Abstract
Histone deacetylases (HDACs) catalyze the removal of acetyl groups on the amino-terminal lysine residues of core nucleosomal histones. This activity is associated generally with transcriptional repression. We have reported previously that inhibition of HDAC activity by hydroxamic acid-based hybrid polar compounds, such as suberoylanilide hydroxamic acid (SAHA), induces differentiation and/or apoptosis of transformed cells in vitro and inhibits tumor growth in vivo. SAHA is a potentially new therapeutic approach to cancer treatment and is in Phase I clinical trials. In several tumor cell lines examined, HDAC inhibitors alter the expression of less than 1% of expressed genes, including the cell cycle kinase inhibitor p21WAF1. In T24 bladder carcinoma cells, SAHA induces up to a 9-fold increase in p21WAF1 mRNA and protein, which is, at least in part, because of an increase in the rate of transcription of the gene. SAHA causes an accumulation of acetylated histones H3 and H4 in total cellular chromatin by 2 h, which is maintained through 24 h of culture. An increase in the accumulation of acetylated H3 and H4 was detected throughout the p21WAF1 promoter and the structural gene after culture with SAHA. The level of histone acetylation did not change in chromatin associated with the actin and p27 genes, and their mRNA expression was not altered during culture of T24 cells with SAHA. Thus, the present findings indicate that the induction of p21WAF1 by SAHA is regulated, at least in part, by the degree of acetylation of the gene-associated histones and that this induced increase in acetylation is gene selective.
Keywords: chromatin, HDAC inhibitor, suberoylanilide hydroxamic acidSAHA
Transformed cells are characterized generally by unregulated cell proliferation. However, the potential for cancer cells to differentiate terminally or to undergo apoptosis is not necessarily lost (1, 2). Hydroxamic acid-based hybrid polar compounds that are inhibitors of histone deacetylase (HDAC) have been shown to induce differentiation and/or apoptosis of transformed cells and cause the accumulation of acetylated core nucleosomal histones in cultured cells (3). These agents are active at nanomolar concentrations as inhibitors of partially purified HDACs; at micromolar concentrations, they are inducers of transformed cells in culture (4–6) and inhibit the growth of tumors in animals (7, 8). The prototype of this class of hydroxamic acid-based hybrid polar compound is suberoylanilide hydroxamic acid (SAHA), which binds directly to the catalytic site of HDAC, inhibiting its enyzmatic activity (9).
Several lines of evidence suggest that histone acetylation plays a role in transcriptional regulation (10), probably by altering chromatin structure. Acetylation of core nucleosomal histones is regulated by the opposing activities of histone acetyltransferases (HATs) and HDACs. HDACs catalyze removal of an acetyl group from the ɛ-amino group of lysine side chains of histone H2A, H2B, H3, and H4, thereby reconstituting the positive charge on the lysine. Transcriptionally silent chromatin is composed of nucleosomes in which the histones have low levels of acetylation on lysine residues of their amino-terminal tails. Acetylation of histone proteins neutralizes the positive charge on lysine residues and disrupts nucleosome structure, allowing unfolding of the associated DNA, access by transcription factors, and changes in gene expression. Chromatin fractions enriched in actively transcribed genes are also enriched in the more highly acetylated isoforms of the core histones (11).
HDAC inhibitors, such as trapoxin (12) and trichostatin A (TSA) (13), appear to be selective with regard to the genes whose expression is altered. Van Lint et al. (13) demonstrated, by differential display, that expression of only 2–5% of genes is changed significantly after treatment of cultured cells with the HDAC inhibitor, TSA.
The expression of the cell cycle kinase inhibitor p21WAF1 is induced in transformed cells by HDAC inhibitors such as phenylbutyrate, trichostatin A, and SAHA (4, 14, 15). Increased expression of p21WAF1 may play a critical role in the growth arrest induced in transformed cells by these agents. In this study, we have examined the effects of SAHA on the acetylation of histones in the chromatin of the p21WAF1 gene. We found that SAHA induces the accumulation of acetylated histones in the chromatin of the p21WAF1 gene and that this increase is associated with an increase in p21WAF1 expression in T24 human bladder transitional cell carcinoma cells. There was no change in acetylation of histones of the chromatin associated with the actin and p27 genes, genes whose transcription is not altered by SAHA. These findings indicate that induction of p21WAF1 by SAHA may be regulated, at least in part, by histone acetylation of the chromatin associated with the p21WAF1 gene and that HDAC inhibitor-induced histone acetylation and gene activation are selective.
Materials and Methods
Cell Culture.
The human bladder carcinoma cell line (T24) was obtained from American Type Culture Collection (Rockville, MD). T24 cells were cultured in Minimal Essential Medium-α with 10% FBS and were incubated at 37°C with 5% CO2 (16). Cells were passed with trypsin-EDTA, and the resulting suspension of cells was centrifuged at 2,500 × g for 5 min. Cells were resuspended in fresh media. SAHA was synthesized as previously described (4).
