Abstract
Background
Genetic and developmental disruption of surfactant protein B (SP-B) expression causes neonatal respiratory distress syndrome (RDS). Objectives: To assess developmental and genetic regulation of SP-B expression in vivo.
Methods
To evaluate in vivo developmental regulation of SP-B, we used immunoblotting to compare frequency of detection of mature and pro-SP-B peptides in developmentally distinct cohorts: 24 amniotic fluid samples, unfractionated tracheal aspirates from 101 infants ≥34 weeks’ gestation with (75) and without (26) neonatal RDS, and 6 nonsmoking adults. To examine genetic regulation, we used univariate and logistic regression analyses to detect associations between common SP-B (SFTPB) genotypes and SP-B peptides in the neonatal RDS cohort.
Results
We found pro-SP-B peptides in 24/24 amniotic fluid samples and in 100/101 tracheal aspirates from newborn infants but none in bronchoalveolar lavage from normal adults (0/6) (p < 0.001). We detected an association (p = 0.0011) between pro-SP-B peptides (Mr 40 and 42 kDa) and genotype of a nonsynonymous single nucleotide polymorphism at genomic position 1580 that regulates amino-terminus glycosylation.
Conclusions
Pro-SP-B peptides are more common in developmentally less mature humans. Association of genotype at genomic position 1580 with pro-SP-B peptides (Mr 40 and 42 kDa) suggests genetic regulation of amino terminus glycosylation in vivo.
Key Words: Neonatal respiratory diseases, Respiratory distress syndrome, Surfactant proteins
Introduction
The pulmonary surfactant is a phospholipid-protein complex synthesized by alveolar type II cells that lowers surface tension and maintains alveolar expansion at end-expiration [1]. A developmentally regulated, quantitative deficiency of pulmonary surfactant phospholipids results in surfactant dysfunction and respiratory distress syndrome (RDS) in premature newborns with increasing risk and severity associated with decreasing gestational age [2, 3]. However, the importance of genetic regulation of surfactant function has been suggested by increased risk of respiratory distress in males and European-descent infants and by disruption of pulmonary surfactant function by completely penetrant, recessive, loss of function mutations in the surfactant protein B gene (SFTPB)[4,5,6,7,8].
Surfactant protein B (SP-B) is a lung-specific, hydrophobic peptide [9] encoded in a 9.7-kb gene (SFTPB; GeneID: 6439; Locus tag: HGNC:10801; MIM: 178640) that directs the synthesis of a 381-amino-acid, 40-kDa pre-pro-protein. Pro-SP-B undergoes co-translational, carboxy-terminus glycosylation, may undergo amino-terminus glycosylation depending upon the presence of a nonsynonymous single nucleotide polymorphism (SNP) in exon 4, SFTPB genomic position (g.) 1580, and is sequentially proteolytically cleaved at amino and carboxy termini to yield the 79-amino-acid mature SP-B peptide [10,11,12].
Developmental regulation of SP-B expression has been documented in human amniotic fluid samples and immunohistochemical studies of human fetal lungs [13,14,15]. Although the amino- and carboxy-terminus cleavage fragments of pro-SP-B have also been detected in amniotic fluid and bronchoalveolar lavage fluid, the functions of these processing fragments are unknown [10, 11, 16]. Studies in mice that express truncated SP-B have demonstrated the importance of SP-B processing and itinerary for function of the pulmonary surfactant [17,18,19].
Genetic regulation of SP-B expression and surfactant function has been demonstrated in human newborn infants with rare, recessive, loss of function SFTPB mutations, in murine lineages with conditionally regulated SFTPB expression, and in cell culture systems with a nonsynonymous SNP at SFTPB g. 1580 [20,21,22]. To characterize developmental and genetic regulation of SP-B expression in vivo, we compared the frequencies of mature and pro-SP-B peptides in developmentally distinct cohorts and interrogated associations between common SFTPB genotypes and SP-B peptides in term and near-term infants with and without neonatal RDS.
