Abstract
Patten recognition receptors, which recognize pathogens or components of injured cells (danger), trigger activation of the innate immune system. Whether and how the host distinguishes between danger- versus pathogen-associated molecular patterns remains unresolved. We report that CD24-deficient mice exhibit increased susceptibility to danger- but not pathogen-associated molecular patterns. CD24 associates with high mobility group box 1 (HMGB1), heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90), negatively regulates their stimulatory activity and inhibits nuclear factor-kappa B (NF-κB) activation. This occurs at least in part through CD24 association with Siglec-10 in humans or Siglec-G in mice. Our results reveal that the CD24-Siglec G pathway protects the host against a lethal response to pathological cell death and discriminates danger- versus pathogen-associated molecular patterns.
Pathogen-associated molecular patterns (PAMPs) interact with Toll-like receptors (TLR) on innate immune cells to initiate protective immune responses (1-3). Danger-associated molecular patterns (DAMPs) (4), which are intracellular components such as HMGB1, HSP70, HSP90 and cellular RNA released during cellular injury, also induce TLR-dependent inflammatory responses (5-8). Whether the host is able to discriminate between DAMPs and PAMPs is not clear.
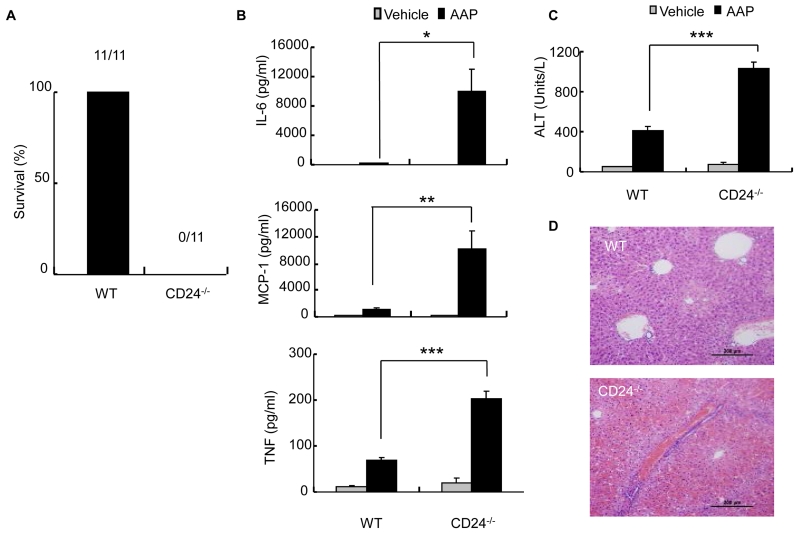
We used an acetaminophen (AAP)-induced liver necrosis model (9) to identify genes that regulate the innate immune response resulting from tissue injury. A sublethal dose of AAP (10 mg/mouse), which is tolerated by wild-type (WT) mice, caused rapid death of CD24-deficient (CD24-/-) mice within 20 hours (Fig. 1A). We then tested whether CD24 regulated the inflammatory response to AAP-induced liver injury because CD24 is expressed on liver oval cells and hematopoeitic cells, but not hepatocytes (10). Indeed, we detected a massive increase in the inflammatory cytokines interleukin-6 (IL-6), monocyte chemotactic protein-1 (MCP-1) and tumor necrosis factor-alpha (TNF-α) after AAP treatment (Fig. 1B). This was accompanied by increased amounts of serum alanine transaminase (ALT), which is indicative of liver damage (Fig. 1C), and liver hemorrhage and necrosis (Fig. 1D). These observations revealed that CD24 protects against AAP-induced hepatoxicity, most likely by regulating the inflammatory response.
Fig. 1.
CD24 negatively regulates the immune response to AAP-induced liver injury. CD24-/- mice or WT mice were treated with AAP (10 mg/mouse, dissolved in H20) or vehicle control. (A) Survival of mice 20 hours after treatment. Numbers on graph indicate the number of viable over total mice used per group. All WT mice remained healthy. (B) Serum levels of IL-6, MCP-1 and TNF-α at 6 hours after AAP injection (mean ± SD, n = 5; *P < 0.02, **P < 0.009; ***P < 0.002, student’s t-test). (C) ALT levels measured at 6 hours after treatment (mean ± SD, n = 5; ***P < 0.00004, student’s t-test). Data shown in (B) and (C) have been repeated 2 times. (D) Livers were isolated at 9 hours after treatment. Representative images (20x) of H&E staining are shown (n = 3).