SDS/PAGE and Immunoblotting.
T24 cells (0.5–1.0 × 107) were cultured as described below without and with inducer. Cells were recovered by centrifugation, washed twice with ice-cold PBS, and resuspended for lysis in 75–150 μl ice-cold ×1 E1A buffer (250 mM NaCl/50 mM Hepes, pH 7.0/0.1% Nonidet P-40/50 mM NaF/5 mM EDTA/0.45 mM PMSF). Samples were kept on ice for 30 min before centrifugation at 15,000 × g for 10 min at 4°C. For histone preparation, T24 cells (1 × 107) were cultured as indicated and harvested and washed with PBS. Histones were isolated by a modification of the method of Yoshida et al. (17). Cells were pelleted and resuspended in 1 ml ice-cold lysis buffer (10 mM Tris⋅HCl, pH 6.5/50 mM sodium bisulfite/1% Triton X-100/10 mM MgCl2/8.6% sucrose) before homogenization with two dounce strokes. Nuclei were centrifuged at 700 rpm in a Beckman GS-6R centrifuge for 5 min and washed 3 times with 1 ml of lysis buffer. The final wash was performed with 1 ml of Tris⋅EDTA solution (10 mM Tris⋅HCl, pH 7.4/13 mM EDTA). Nuclei were pelleted and resuspended in 100 μl of ice-cold water. Sulfuric acid was added to the samples to a final concentration of 0.2 M; samples were vortexed and incubated on ice for 1 h. Samples were centrifuged at 15,000 × g for 10 min at 4°C, and the supernatant was precipitated with 1 ml of acetone overnight at −20°C. Precipitated protein was collected by centrifugation at 15,000 × g for 10 min at 4°C, air dried, and resuspended in 50–100 μl of water. Protein concentrations of lysates and histone preparations were determined by using the Bio-Rad Protein Assay kit with BSA as the standard. Proteins (1 to 25 μg protein) were denatured at 100°C in loading buffer for 10 min and electrophoresed in 15% polyacrylamide gels. After electrophoresis, samples were transferred onto nitrocellulose (0.45 μm) in buffer containing 0.25 M Tris-base (pH 8.3), 1.86 M glycine, and 20% methanol for 2 h at 4°C, as previously described (18). To verify equal protein loading, a parallel protein gel was run and stained with Gelcode blue.
Nitrocellulose membranes were incubated in PBS with 3% milk for 30 min at room temperature. Buffer was replaced with PBS containing the specific rabbit polyclonal antiserum diluted 1:1,000 in 3% milk and incubated at 4°C for 16 h. The following antibodies were used: mouse monoclonal anti-p21WAF1 antibody F5 (Santa Cruz Biotechnology) and rabbit antiacetylated histone H3 and rabbit anti-acetylated histone H4 (Upstate Biotechnology, Upstate, NY). Nitrocellulose blots were washed in distilled water and then incubated in PBS containing 3% milk and a 1:5,000 dilution of goat anti-rabbit horseradish peroxidase for 2 h at room temperature. Blots were washed in distilled water, rinsed, and soaked in Pierce Super Signal chemiluminescent substrate for 5 min. Membranes were then exposed to Kodak BioMax film for 1–5 min. Differences in band density on autoradiograms were quantified by using the IS-1000 Digital Imaging System (Alpha Innotech, San Leandro, CA).
RNA Isolation and Northern Blotting.
RNA was prepared from T24 cells by using the guanididium thiocyanate method (19). Total RNA (10 μg) was denatured by using glyoxyl, fractionated on agarose gels, and transferred overnight to a Nytran N+ nylon membrane (Amersham Pharmacia Biotech) according to Ausubel et al. (20). Northern blot hybridizations were performed by using the 2,120-bp human p21WAF1 cDNA fragment from pZL-WAF1 (21) and with a 2.0-kb chicken γ-actin cDNA. The cDNA inserts were labeled with [α32P]-dCTP by using the random-primed DNA labeling method (20). Northern blots were prehybridized at 62°C for at least 3 h in a 1 M NaCl/1% SDS solution containing Background Quencher (Molecular Research Center, Cincinnati). Hybridization was completed in a High-Efficiency Hybridization Solution (Molecular Research Center), containing the 32P-labeled probe at 62°C, for at least 16 h. Blots were washed three times at room temperature (5 min) in ×1 SSC/1% SDS and three times at room temperature (10 min) in ×0.1 SSC/0.1% SDS. RNA/cDNA hybrids were visualized on BioMax film by using two intensifying screens and a 12- to 48-h exposure period. Blots were stripped and reprobed with the γ-actin cDNA. Densitometric analysis was performed on the 2.0-kb γ-actin transcript for the standardization of RNA loading. 18S rRNA-specific 50-mer oligodeoxyribonucleotide was end labeled with [γ32P]-ATP by using T4 polynucelotide kinase (22).
Run-On Analysis.