Materials and Methods
Patient Populations and Sample Acquisition
To evaluate developmental regulation of SP-B, we obtained 24 archived, anonymized amniotic fluid samples from pregnancies at gestational ages ranging from 33 to 39 weeks (mean 37 ± 2 weeks gestation), tracheal aspirates from a neonatal RDS cohort which included infants ≥34 weeks gestation with (n = 75; obtained >24 h after the last dose of surfactant replacement therapy) and without (n = 26) neonatal RDS (median 5 days, range 1–222 days, chronological age; table 1), and bronchoalveolar lavage samples from 6 nonsmoking adult volunteers (the gift of Tomoko Betsuyaku, MD, Hokkaido University School of Medicine, Kita-ku, Japan). Tracheal aspirate sample acquisition and processing were performed as previously described [23, 24]. Infants with RDS had abnormal chest radiographs and need for mechanical ventilation and supplemental oxygen, while those without RDS required mechanical ventilation for nonpulmonary reasons with normal chest radiographs and no requirement for supplemental oxygen. To confirm that SP-B peptide detection is independent of sample acquisition site, we compared SP-B peptides in 6 simultaneously acquired tracheal aspirate and BAL fluid specimens from older children who were undergoing diagnostic bronchoscopy (fig. 1).
Table 1.
Neonatal RDS cohort characteristics
| RDS (n = 75) | Control (n = 26) | p value | |
|---|---|---|---|
| Race | |||
| African descent | 14 (19%) | 5 (19%) | 0.9 |
| European descent | 61 (81%) | 21 (81%) | |
| Gender | 0.3 | ||
| Male | 47 (63%) | 19 (73%) | |
| Female | 28 (37%) | 7 (27%) | |
| Gestational age, weeks1 | 37±2 | 37±1.5 | 0.4 |
| Birth weight, kg1 | 3.0±0.7 | 3.0±0.6 | 0.9 |
| Age at tracheal aspirate | |||
| acquisition, days2 | 4 (1–48) | 6 (2–222) | 0.008 |
| Duration of mechanical | |||
| ventilation, days2 | 6 (1−193) | 0 (0−55) | <0.001 |
| Duration of supplemental | |||
| oxygen, days2 | 8 (1−193) | ||
| Survived/died | 67/4 | 23/0 | 0.2 |
p values of χ2 tests of RDS vs. control.
Mean ± SD.
Median (range).
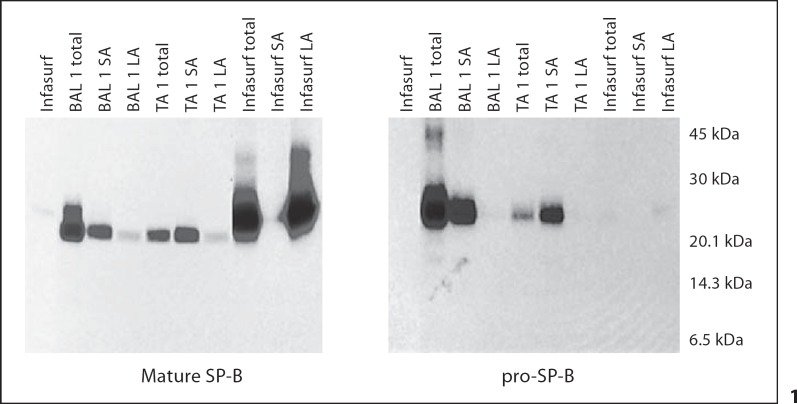
Fig. 1.
Western blots of mature and pro-SP-B in the unfractionated (total), large aggregate (pellet; LA), and small aggregate (supernatant; SA) surfactant fractions from simultaneously obtained trachéal aspirate (TA) and bronchoalveolar lavage (BAL) specimens from 1 older child. Mature and pro-SP-B peptides were similar in both the BAL and TA: mature SP-B was present in both SA fraction and the surface active LA fraction, while pro-SP-B was present in the SA fraction only. In Infasurf, only mature SP-B was detected, and only in the LA fraction.
To evaluate genetic regulation of SFTPB expression, we obtained DNA from peripheral blood white cells from infants in the neonatal RDS cohort (n = 101). In view of the limited statistical power of the RDS cohort, we used a replication cohort (225 race and gestational age-matched anonymized DNA samples, 3 matched to each infant with RDS, from infants without RDS in a previously reported cohort) [25] to interrogate SFTPB genotype associations with RDS. The Washington University Human Research Protection Office approved this study.
