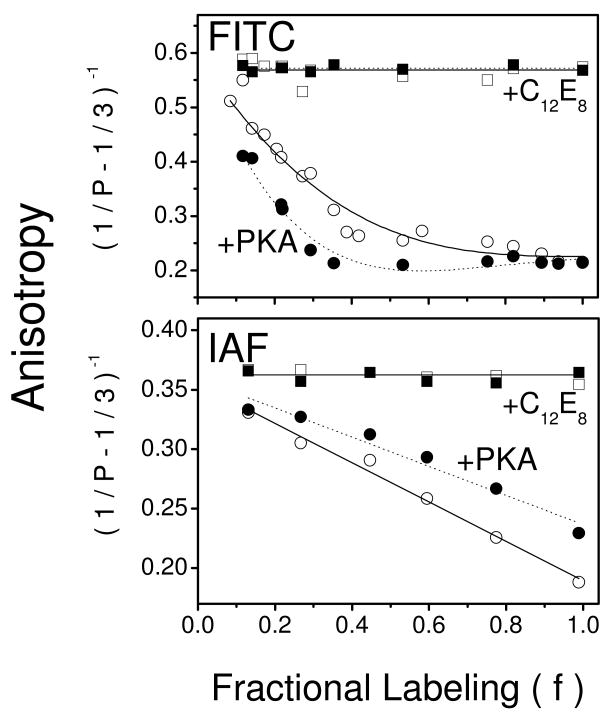
Figure 5. PLB Modulates Spatial Arrangement of Ca-ATPase Polypeptide Chains.
Anisotropy (1/P – 1/3)-1 of FITC- (top panel) or IAF- (bottom panel) labeled Ca-ATPase in cardiac SR microsomes (0.1 mg/mL) as a function of fractional labeling (f) of the probe sites (i.e., Lys514 or Cys674) (25). The maximal labeling stoichiometry is 3.4 ± 0.1 nmol FITC or 3.7 ± 0.2 nmol IAF per mg of microsomal protein (see Figure S1 in Supplemental Materials). Anisotropy was measured for 100 μg microsomes /ml in 20 mM MOPS (pH 7.0), 0.1 M KCl, 5 mM MgCl2, 0.1 mM EGTA, and 0.11 mM CaCl2 (Cafree is 0.5 μM) in the absence (open symbols) or presence (closed symbols) of PKA (80 μg/mL), cAMP (1 μM), and ATP (0.1 mM) before (circles) and after (squares) solubilization with C12E8 (6 mM). Excitation was at 485 nm and fluorescence emission was measured using a Schott OG530 cutoff filter. Lines represent nonlinear least squares fits to the data according to the models for interactions between neighboring fluorophores (see Eq. 2 in Experimental Procedures).