Nuclei were prepared and run-on transcription assays were performed as previously described (23). For the run-on assay, 20 μl of packed nuclei was added to 40 μl of run-on buffer (80 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/1 mg/ml heparin/0.6% (wt/vol) Sarkosyl/350 mM (NH4)2SO4/400 μM ATP/400 μM GTP/400 μM UTP/250 μCi [α32P]-CTP) and incubated at 37°C for 30 min. The reactions were stopped by placement on ice and addition of 10 μg of tRNA, 175 μl of 20 mM Tris⋅HCl containing 10 mM CaCl2, and 25 μl of a solution containing 1 mg/ml proteinase K and DNase I. The resulting mixture was incubated at 37°C for 30 min, and the reaction was stopped by addition of 25 μl of 10% (wt/vol) SDS and 25 μl of 0.2 M EDTA. The labeled RNA was then extracted with phenol/chloroform/isoamyl alcohol (25:24:1). Equal amounts of 32P-labeled RNA, (approximately 5 × 107 cpm) were then hybridized to nylon membranes containing 5 μg of p21WAF1 and γ-actin cDNA. Hybridization and washings were as described for Northern blotting analysis. The filters were exposed to BioMax film at −70°C for 2–10 days with intensifying screens. The relative rate of transcription was determined by densitometric scanning and dividing by the γ-actin signal.
Chromatin Immunoprecipitation (ChIP) Assay.
Cells were plated at a density of 4 × 106 cells/15-cm dish and incubated overnight at 37°C with 5% CO2. The next day, cells were cultured with 0 or 7.5 μM SAHA for 2, 6, 15, or 24 h. Formaldehyde was then added to the cells to a final concentration of 1%, and the cells were incubated at 37°C for 10 min. The medium was removed, and the cells were suspended in 1 ml of ice-cold PBS containing protease inhibitors (Complete, Boehringer Mannheim). Cells were pelleted, resuspended in 0.5 ml of SDS lysis buffer (1% SDS/0 mM EDTA/50 mM Tris⋅HCl, pH 8.1), and incubated on ice for 10 min. Lysates were sonicated with 15 10-sec bursts. Debris was removed from samples by centrifugation for 10 min at 15,000 × g at 4°C. An aliquot of the chromatin preparation (200 μl) was set aside and designated as the Input Fraction. Supernatants were diluted 5-fold in immunoprecipitation buffer (0.01% SDS/1.1% Triton X-100/1.2 mM EDTA/16.7 mM Tris⋅HCl, pH 8.1/16.7 mM NaCl), and 80 μl of a 50% protein A Sepharose slurry containing 20 μg sonicated salmon sperm DNA and 1 mg/ml BSA in TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA) was added and incubated, rocking for 30 min at 4°C. Beads were pelleted by centrifugation, and supernatants were placed in fresh tubes with 5 μg of antiacetylated histone H3 antibody, antiacetylated histone H4 antibody, or normal rabbit serum and incubated overnight at 4°C. Protein A Sepharose slurry (60 μl) was added, and samples were rocked for 1 h at 4°C. Protein A complexes were centrifuged and washed 5 times for 5 min each, according to the manufacturer's protocol. Immune complexes were eluted twice with 250 μl of elution buffer (1% SDS/0.1 M NaHCO3) for 15 min at room temperature. Twenty microliters of 5 M NaCl was added to the combined eluates, and the samples were incubated at 65°C for 4 h. EDTA, Tris⋅HCl, pH 6.5, and proteinase K were then added to the samples at a final concentration of 10 mM, 40 mM, and 0.04 μg/μl, respectively, and the samples were incubated at 45°C for 1 h. Immunoprecipitated DNA (both immunoprecipitation samples and Input) was recovered by phenol/chloroform extraction and ethanol precipitation and analyzed by PCR. Actin-, p21WAF1-, or p27-specific primers were used to carry out PCR from DNA isolated from ChIP experiments and Input samples. The optimal reaction conditions for