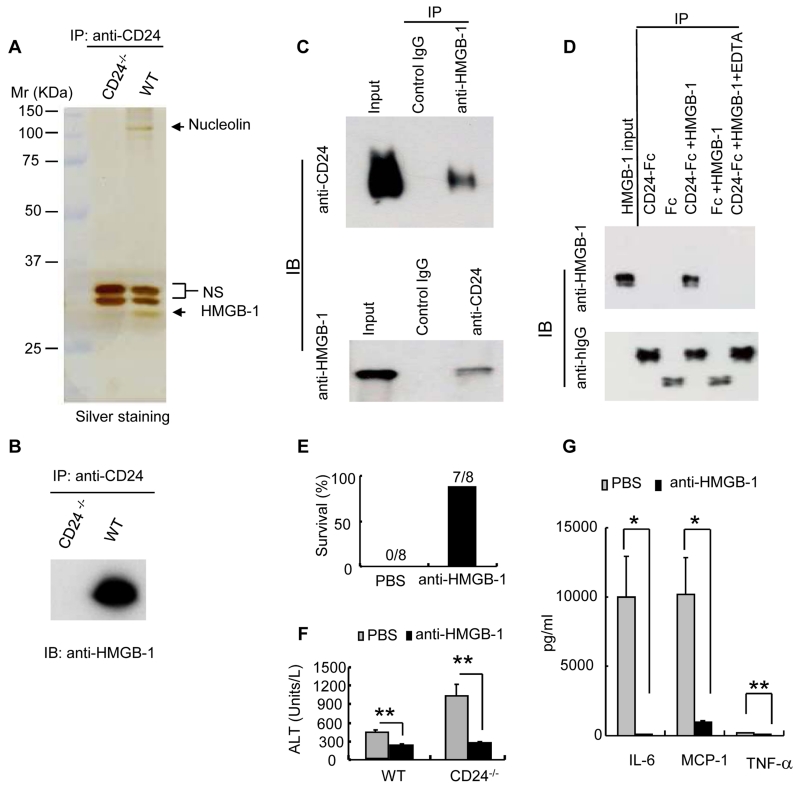
CD24 is a small glycosyl-phosphoinositol-anchored protein that is able to provide costimulatory signals to T cells and has been implicated in the development of autoimmune disease (11-15). We set out to identify proteins that associate with CD24 because none of the known CD24 ligands provided insight into its protective effect in our liver injury model. We focused on proteins whose interactions can be disrupted by the cation chelator, EDTA, because more than 90% of the mass of CD24 is estimated to be derived from glycosylation (12), and because protein-polysaccharide interactions largely depend on cations. Briefly, we immunoprecipitated CD24 and its associated proteins from lysates of mouse splenocytes. The proteins eluted by EDTA were subjected to high throughput mass spectrometry analysis and SDS-PAGE. HMGB1, a prototypical DAMP molecule that activates the immune response following tissue damage (16), was among the most prominent proteins that we identified (Fig. 2A and table S1). HMGB1 coimmunoprecipitated with CD24 and this interaction was specific (Fig. 2B and C). A recombinant CD24-Fc fusion protein specifically coimmunoprecipitated recombinant HMGB1, demonstrating that the interaction between CD24 and HMGB1 was direct (Fig. 2D).
Fig. 2.
