCVD442-lac Δhfq (deletes codons 1-100) | |
| pES31 | pCVD442-lac Δler (deletes codons 2-123) | |
| pES145 | pCVD442-lac ΔescN (deletes codons 19-440) | |
| pES144 | pCVD442-lac ler-his6 (replaces ler with ler-his6) | |
| pES78 | pCVD442-lac map-his6 (replaces map with map-his6) | |
| pES123 | pCVD442-lac tir-his6 (replaces tir with tir-his6) | |
| pCHfq | pCVD442Δhfq (deletes codons 13-77 in V.c.) | (Ding et al., 2004) |
| pEXG2 | allelic exchange vector for P. aeruginosa. | (Rietsch et al., 2005) |
| pES106 | pEXG2Δhfq (deletes codons 4-82 in P.a.) | |
| pVIK112 | suicide vector for creating chromosomal lacZ fusions | (Kalogeraki and Winans, 1997) |
| pES85 | Pler (−675 - + 289)b cloned into pVI112 | |
| pES88 | Ptir (−558 - + 369)b cloned into pVIK112 | |
| pES94 | Pmap (−669 - +378)b cloned into pVIK112 | |
| pUC19 | high copy vector | (Yanisch-Perron et al., 1985) |
| pES153 | Phfq and hfq (−738 - +322)b cloned into pUC19 | |
| pCB192 | transcriptional fusion vector | (Schneider and Beck, 1986) |
| pES147 | PLtetOc cloned into pCB 192 | |
| pES154 | PLtetOc and ler 5′UTR cloned into pCB192 |
No reference denotes plasmid was created for this study
Relative to ATG
This is a truncated PLtetO and is not repressible by the Tet repressor
RNA analyses
RNA was harvested from early stationary phase cultures (OD600 0.6-0.7) grown in LB. RNA was extracted with heated Trizol (Invitrogen) and then subjected to DNAseI digestion (Ambion). For microarray analysis, 10μg of RNA was submitted to the Biopolymers facility at Harvard Medical School for bioanalyzer analysis, labeling, and hybridization to Affymetix GeneChip® E. coli Genome 2.0 Array. Microarray data analysis is described below. For Northern blots, 6μg of RNA was electrophoresed on agarose gels using glyoxal buffer and loading dye (Ambion) and transferred to Bright Star Plus nylon membranes (Ambion). Blots were hybridized to 32P-labelled in vitro transcribed probes (Ambion Strip-EZ kit) at 68°C and washed according to the manufacturer’s instructions. For 5′RACE, 5μg of RNA isolated from EDL933Δhfq was used in a reverse transcriptase reaction with a ler-specific primer, followed by TdT tailing of cDNA, and PCR amplification all according to manufacturer’s instructions (Invitrogen). The resulting products were TOPO-cloned (Invitrogen) and sequenced.
Microarray analysis
Microarray analysis was performed using the Affymetix GeneChip® E. coli Genome 2.0 Array, which allows quantitation of transcripts from E. coli K12 (MG1655), UPEC (CFT073), and EHEC (EDL933 and Sakai). Probes are present to monitor expression of 5303 of the 5349 genes in the EDL933 genome. Genes were scored as upregulated or downregulated if the ratio of the hfq/wt signal was ≥2.0 or ≤0.5, respectively. Genes represented by multiple probes within the array were automatically scored as differentially regulated if all probes showed a 2-fold difference in expression. Genes with discordant results among their probe sets (199 genes) were individually examined. Of these, the majority (185 genes) corresponded to probes with absent or marginal signals, these genes were not scored as differentially regulated. Genes with adequate signals but discordant results (14 genes) were scored as differentially regulated only if all probes were altered at least 1.8-fold and the average change was ≥3 fold.