PCR were determined for each primer pair. Parameters were denaturation at 95°C for 1 min and annealing at 6°C for 1 min, followed by elongation at 72°C for 1 min. PCR products were analyzed by 2.5% agarose/ethidium bromide gel electrophoresis. The primer pairs used for p21WAF1 ChIP analysis were: 5′-ACC AAC GCA GGC GAG GGA CT-3′ (uP1), 5′-CCG GCT CCA CAA GGA ACT GA-3′ (dP1), 5′-GGT GTC TAG GTG CTC CAG GT-3′ (uP2), 5′-GCA CTC TCC AGG AGG ACA CA-3′ (dP2), 5′-CGT GGT GGT GGT GAG CTA GA-3′ (uP3), 5′-CTG TCT GCA CCT TCG CTC CT-3′ (dP3), 5′-GAG GCC CAC AAG GAC TCT CA-3′ (uP4), 5′-GCT GAG ATC ATG CCA CCT GC-3′ (dP4), 5′-CGG TGC TTG GTC TCT ATG AA-3′ (uP5), 5′-TGG CCA CAC TGA GGA ATG AT-3′ (dP5), 5′-AGG AAT CCC TGG TCA CGC TC-3′ (uI1a), 5′-GTG GTG GAC ACA GTG GCG TA-3′ (dI1a), 5′-CCA TCG GCA CAG TGA CCT AT-3′ (uI1b), 5′-CCC CTC AAA GAC ATG AAC CC-3′ (dI1b), 5′-GCA CTC CTT AGG TTC AAC AG-3′ (uI1c), 5′-TAG GGC TTC ATC AAG GTG CT-3′ (dI1c), 5′-AGA CAC TGC ACC CAA CCA CC-3′ (uI1d), 5′-GGC ACA GCC AGG ATC TGA AC-3′ (dI1d), 5′-TGA AGG AGG TAG TTG GCA GG-3′ (uI1e), 5′-GCT CTG GAG CCA GAT CAC CT-3′ (dI1e), 5′-AGC TGA GCC GCG ACT GTG AT-3′ (uE2), 5′-CTG AGC GAG GCA CAA GGG TA-3′ (dE2), 5′-GCA CAC AGA GCT AGT AGT GG-3′ (uI2), 5′-CTG AGG AGA GAC AGC AGA AG-3′ (dI2), 5′-AAT CGT CCA GCG ACC TTC CT-3′ (uE3), and 5′-ACA TGG GGA GCC GAG AGA AA-3′ (dE3), 5′-ACA TCC TTC CTG CAC CTG CT-3′. The primers used for actin ChIP analysis were: 5′-GCC AGC TCT CGC ACT CTG TT-3′ (uGACP1), 5′-AGA TCG CAA CCG CCT GGA AC-3′ (dGACP1), 5′-CAT GTA CGT GGC CAT CCA GG-3′ (uGACE), and 5′-GTG GCC ATC TCC TGC TCG AA-3′ (dGACE). The primers used for p27 ChIP analysis were: 5′-CTC GCC GTG TCA ATC ATT TTC-3′ (up27P1), 5′-GAT CAA ATG GAC TGG CGA GCG 3′ (dp27P1); 5′-CAG GTT TGT TGG CAG CAG TAC-3′ (up27P2); 5′-GAA AAT GAT TGA CAC GGC GAG 3′ (dp27P2).
Results
SAHA Inhibits T24 Cell Proliferation and Induces Accumulation of Acetylated Histones.
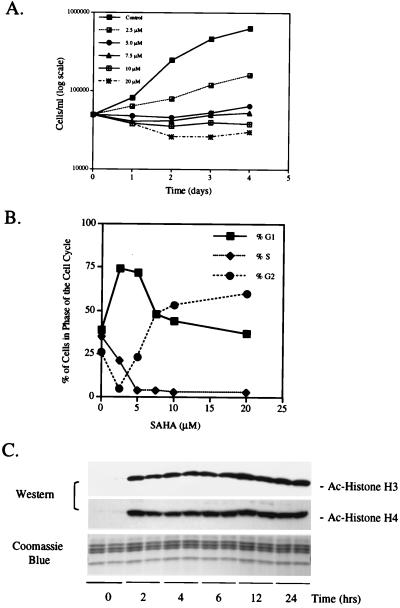
We examined the effect of the HDAC inhibitor SAHA on the proliferation of T24 cells. Cells were cultured without and with 2.5, 5, 7.5, 10, and 20 μM SAHA for 1–4 days. SAHA partially inhibited proliferation at 2.5 μM, and at higher levels (5 to 20 μM) no increase in cell density was detected over the duration of culture (Fig. 1A). SAHA at 5 and 7.5 μM inhibited proliferation without loss of cell viability (>90% viable), as determined by trypan blue exclusion. SAHA at 10 and 20 μM caused a 50% loss of cell viability (data not shown). At SAHA concentrations of 5 and 7.5 μM SAHA, inhibition of cell proliferation was associated with a marked decrease in the proportion of cells in S phase of the cell cycle (Fig. 1B). At concentrations of 2.5 and 5 μM SAHA, cells arrested predominately in G1, whereas at higher concentrations, cells arrested in both G1 and G2 (Fig. 1B).
Figure 1.
SAHA inhibits proliferation and causes accumulation of acetylated histones in T24 cells. (A) SAHA time course and dose response. Cells were harvested and cell number and viability determined as indicated. Each time point was performed in triplicate, and data are presented as means. (B) T24 cells were cultured with the indicated concentrations of SAHA for 24 h. Cells were harvested and DNA content was assessed by FACS analysis of propidium iodide-stained nuclei. (C) Western blot analysis of acetylated histones H3 and H4 in T24 cells. Histones were isolated by acid extraction from cells cultured for indicated times with 7.5 μM SAHA. Acetylation was detected by using antiacetylated H3 and H4 antibodies. A parallel gel was stained with Coomassie blue for a loading control.