CD24 associates with, and negatively regulates the immune response to HMGB1. (A) Identification of CD24-associated proteins by coimmunoprecipitation. Silver-staining of the SDS-PAGE gel is shown. Arrows indicate the positions of HMGB1 and nucleolin, two abundant CD24-associated DAMP molecules. NS: proteins that coimmunoprecipitated with anti-CD24 non-specifically. (B) Confirmation of CD24-HMGB1 association by Western blot of EDTA-disassociated proteins. (C) Reciprocal immunoprecipitations of CD24 and HMGB1 were performed with splenocyte lysates isolated from WT mice. (D) Direct, cation-dependent interaction between CD24 and HMGB1. Coimmunoprecipitation of recombinant HMGB1 protein with CD24-Fc fusion protein or control IgG-Fc. The requirement for cations was confirmed by disruption of the complex with EDTA. This experiment has been repeated 3 times. (E) Mice received i.v. injections with either vehicle (PBS) or mouse HMGB1 mAb (clone 3B1, 150 μg/mouse) 30 minutes prior to i.p. injection of AAP. Composite data from two independent experiments are shown (n = 8). (F) Serum ALT at 6 hours after treatment with AAP and HMGB1 antibodies (mean ± SD, n = 5, **P < 0.005). (G) Serum cytokine levels at 6 hours after treatment with AAP and HMGB1 antibodies (mean ± SD, n = 5, *P, 0.03, **P < 0.004). Samples in (F) and (G) represent two independent experiments, the statistical significance determined by student’s t-test.
To determine whether the hypersensitivity to AAP observed in CD24-/- mice was the result of an enhanced immune response to HMGB1, we injected AAP-treated mice with antibodies to HMGB1 (fig. S1). In one representative experiment, blockade of HMGB1 rescued 87.5% of the mice that received AAP (Fig. 2E). Treated mice exhibited decreased ALT abundance, indicating reduced hepatocyte destruction (Fig. 2F). The production of IL-6, MCP-1 and TNF-α were also greatly reduced (Fig. 2G). Thus, CD24 protects against AAP-induced lethal hepatoxicity by dampening the immune response against HMGB1.
HMGB1 can be divided into two domains: an inhibitory A box and a stimulatory B box (17). To determine whether CD24 inhibits HMGB1 by binding to the inhibitory A box, we produced deletion mutants lacking either the A box or the B box. CD24-Fc immunoprecipitated full-length HMGB1 and the box B-containing mutant, but not the box A-containing mutant (fig. S2). Thus, inhibition of HMGB1 by CD24 does not require direct interaction with box A.
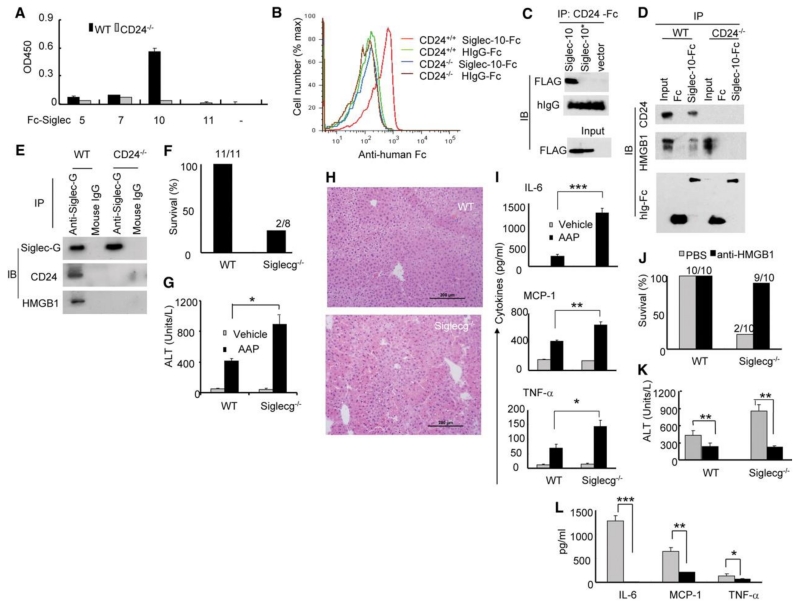
CD24 has no known mechanism for signal transduction. In order to understand how CD24 negatively regulates HMGB1, we searched for a potential CD24 receptor that may transduce signals downstream of CD24. We were particularly interested in sialic acid-binding Ig-like lectins (Siglecs), which are cell surface receptors of the immunoglobulin super-family that recognize sialic acid-containing proteins (18). Siglecs are primarily expressed by cells of hematopoietic origin (18). Most Siglecs are considered to be negative regulators of the immune system because they contain one or more cytosolic immune receptor tyrosine-based inhibitory motifs (ITIMs) (18). To determine whether CD24 interacts with Siglecs, we incubated splenocytes on plates coated with the recombinant extracellular domains of ITIM-containing Siglec-5, -7, -10 or -11. Siglec-10, but not Siglecs -5, -7 or -11, bound to CD24 (Fig. 3A). Flow cytometic analysis indicated that CD24 is the primary receptor for Siglec-10 because WT but not CD24-/- splenocytes showed detectable binding to soluble Siglec-10-Fc (Fig. 3B). Furthermore, in COS cells, FLAG-tagged Siglec-10 coimmunoprecipitated with Siglec-10-Fc whereas the inactivating R119A mutation of Siglec-10 (analogous to the R97A in sialoadhesin (19)), abrogated the interaction (Fig. 3C).