Study Design and Methods
Developmental Analysis and Immunoblotting
To evaluate developmental regulation of SP-B in vivo, we used immunoblotting of samples on nitrocellulose membranes transferred after SDS polyacrylamide gel electrophoresis under nonreduced and reduced (2–10% β-mercaptoethanol) conditions as previously described [19]. We probed first with anti-serum to mature SP-B (Chemicon, Inc., Temecula, Calif., USA; No. AB3780), and, secondly, after stripping nitrocellulose membranes using Re-Blot Plus Strong Antibody Stripping Solution (Chemicon No. 2504), reprobed with primary anti-serum to pro-SP-B (Chemicon No. AB3430). We confirmed specificity of mature SP-B peptide detection by abrogation of mature peptide signal after preincubation of the primary antibody with excess purified human SP-B (the gift of T.E. Weaver from the University of Cincinnati) and specificity of pro-SP-B antibody by failing to detect any peptides in purified human SP-B or in Infasurf® (fig. 2). We further confirmed mature and pro-SP-B antibody specificity by failing to detect mature or pro-SP-B peptides with nonimmune serum or in tracheal aspirate or BAL samples from infants with genetic SP-B deficiency (data not shown). Each sample was adjusted to contain a mean of 3.0 ng (±2.6 ng SD) of SP-B (median 2.6 ng, range 0.2–18.4 ng) based on prior slot blot analysis with antibody to mature SP-B. Immunoblotting of amniotic fluid, tracheal aspirate, and bronchoalveolar lavage samples fractionated by high-speed centrifugation to separate surface active components (27,000 g for 60 min) [24] revealed SP-B peptides in both small aggregate and large aggregate fractions (fig. 1). To ensure comprehensive detection of SP-B peptides, we performed immunoblotting on unfractionated samples.
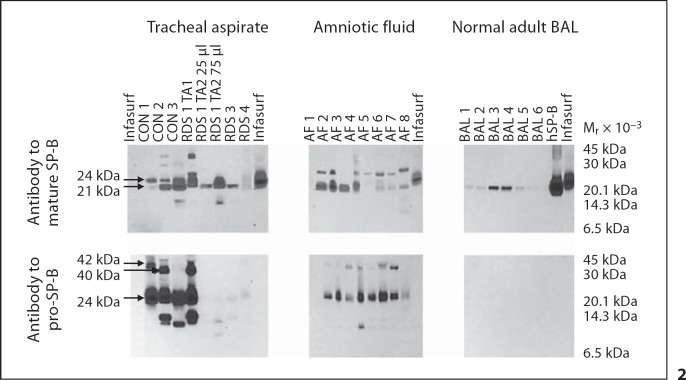
Fig. 2.
Mature and pro-SP-B peptides from trachéal aspirate, amniotic fluid, and normal adult bronchoalveolar lavage (BAL) under nonreducing conditions. Immunoblots of TA samples from 3 newborn infants with RDS (RDS 1, 3, 4), 3 control infants (CON 1, 2, 3), 8 amniotic fluid samples (AF 1-8), 6 BAL specimens from adults (BAL 1-6), purified human SP-B (hSP-B), and Infasurf were probed with antibody to mature or pro-SP-B. Locations of common mature and pro-SP-B peptides (Mr 42, 40, 24, and 21 kDa) are indicated. TA samples from RDS 1 obtained 1 week apart (TA1 vs. TA2) demonstrate changes in mature and pro-SP-B peptide mobilities over time in the same infant. Antibody to pro-SP-B does not detect any pro-SP-B peptides in purified human SP-B or in Infasurf.
Genetic Analysis and Sequencing
To evaluate genetic regulation of SFTPB expression in vivo, we prepared genomic DNA from blood samples of the neonatal RDS cohort, amplified and resequenced SFTPB (g. −880 in the 5′ promoter region through g. 7022 in intron 10), and assembled, analyzed, and edited chromatograms as previously described [25]. We omitted 380 bps in intron 4 due to inability of BigDye terminator sequencing chemistry to resolve multiple CA dinucleotide repeats in this region [26] as well as intron 10, untranslated exon 11, and the 3′ untranslated region.