We determined next the level of histone acetylation in T24 cells after culture with SAHA. Histones were isolated from cells cultured with SAHA (7.5 μM) for 0, 2, 4, 6, 12, and 24 h. Western blot analysis showed that before incubation with SAHA (0 h), the levels of acetylated histone H3 and H4 in T24 cells were low (Fig. 1C). Incubation for 2 h with SAHA resulted in the accumulation of acetylated histones H3 and H4, which levels then remained unchanged during the 24-h incubation period.
SAHA Induces Expression of p21WAF1 mRNA and p21WAF1 Protein.
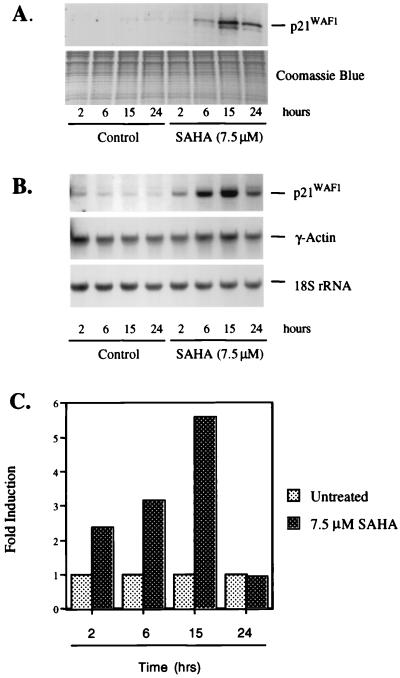
The effect of SAHA on p21WAF1 protein levels was determined by Western blot analysis. T24 cells were cultured without and with SAHA for 2, 6, 15, and 24 h. At each time point, proteins and RNA were prepared from the cells. After culture with SAHA, the p21WAF1 protein levels were slightly, if at all, increased at 2 h, increased 3-fold by 6 h, 9-fold by 15 h, and decreased to about 6-fold at 24 h, compared with p21WAF1 protein levels in cells not cultured with SAHA (Fig. 2A).
Figure 2.
SAHA induces p21WAF1 mRNA and protein in T24 cells. T24 cells were cultured with SAHA (7.5 μM) for the indicated times. (A) Western blot analysis of p21WAF1 protein expression. Protein extracts were prepared and resolved (25 μg) on 15% SDS/PAGE. p21WAF1 protein was detected by using the mouse monoclonal anti-p21WAF1 F5 antibody. A parallel gel was stained with Coomassie blue for a loading control. (B) Northern blot analysis of p21WAF1 mRNA expression. Total RNA (10 μg) was isolated and fractionated on 1.2% agarose and transferred to a nylon membrane. The membrane was probed by using 32P-random-labeled human p21WAF1 cDNA, the γ-actin cDNA, and a 50-mer oligonucleotide specific for 18S rRNA. (C) Run-on transcription assay by using nuclei from SAHA-induced T24 cells. Nuclei were prepared and labeled with [α32P]-CTP. Equal levels of radioactivity (approximately 5 × 107 cpm) were incubated with filters containing 5 μg of the indicated plasmid cDNA. Radioactivity bound to the filters was quantified and normalized to actin at each time point examined. The actin-normalized value for the untreated sample was adjusted to a value of 1 to determine fold increase after culture with SAHA.
p21WAF1 mRNA levels also increased in cells cultured with SAHA in a time-dependent manner, as determined by Northern blot analysis (Fig. 2B). The level of p21WAF1 mRNA was increased by 2 h and more markedly by 6 and 15 h after onset of culture with SAHA. By 24 h, the level of p21WAF1 mRNA decreased but remained elevated above the level detected in cultures without inducer. The p21WAF1 mRNA levels were normalized with respect to the level of γ-actin mRNA, an mRNA whose level did not change during culture with SAHA (Fig. 2B).
The transcription rate of the p21WAF1 gene during culture with SAHA was determined by nuclear run-on measurements. T24 cells were cultured with 7.5 μM SAHA, nuclei prepared after 2, 6, 15 and 24 h of culture, and nuclear run-on analyses performed as described in Materials and Methods. The relative rate of transcription was normalized to that of γ-actin. The rate of p21WAF1 gene transcription was increased 2.5-fold by 2 h of culture with SAHA. By 6 and 15 h, p21WAF1 transcription was increased 3- and 5.5-fold, respectively. By 24 h, the rate of p21WAF1 gene transcription had returned to control levels (Fig. 2C). Both the accumulation of p21WAF1 mRNA and the transcription rate peaked at about 15 h of culture with SAHA. By 24 h, both the steady-state p21WAF1 mRNA level and the transcription rate of the p21WAF1 gene were decreased.
SAHA Induces Accumulation of Acetylated Histones in Chromatin Associated with the p21WAF1 Gene.