Fig. 3.
The Siglec 10/G-CD24-HMGB1 axis negatively regulates immune responses to AAP-induced liver injury. (A) Interaction between CD24 and Siglec-Fc fusion proteins. Data shown are optical density and have been repeated 3 times. (B) Flow cytometric analysis of CD24 interaction with Siglec-10. Representative histograms of two independent experiments are shown. (C) COS cells were transfected with FLAG-tagged WT or mutant (*, R119A) Siglec-10 cDNA or a vector control. Coimmunoprecipitations were performed 48 hours later. (D) Lysates from WT or CD24-/- splenocytes were used to coimmunoprecipitate Siglec-10-Fc, CD24 and HMGB1. (E) Lysates from WT and CD24-/- spleen cells were precipitated with either Siglec-G antibodies or control mouse Ig. The precipitates were probed with Siglec-G antisera and mAbs specific for CD24 and HMGB1. (F) Percent survival 20 hours after AAP treatment. Numbers on graph represent the number of surviving mice over total mice used. (G) ALT release in serum 6 hours after AAP treatment (mean ± SD, *P < 0.005, n=5). (H) 20x images of H&E staining of livers harvested 6 hours after AAP injection. (I) Cytokine production in blood measured 6 hours after AAP treatment (mean ± SD, n=5. *P <0.05, **P < 0.009, ***P < 0.002). (J) Survival of WT and Siglecg-/- mice 20 hours after treatment. (K) ALT release in the blood 6 hours after treatment (mean ± SD, n = 5, *P < 0.006). (L) Cytokine release in the blood 6 hours after treatment (mean ± SD, n = 5, *P < 0.03, **P < 0.0006, ***P < 0.0004). (K-L) are representative of two independent experiments. Statistical significance was determined by the student’s t-test.
We hypothesized that CD24, Siglec-10 and HMGB1 might form a tri-molecular complex because CD24 can interact with both HMGB1 and Siglec-10. Indeed, Siglec-10-Fc was able to immunoprecipitate HMGB1 from lysates of WT but not CD24-/- splenocytes (Fig. 3D), indicating that their interaction was strictly dependent on CD24 expression.
The likely murine homologue of Siglec-10 is Siglec-G (18). We prepared anti-Siglec-G anti-sera by immunizing Siglecg-/- mice (20) with WT spleen cells (fig. S3). Using this antisera, Siglec-G coimmunoprecipitated CD24 (Fig. 3E). CD24-Fc showed stronger binding to WT splenocytes in comparison to Siglecg-/- splenocytes, indicating that Siglec-G contributed to CD24-Fc binding; however, consistent with previous reports of multiple CD24 receptors (12), Siglec-G-deficiency did not abrogate CD24-Fc splenocyte binding (fig. S4). We next determined if the absence of Siglec-G would also convey hypersensitivity to AAP. Indeed, only 25% of Siglecg-/- mice survived a sublethal dose of AAP (Fig. 3F). The enhanced susceptibility was accompanied by increased release of ALT (Fig. 3G), liver necrosis and hemorrhage (Fig. 3H), as well as increased amounts of inflammatory cytokines in the blood (Fig. 3I). To test whether the enhanced liver toxicity was mediated by HMGB1, we treated Siglecg-/- mice with antibodies to HMGB1. Inhibition of HMGB1 prevented mortality in 90% of AAP-treated Siglecg-/- mice (Fig. 3J). Serum ALT and inflammatory cytokines were also largely diminished (Fig. 3K and L).