Analysis
We linked all clinical, genotype, and immunoblot data and performed data analyses using the Statistical Analysis Package (SAS version 9.1.3 Service Pack 4; SAS Institute, Cary, N.C., USA). We used χ2 analysis to compare detected allele frequencies with predictions by the Hardy-Weinberg equilibrium. We used exact logistic regression to assess associations between SFTPB variants and SP-B peptides in the entire neonatal RDS cohort and between SFTPB variants and RDS status separately in African- and European-descent infants and included gestational age and gender as covariates. We used the Bonferroni method to correct for multiple independent testing and p values ≤0.01 to declare significant statistical inferences. To achieve sufficient statistical power (>0.8) to detect associations between SFTPB variants and specific SP-B peptides and also to detect a minimum twofold difference in allele frequencies between RDS and control infants, we analyzed associations between 5 SNPs with minor allele frequencies (MAFs) ≥0.3 and the 6 most commonly detected (frequencies >0.4) SP-B peptides.
Results
Developmental Regulation of SP-B Peptide Expression
To assess developmental regulation of SP-B in vivo, we compared frequencies of SP-B peptides in developmentally distinct groups. We detected both mature and pro-SP-B peptides (Mr 10–42 kDa) in all 24 amniotic fluid samples and in 100 of 101 tracheal aspirate samples from the RDS cohort (fig. 2; table 2). The diverse SP-B peptides detected in tracheal aspirates did not result from incompletely metabolized SP-B after surfactant replacement therapy: these peptides were detected by antibodies against both mature and pro-SP-B, while the antibody to pro-SP-B did not detect any peptides from Infasurf (fig. 2). In contrast to the amniotic fluid and tracheal aspirate samples, we detected only mature SP-B peptides and no pro-SP-B peptides in the 6 bronchoalveolar samples from adults (fig. 2; probability of observed distribution by chance p < 0.0001; Fisher's exact test). We also found that presence of each of the 6 most common SP-B peptides was significantly associated with gestational age (p < 0.01). These observations suggest that proteolytic processing of pro-SP-B in vivo differs in developmentally distinct cohorts. To assess the possibility that mobilities of mature or pro-SP-B peptides resulted from disulfide mediated multimers or postacquisition proteolysis, we used reducing conditions (2–10% β-mercaptoethanol) or incubation at room temperature or 37°C for 1, 3, and 6 h. While purified, human SP-B migrated with Mr of 21 kDa under nonreducing and Mr of 8 kDa under reducing conditions, most mature and pro-SP-B peptides from tracheal aspirates failed to resolve to an 8-kDa peptide (fig. 3). Failure of mature SP-B peptides to resolve to 8 kDa suggests that mature SP-B peptide multimerization in unfractionated tracheal aspirates is not mediated via disulfide bonds or that inter-SP-B disulfide bonds are resistant to reduction in unfractionated tracheal aspirates. In addition, incubation of tracheal aspirates at varying time intervals failed to alter Mr of the pro-SP-B or mature SP-B peptides (data not shown), an observation that suggests that postacquisition proteolysis did not contribute to the diversity of SP-B peptides we detected.
Table 2.
Frequencies of all detected SP-B peptides
| SP-B peptide | CON |
RDS |
||
|---|---|---|---|---|
| AD (n = 5) | ED (n = 21) | AD (n = 14) | ED (n = 61) | |
| M42 | 2 | 7 | ||
| M40 | 1* | 4 | 3 | 8 |
| M38 | 1 | 1 | ||
| M36 | 2 | |||
| M34 | 1 | 1 | 1 | 2 |
| M32 | 1 | 3 | 3 | |
| M30 | 1 | 2 | ||
| M29 | 2 | 3 | ||
| M28 | 2 | 3 | 12 | |
| M27 | 1 | 2 | 8 | |
| M26 | 1 | 1 | 3 | |
| M25 | 2 | 1 | 3 | |
| M24 | 2 | 15 | 10 | 55 |
| M23 | 1 | 6 | ||
| M22 | 1 | |||
| M21 | 5 | 21 | 9 | 44 |
| M19 | ||||
| M17 | 1 | 1 | 4 | |
| M16 | 1 | 5 | ||
| M15 | 1 | 1 | 2 | |
| M14 | 1 | 1 | 1 | |
| M13 | 1 | 1 | ||
| P42 | 1 | 6 | 6 | 28 |
| P40 | 2 | 10 | 7 | 26 |
| P36 | 1 | |||
| P28 | 2 | |||
| P27 | 1 | |||
| P26 | 1 | |||
| P25 | 2 | 1 | ||
| P24 | 4 | 21 | 14 | 59 |
| P21 | 1 | 1 | ||
| P20 | 1 | 2 | 15 | |
| P19 | 1 | 2 | 2 | 18 |
| P18 | 1 | 2 | ||
| P17 | 5 | 13 | 3 | 28 |
| P16 | 1 | 1 | 2 | |
| P15 | 3 | 14 | 8 | 39 |
| P13 | 1 | 1 | ||
| P12 | 1 | 3 | ||
| P11 | 1 | 2 | 4 | |
| P10 | 2 | |||
SP-B peptides are designated by M (detected by antibody to mature SP-B) or P (detected by antibody to pro-SP-B) and their molecular mass (Mr) in kDa.