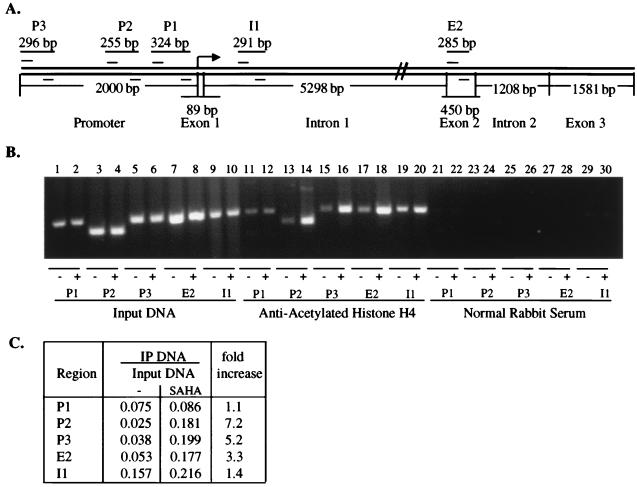
The effect of HDAC inhibition on the acetylation of histone H4 associated with the p21WAF1 gene promoter was examined by using ChIP. Chromatin fragments from cells cultured without and with SAHA (7.5 μM) for 2 h were immunoprecipitated with antibody to acetylated histone H4. DNA from the immunoprecipitate was isolated. From this DNA, a 255-bp fragment of the p21WAF1 promoter region was amplified (see P2, Fig. 3A). After culture with SAHA, approximately 7-fold more p21WAF1 promoter DNA was associated with highly acetylated histone, compared with the same region isolated from cells cultured without SAHA (Fig. 3 B and C).
Figure 3.
SAHA induces accumulation of acetylated histone H4 in chromatin associated with the p21WAF1 gene. (A) Schematic representation of the human p21WAF1 gene. Primer sets are indicated as P3, P2, P1, I1, and E2. Sp1 sites are at −82 and −69. (B) Soluble chromatin was immunoprecipitated with antiacetylated histone H4 antibodies from T24 cells cultured without (odd-numbered lanes) and with (even-numbered lanes) 7.5 μM SAHA for 2 h. PCR primers for the regions of the p21WAF1 gene as indicated above were used to amplify the DNA isolated from the immunoprecipitated chromatin. (C) The figure in B was scanned and quantified by using nih image 1.62 analysis software. The ratio between input DNA and precipitated DNA was calculated for each treatment and primer set. The fold increase after treatment with SAHA was calculated from the indicated ratios.
We examined next the extent of acetylation within the p21WAF1 gene-associated chromatin to determine whether the increase in acetylation was localized to promoter-associated chromatin or was present throughout the gene. Primer sets encompassing the p21WAF1 promoter and the coding gene (Fig. 3A) were used to assess the extent of histone acetylation (Fig. 3B). PCR analysis with the different primer sets demonstrated that increased accumulation of acetylated histones in the chromatin of the p21WAF1 gene was present in all regions examined after culture with SAHA (Fig. 3 B and C). The fold increase in accumulation of acetylated histones was not the same for all areas of the p21WAF1 gene examined, possibly reflecting localized areas of acetylation within the gene or, alternatively, regions with a higher density of nucleosomes. Similar results were also obtained by using antiacetylated H3 antibodies (data not shown).
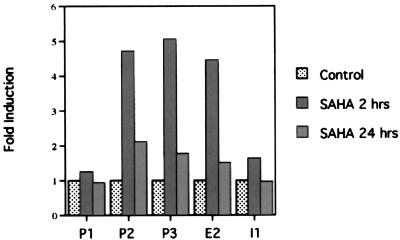
As indicated above, within 2 h of culture with SAHA there was an increase in the transcription rate of the p21WAF1 gene, whereas after 24 h the transcription rate of the gene decreased to the level found in control cultures. We determined whether these changes in transcription rates were associated with changes in the histone acetylation status of the chromatin of the p21WAF1 gene (Fig. 4). At 2 h, there was a 3- to 7-fold increase in p21WAF1 DNA corresponding to the promoter regions P2 and P3 and E2 and a 1.1- to 1.4-fold increase in the regions P1 and I1. By 24 h, the increase in acetylation of histone H4 associated with the p21WAF1 gene observed at 2 h was no longer detectable. The increase at 2 h and fall at 24 h in transcription rate of the p21WAF1 gene correlated directly with the increase and fall in level of gene-associated histone acetylation.
Figure 4.
SAHA induces a transient increase in acetylated histones in the chromatin associated with the p21WAF1 gene. Chromatin from cells cultured without and with 7.5 μM SAHA for 2 or 24 h was isolated and ChIP assay performed as described above. Gels were scanned and fold induction was determined and graphed for each region indicated of the p21WAF1 gene.