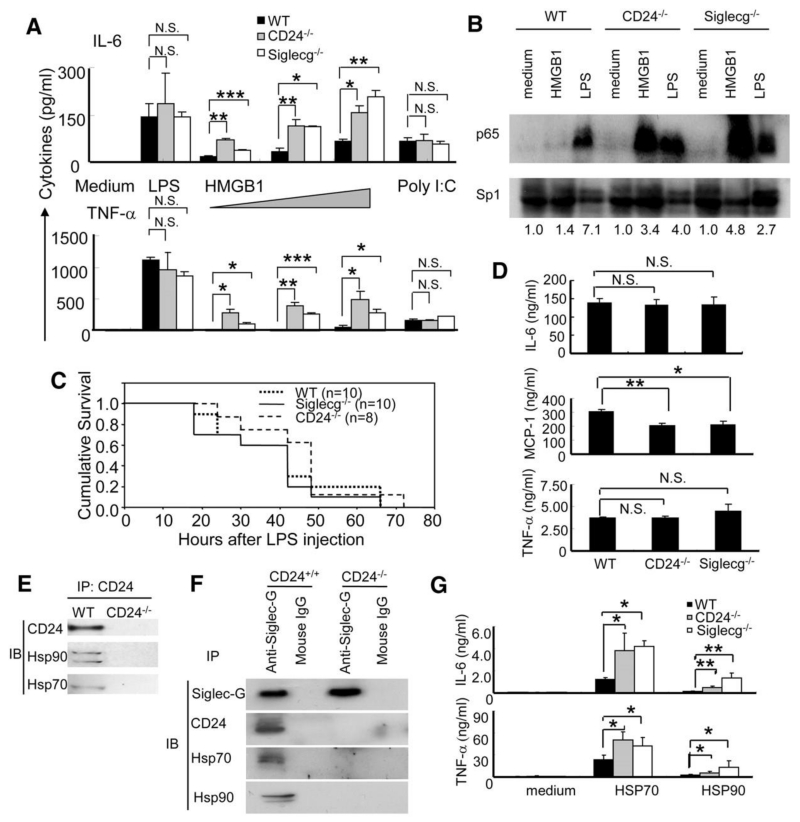
CD24 and Siglec-10 are unlikely to function by acting directly on hepatocytes because they are not expressed by these cells (10, 18). Dendritic cells (DCs), however, respond to HMGB1 (21) and express both CD24 (22) and Siglec-G (20). To test whether DCs can respond to HMGB1, we cultured bone marrow-derived DCs isolated from WT, CD24-/- or Siglecg-/- mice and stimulated them with HMGB1 or the TLR ligands LPS or poly I:C. HMGB1 stimulation resulted in significantly greater production of IL-6 and TNF-α by CD24-/- or Siglecg-/- DC than by WT DC (Fig. 4A). In contrast, CD24 or Siglec-G-deficiency did not affect the production of inflammatory cytokines by DCs in response to LPS or poly I:C (Fig. 4A).
Fig. 4.