Protein analyses
Single colonies from plates were inoculated into 1mL of LB containing appropriate antibiotics and grown with shaking at 37°C for a minimum of 8 hrs. Cultures were diluted 1:50 and grown for 16-19 hrs at 37°C without shaking. The cultures were then adjusted to have equal densities (OD600). Supernatant fractions were prepared by filtering supernatants through a .22μM filter (Millipore) followed by TCA precipitation. Precipitated proteins were washed with cold acetone and resuspended in loading buffer. Pellet samples were prepared by resuspending centrifuged bacterial pellets in loading buffer and sonicating briefly. Protein samples were separated on 4-12% bis-tris gradient midi gels (Invitrogen) and either silver stained or transferred to PVDF membranes using an iBlot kit (Invitrogen). For western blots, α-His6 (Genetex) was used at 1:5,000, α-RNAP (Sigma) was used at 1:5,000, and α-H-NS was used at 1:1,000. All primary antibodies were used in 3% milk. HRP-conjugated secondary α-rabbit and α-mouse antibodies were used at 1:20,000 in 1% BSA. Western blots were visualized using Super signal West Pico Kit (Pierce).
β-galactosidase assays
Assays were performed on triplicate overnight cultures as described previously (Miller, 1992). Multiple exconjugants or transformants were assayed for each genotype assayed.
Acknowledgements
We are grateful to Dr. J. Mekalanos for supplying the α-RNAP antibody and reagents for work with P. aeruginosa, Dr. I. Rosenshine for supplying the α-H-NS antibody, and Dr. J. Kaper for supplying EPEC strain 2348/69. This work was supported by NIH AI42347 and HHMI (MKW) and NRSA AI081483 (ES).
References
- Berdichevsky T, Friedberg D, Nadler C, Rokney A, Oppenheim A, Rosenshine I. Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J Bacteriol. 2005;187:349–357. doi: 10.1128/JB.187.1.349-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel ED, Yahr TL. Control of gene expression by type III secretory activity. Curr Opin Microbiol. 2008;11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante VH, Santana FJ, Calva E, Puente JL. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39:664–678. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A. 2004;101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Roe AJ, Lobner-Olesen A, Murphy KC, Magoun L, Brady MJ, Donohue-Rolfe A, Tzipori S, Gally DL, Leong JM, Marinus MG. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2007;63:1468–1481. doi: 10.1111/j.1365-2958.2007.05602.x. [DOI] [PubMed] [Google Scholar]
- Caprioli A, Morabito S, Brugere H, Oswald E. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MB, Giannella RA. Hemorrhagic colitis associated with Escherichia coli O157:H7. Adv Intern Med. 1992;37:173–195. [PubMed] [Google Scholar]
- Creasey EA, Delahay RM, Daniell SJ, Frankel G. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology. 2003;149:2093–2106. doi: 10.1099/mic.0.26355-0. [DOI] [PubMed] [Google Scholar]
- Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Davis BM, Waldor MK. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P, Kenny B. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol. 2009;12:101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, Gruenheid S, Strynakda NC, Puente JL, Finlay BB. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun. 2005;73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A. 2005;102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantappie L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun. 2009;77:1842–1853. doi: 10.1128/IAI.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- Garmendia J, Frankel G, Crepin VF. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun. 2005;73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Sekirov I, Thomas NA, Deng W, O’Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2004;51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Regnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci U S A. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AM, Kaper JB. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartland EL, Batchelor M, Delahay RM, Hale C, Matthews S, Dougan G, Knutton S, Connerton I, Frankel G. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol Microbiol. 1999;32:151–158. doi: 10.1046/j.1365-2958.1999.01338.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hirakawa H, Kodama T, Takumi-Kobayashi A, Honda T, Yamaguchi A. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 2009;155:541–550. doi: 10.1099/mic.0.020420-0. [DOI] [PubMed] [Google Scholar]
- Huang LH, Syu WJ. GrlA of enterohemorrhagic Escherichia coli O157:H7 activates LEE1 by binding to the promoter region. J Microbiol Immunol Infect. 2008;41:9–16. [PubMed] [Google Scholar]
- Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157 : H7 to HEp-2 cells. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- Iyoda S, Koizumi N, Satou H, Lu Y, Saitoh T, Ohnishi M, Watanabe H. The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J Bacteriol. 2006;188:5682–5692. doi: 10.1128/JB.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschlager T, Hacker J. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun. 1999;67:5994–6001. doi: 10.1128/iai.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- Levine E, Hwa T. Small RNAs establish gene expression thresholds. Curr Opin Microbiol. 2008;11:574–579. doi: 10.1016/j.mib.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- Lodato PB, Kaper JB. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009;71:273–290. doi: 10.1111/j.1365-2958.2008.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Miller J. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Nakano M, Takahashi A, Su Z, Harada N, Mawatari K, Nakaya Y. Hfq regulates the expression of the thermostable direct hemolysin gene in Vibrio parahaemolyticus. BMC Microbiol. 2008;8:155. doi: 10.1186/1471-2180-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Ooka T, Asadulghani, Terajima J, Nougayrede JP, Kurokawa K, Tashiro K, Tobe T, Nakayama K, Kuhara S, Oswald E, Watanabe H, Hayashi T. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 2007;8:R138. doi: 10.1186/gb-2007-8-7-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun. 2003;71:7129–7139. doi: 10.1128/IAI.71.12.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JM, Waldor MK. The locus of enterocyte effacement-encoded effector proteins all promote enterohemorrhagic Escherichia coli pathogenicity in infant rabbits. Infect Immun. 2005;73:1466–1474. doi: 10.1128/IAI.73.3.1466-1474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Roop RM., Jr. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- Roe AJ, Yull H, Naylor SW, Woodward MJ, Smith DG, Gally DL. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect Immun. 2003;71:5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AJ, Naylor SW, Spears KJ, Yull HM, Dransfield TA, Oxford M, McKendrick IJ, Porter M, Woodward MJ, Smith DG, Gally DL. Co-ordinate single-cell expression of LEE4- and LEE5-encoded proteins of Escherichia coli O157:H7. Mol Microbiol. 2004;54:337–352. doi: 10.1111/j.1365-2958.2004.04277.x. [DOI] [PubMed] [Google Scholar]
- Roe AJ, Tysall L, Dransfield T, Wang D, Fraser-Pitt D, Mahajan A, Constandinou C, Inglis N, Downing A, Talbot R, Smith DG, Gally DL. Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157 : H7. Microbiology. 2007;153:1350–1360. doi: 10.1099/mic.0.2006/003707-0. [DOI] [PubMed] [Google Scholar]
- Russell RM, Sharp FC, Rasko DA, Sperandio V. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J Bacteriol. 2007;189:5387–5392. doi: 10.1128/JB.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Beck CF. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- Sharma VK, Zuerner RL. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2004;186:7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol. 2001;41:1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x. [DOI] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Li CC, Kaper JB. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect Immun. 2002;70:3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Thompson KM, Rhodius VA, Gottesman S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol. 2007;189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Matthews S, Marches O, Frankel G, Hayashi T, Pallen MJ. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Wu YC, Chen JW, Wu CF, Tzeng CM, Syu WJ. Multiple functions of l0036 in the regulation of the pathogenicity island of enterohaemorrhagic Escherichia coli O157:H7. Biochem J. 2006;393:591–599. doi: 10.1042/BJ20051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Blasi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder-Olsson C, Kaijser B. Enterohemorrhagic Escherichia coli (EHEC) Scand J Infect Dis. 2005;37:405–416. doi: 10.1080/00365540510038523. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yona-Nadler C, Umanski T, Aizawa S, Friedberg D, Rosenshine I. Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohaemorrhagic Escherichia coli. Microbiology. 2003;149:877–884. doi: 10.1099/mic.0.25970-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chaudhuri RR, Constantinidou C, Hobman JL, Patel MD, Jones AC, Sarti D, Roe AJ, Vlisidou I, Shaw RK, Falciani F, Stevens MP, Gally DL, Knutton S, Frankel G, Penn CW, Pallen MJ. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect Immun. 2004;72:7282–7293. doi: 10.1128/IAI.72.12.7282-7293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]