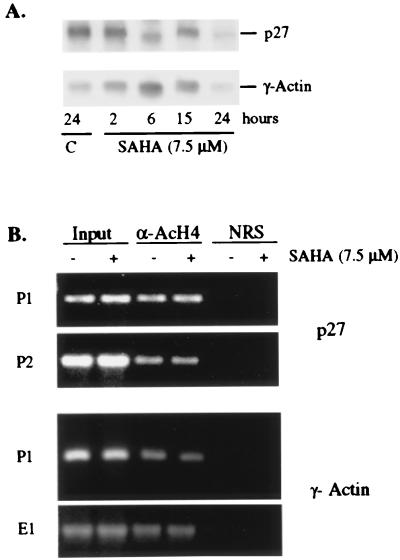
SAHA Does Not Induce Accumulation of Acetylated Histones in Chromatin Associated with γ-Actin or p27 Genes.
To determine whether this effect was selective for a limited number of genes, we examined the level of histone H4 acetylation in the γ-actin and p27 genes. The expression of p27 and actin RNA was not altered by culture with SAHA (see Figs. 2B and 5A). No change in the levels of histone H4 acetylation was detected after culture with SAHA (Fig. 5B). Taken together, these results suggest that the SAHA-induced changes in histone acetylation are localized to specific areas of the chromatin.
Figure 5.
SAHA does not induce accumulation of acetylated histone H4 in the chromatin associated with actin and p27 genes. (A) Northern blot analysis of p27 mRNA expression. Total RNA (10 μg) was isolated and fractionated on 1.2% agarose and transferred to a nylon membrane. The membrane was probed by using 32P-random labeled human p27 cDNA, the γ-actin cDNA, and a 50-mer oligonucleotide specific for 18S rRNA. (B) Soluble chromatin was immunoprecipitated with antiacetylated H4 antibodies from T24 cells cultured without and with 7.5 μM SAHA for 2 h. PCR primers for the regions of the actin and p27 genes as indicated in Materials and Methods were used to amplify the regions from the acetylated chromatin.
Discussion
HDAC inhibitors, such as SAHA, are potent inducers of differentiation and/or apoptosis of transformed cells in culture and are inhibitors of tumor growth in animals (3–8). In this study, we show that SAHA induces the transcription of p21WAF1 and accumulation of acetylated histones associated with the promoter and coding regions of that gene. We show that both histone acetylation and increased transcription are induced after 2 h and fall by 24 h in cells after onset of culture with inducer. The accumulation of acetylated histones by SAHA does not appear to be global. The γ-actin and p27 genes are not transcriptionally activated, and there is no change in the level of acetylated histone in chromatin associated with these genes in response to SAHA.
Several models have been proposed for the relationship between histone acetylation and transcription (24). In these models, histone acetylation can, (i) be untargeted and occur at both promoter and nonpromoter regions; (ii) be targeted generally to promoter regions; or (iii) be targeted to specific promoters by gene-specific activator proteins. In the first and second models, selective gene expression does not depend on histone acetylation, whereas in the third model, gene-selective acetylation leads to transcriptional activation. The present findings support a model in which increased histone acetylation is targeted to specific genes and occurs throughout the entire gene, not just the promoter region. These results suggest that the activities of HDACs and HATs are exerted throughout the p21WAF1 gene and that the HDAC activity is inhibited by SAHA. Some studies suggest that acetylation might be limited to a region of only one or two nucleosomes (25), evoking a model in which multiple HDACs are recruited to the p21WAF1 gene. Other studies suggest that although HATs and HDACs may be targeted to a single site, they can acetylate histones up to seven nucleosomes away (26). In this example, an expanded region of histone acetylation of the HO locus was noted when the Sin3p/Rpd3p deacetylase complex was inactivated by mutation of Sin3p (26). In any case, SAHA appears to inhibit all HDACs recruited to the p21WAF1 gene.
HDACs and HATs do not bind directly to DNA but are recruited to specific locations by forming complexes with sequence-specific transcription factors. The Sp1 transcription factor has been shown to play a role in the induced transcription of p21WAF1 expression by HDAC inhibitors (14, 27). Trichostatin A induces the p21WAF1 promoter through the Sp1 sites at −82 and −69 relative to the transcription start site in a p53-independent fashion (27). Sp1 has been shown to repress directly transcription of other promoters, such as the murine tyrosine kinase promoter, by interaction with HDAC1 (27). It is possible that Sp1 represses p21WAF1 transcription by recruitment of HDAC to the promoter. HDAC inhibitors may therefore change the balance of histone acetylation in the p21WAF1 promoter at the Sp1 site. Although in our study we find a consistent increase in acetylation by using the P1 primers, which includes the Sp1 site, it is only an approximately 1.1-fold increase compared with an approximately 5-fold increase at other sites in the promoter region. It is possible that the relatively small increase in acetylation reflects a lower density of nucleosomes in this region.