CD24 and Siglec-G negatively regulate immune responses to HMGB1, HSP70 and HSP90, but not to LPS and poly I:C. (A) Production of cytokines by DCs. DCs cultured from WT, CD24-/- or Siglecg-/- bone marrow were stimulated with LPS (100 ng/ml), polyI:C (10 μg/ml) or increasing doses (5, 10 and 20 μg/ml) of HMGB1 for 6 hours, the supernatants were analyzed for the levels of inflammatory cytokines, using cytokine beads array. Data represents the mean ± SD for three independent cultures of DCs in each genotype and have been repeated at least three times. (B) BMDCs isolated from WT, CD24-/- or Siglecg-/- mice were stimulated under the indicated conditions for 6 hours. The nuclear lysates were prepared and the activation of NF-κB was assessed by blotting for the p65 subunit of NF-κB. The loading of nuclear protein was determined by amounts of Sp1 protein. Fold induction over medium control are provided underneath the photograph. Data are representative of two independent experiments. (C) Age-matched male mice received i.p. injections of LPS (450μg/mouse). Kaplan Meier survival plots are shown. No statistical significance was found by log-rank tests. (D) Cytokine production in the serum 4 hours after LPS injection (mean ± SD, the statistical significance of the differences between the control and one of the treated groups were determined by student’s t-test. *P < 0.03, **P < 0.002). The numbers of mice used are the same as (C). (E) Coimmunoprecipitation of CD24 and Hsp70 and Hsp90. (F) Siglec-G associates with Hsp70 and Hsp90 through CD24. The same precipitates used in Fig. 3E were analyzed for Hsp70 and Hsp90 by immunoblot. (G) Deficiencies in CD24 and Siglec-G enhanced production of IL-6 and TNF-α at 6 hours after stimulation with HSP70 and HSP90. Data shown represent the mean ± SD of cytokines from 4 independent isolates of DCs from each genotype and have been repeated twice.
Siglec-10 associates with the tyrosine phosphatase SHP-1, a known negative regulator of NF-κB activation (23). In a subpopulation of B cells that reside in the peritoneum (20), the absence of Siglec-G results in the constitutive activation of NF-κB. To test whether activation of NF-κB by HMGB1 or LPS is affected by the absence of CD24 or Siglec-G, we assayed the nuclear translocation of the NF-κB subunit p65 in WT, CD24-/- and Siglecg-/- DCs. Both LPS and to a much lesser extent, HMGB1, induced nuclear translocation of p65 in WT DCs; however, in CD24 or Siglecg-deficient DCs, HMGB1 caused even greater increases in nuclear translocation of p65 than did LPS (Fig. 4B). These data suggest that the CD24-Siglec-G pathway may serve to decrease the host response to DAMPs, such as HMGB1, but not to TLR ligands of microbial origin (PAMPs), by selective repression of NF-κB.
To substantiate this hypothesis, we administered a lethal dose of LPS to WT, CD24-/- or Siglecg-/- mice. Neither the absence of Siglec-G nor CD24 affected the kinetics of LPS-induced lethality (Fig. 4C) or production of inflammatory cytokines (Fig. 4D). Despite an established contribution of HMGB1 to the late stage of sepsis (24), potential amplification of HMGB1 signaling by mutation of CD24 or Siglecg did not affect host survival in response to LPS. Therefore, CD24 and Siglec-G are selective modulators of the host response to HMGB1, but not to TLR ligands such as LPS, despite their potential to induce release of HMGB1 (24, 25).
In addition to nuclear DAMPs, such as HMGB1, DCs also respond to cytoplasmic DAMPs such as HSP70 and 90 by TLR-dependent mechanisms (6). To determine if the CD24-Siglec-G pathway also regulates host responses to HSP70 and 90, we first evaluated whether HSP70 and 90 associate with CD24 and Siglec-G. Coimmunoprecipitations revealed that CD24 associates with both HSP70 and HSP90 (Fig. 4E). Similar to HMGB1, Siglec-G association with HSP70 and HSP90 was CD24-dependent (Fig. 4F) and CD24-/- and Siglecg-/- DCs produced significantly higher IL-6 and TNF-α in response to recombinant HSP70 and HSP90 (Fig. 4G) compared to WT DCs. These data reveal a critical role for CD24 and Siglec-G in negative regulation of DC response to multiple DAMPs.
Our results suggest that CD24 partners with Siglec-10 in humans or Siglec-G in mice to negatively regulate the immune response to proteins released by damaged cells, but not to ligands of microbial origin. Pattern recognition receptors such as TLRs and RAGE mediate activation induced by DAMP (7, 8). Our data indicate that repression of response to HMGB1 may be achieved by inhibition of NF-κB activation. Inhibition may be mediated by SHP-1. SHP-1 associates with Siglec-10 via its ITIM motif (26) and deficiency of either Siglec-G or SHP-1 enhances NF-κB activation (20, 23). Given the role of HMGB1 in the pathogenesis of a number of diseases, including drug toxicity (9) and liver and cardiac ischemia and reperfusion (27, 28), this pathway may uncover new targets for disease intervention.