We find that accumulation of acetylated total cellular histone is maintained in cells cultured with SAHA, whereas the increased histone acetylation localized to the p21WAF1 gene is transient and correlates directly with the level of transcriptional activation of that gene. Chen et al. (28) have shown that increased acetylation of histones is associated with genes that are transcriptionally activated by hormone receptors. The increased histone acetylation parallels the increased transcription, and both are transient. Chen et al. show that on recruitment of a coactivator complex by a hormone receptor, in addition to acetylation of the histones, HATs also acetylate additional proteins in the complex. This acetylation leads to a loss of binding of the coactivator complex and attenuation of transcription.
In summary, this study shows that inhibition of HDAC activity by SAHA increases the transcription of the p21WAF1 gene selectively. SAHA-induced expression of p21WAF1 is associated with arrest of growth of the T24 bladder carcinoma cells. The induced activation of the p21WAF1 gene is accompanied by an accumulation of acetylated histones H3 and H4 associated with the p21WAF1 gene. The effect of SAHA is selective at the gene level, because we find that the HDAC inhibitor alters neither the expression of the γ-actin or p27 genes nor the level of acetylated histones in the chromatin associated with these two genes. The mechanism leading to gene selectivity of SAHA-induced histone acetylation is not understood. We have reported previously that SAHA is a potent inhibitor of tumor cell growth both in culture (3) and in tumor-bearing animal models (7, 8), and SAHA is currently in Phase I clinical trials as a potential anticancer therapy.
Acknowledgments
We are grateful to Gisela Venta-Perez for excellent technical assistance. We thank Drs. Lisa Butler and Xianbo Zhou for many useful discussions and critical reading of the manuscript. T.W.S. is a Cohen Fellow in Biomedical Research. These studies were supported in part by funds granted by the Michael and Ethel Cohen Foundation, Grant CA-0974823 from the National Cancer Institute, and grants from the Japanese Foundation for Promotion of Cancer Research Fund and the DeWitt Wallace Fund for Memorial Sloan–Kettering Cancer Center.
Abbreviations
- HDAC
histone deacetylase
- SAHA
suberoylanilide hydroxamic acid
- HAT
histone acetyltransferase
- ChIP
chromatin immunoprecipitation
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180316197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180316197
References
- 1.Marks P A, Richon V M, Breslow R, Rifkind R A. C R Acad Sci Ser III. 1999;322:161–165. doi: 10.1016/s0764-4469(99)80039-0. [DOI] [PubMed] [Google Scholar]
- 2.Marks P A, Richon V M, Rifkind R A. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 3.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richon V M, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1996;93:5705–5708. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glick R D, Swendeman S L, Coffey D C, Rifkind R A, Marks P A, Richon V M, La Quaglia M P. Cancer Res. 1999;59:4392–4399. [PubMed] [Google Scholar]
- 6.Vrana J A, Decker R H, Johnson C R, Wang Z, Jarvis W D, Richon V M, Ehinger M, Fisher P B, Grant S. Oncogene. 1999;18:7016–7025. doi: 10.1038/sj.onc.1203176. [DOI] [PubMed] [Google Scholar]
- 7.Cohen L A, Amin S, Marks P A, Rifkind R A, Desai D, Richon V M. Anticancer Res. 1999;19:4999–5006. [PubMed] [Google Scholar]
- 8.Butler, L. M., Agus, D. B., Scher, H. I., Higgins, B., Rose, A., Cordon-Cardo, C., Thaler, H. T., Rifkind, R. A., Marks, P. A. & Richon, V. M. (2000) Cancer Res., in press. [PubMed]
- 9.Finnin M S, Donigian J R, Cohen A, Richon V M, Rifkind R A, Marks P A, Breslow R, Pavletich N P. Nature (London) 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 10.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 11.Turner B M. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 12.Sambucetti L C, Fischer D D, Zabludoff S, Kwon P O, Chamberlin H, Trogani N, Xu H, Cohen D. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 13.Van Lint C, Emiliani S, Verdin E. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao H, Hasegawa T, Isobe K. J Cell Biochem. 1999;73:291–302. [PubMed] [Google Scholar]
- 15.DiGiuseppe J A, Weng L J, Yu K H, Fu S, Kastan M B, Samid D, Gore S D. Leukemia. 1999;13:1243–1253. doi: 10.1038/sj.leu.2401471. [DOI] [PubMed] [Google Scholar]
- 16.Richon V M, Russo P, Venta-Perez G, Cordon-Cardo C, Rifkind R A, Marks P A. Cancer Res. 1997;57:2789–2798. [PubMed] [Google Scholar]
- 17.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 18.Burnette W N. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 21.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Richon V M, Ngo L, Rifkind R A, Marks P A. Gene. 1999;233:13–19. doi: 10.1016/s0378-1119(99)00170-5. [DOI] [PubMed] [Google Scholar]
- 23.Mano H, Mori R, Ozawa T, Takeyama K, Yoshizawa Y, Kojima R, Arao Y, Masushige S, Kato S. J Biol Chem. 1994;269:1591–1594. [PubMed] [Google Scholar]
- 24.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 25.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebs J E, Kuo M H, Allis C D, Peterson C L. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]