Although it is well established that the host can recognize “danger” induced by damaged tissue (4), it is unclear whether or how an immune responses triggered by tissue damage is regulated. By identifying the CD24-Siglec-G pathway that selectively suppresses the immune response to DAMPs, our data demonstrate a mechanism by which tissue injury and infection are distinguished, even though they both use the evolutionally conserved TLR (5-8).
Supplementary Material
Acknowledgments
This study is supported by grants from the National Institute of Health (AI064350, CA58033 and CA112001), US department of Defense (W81XWH-08-1-0036) and those from Natural Science Foundation of China (30640420558) and Ministry of Science and Technology of China (2006CB910901).
Footnotes
The authors have no conflict of interest. We dedicate this study to Dr. Charles A. Janeway, Jr.
References and Notes
- 1.Janeway CA. Cold Spring Harbor Symposia on Quantitative Biology. 1989;54:1. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Janeway CA., Jr. International Immunology. 1991;3:323. doi: 10.1093/intimm/3.4.323. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. Nature. 1997 Jul 24;388:394. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 4.Matzinger P. Annual Review of Immunology. 1994;12:991. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 5.Cavassani KA, et al. Journal of Experimental Medicine. 2008 Oct 27;205:2609. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millar DG, et al. Nature Medicine. 2003 Dec;9:1469. doi: 10.1038/nm962. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, et al. Journal of Biological Chemistry. 2004 Feb 27;279:7370. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 8.Tian J, et al. Nat Immunol. 2007 May;8:487. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 9.Scaffidi P, Misteli T, Bianchi ME. Nature. 2002 Jul 11;418:191. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 10.Ochsner SA, et al. Stem Cells. 2007 Oct;25:2476. doi: 10.1634/stemcells.2007-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, et al. Journal of Experimental Medicine. 1992;175:437. doi: 10.1084/jem.175.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Zheng P. Trends Immunol. 2007 Jul;28:315. doi: 10.1016/j.it.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Bai XF, et al. Journal of Clinical Investigation. 2000;105:1227. doi: 10.1172/JCI9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. PLoS Genetics. 2007;3:e49. doi: 10.1371/journal.pgen.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, et al. Proceedings of the National Academy of Sciences of the United States of America. 2003 Dec 9;100:15041. doi: 10.1073/pnas.2533866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris HE, Raucci A. EMBO Rep. 2006 Aug;7:774. doi: 10.1038/sj.embor.7400759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, et al. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jan 6;101:296. [Google Scholar]
- 18.Crocker PR, Paulson JC, Varki A. Nat Rev Immunol. 2007 Apr;7:255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 19.Vinson M, et al. Journal of Biological Chemistry. 1996 Apr 19;271:9267. doi: 10.1074/jbc.271.16.9267. [DOI] [PubMed] [Google Scholar]
- 20.Ding C, et al. PLoS ONE. 2007;2:e997. doi: 10.1371/journal.pone.0000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apetoh L, et al. Nature Medicine. 2007 Sep;13:1050. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 22.Li O, et al. Journal of Experimental Medicine. 2006 Jul 10;203:1713. doi: 10.1084/jem.20052293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pao LI, et al. Immunity. 2007 Jan;27:35. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, et al. Science. 1999 Jul 9;285:248. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 25.Abraham E, Arcaroli J, Carmody A, Wang H, Tracey KJ. Journal of Immunology. 2000 Sep 15;165:2950. doi: 10.4049/jimmunol.165.6.2950. [DOI] [PubMed] [Google Scholar]
- 26.Whitney G, et al. European Journal of Biochemistry. 2001 Dec;268:6083. doi: 10.1046/j.0014-2956.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 27.Andrassy M, et al. Circulation. 2008 Jun 24;117:3216. doi: 10.1161/CIRCULATIONAHA.108.769331. [DOI] [PubMed] [Google Scholar]
- 28.Tsung A, et al. Journal of Experimental Medicine. 2005 Apr 4;201:1135. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.