CON = Control infants;
RDS = RDS infants;
AD = African descent;
ED = European descent.
* Denotes number of individuals with each peptide.
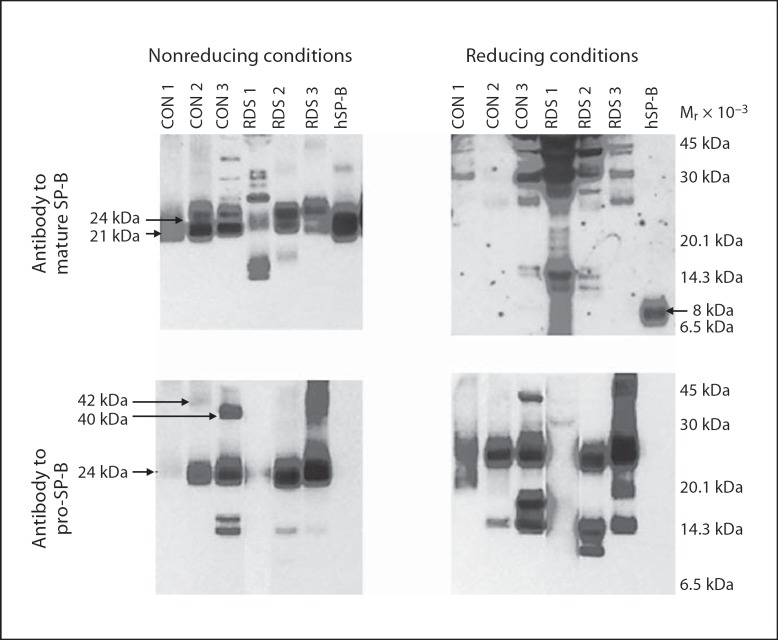
Fig. 3.
Mature and pro-SP-B peptides from trachéal aspirates under nonreducing and reducing conditions. Immunoblots of trachéal aspirates from 3 infants without (CON 1, 2, 3) and with (RDS 1, 2, 3) respiratory distress syndrome and purified hSP-B performed under nonreducing and reducing conditions (10% β-mercaptoethanol). Locations of common mature and pro-SP-B peptides (Mr 42, 40, 24, and 21 kDa) and reduced, mature SP-B (8 kDa) are indicated. Trachéal aspirates contain multiple mature and pro-SP-B peptides that do not resolve to an 8-kDa peptide under reducing conditions.
Genetic Regulation of SP-B Expression
Table 3 displays all polymorphic sites (SNPs and insertion/deletions) identified in the neonatal RDS cohort. To assess genetic regulation of SP-B expression in vivo with adequate statistical power (>0.8) to detect associations between SP-B peptides and SFTPB genotypes, we analyzed the 5 SFTPB genotypes with MAFs ≥0.3 (g. −18, g. 1013, g. 1580, g. 5028, and g. 5796) and 6 of the most commonly detected (frequencies >0.4) SP-B peptides (Mr 15, 17, 21, 24, 40, and 42 kDa) in the RDS cohort (n = 101). Because of small cohort size, overrepresentation of rare SFTPB variants in this cohort (23 of 40 polymorphic sites, 36 SNPs and 4 insertions or deletions, were detected in <3 infants; table 3) and multiple SP-B peptides, we had sufficient statistical power to interrogate only these associations. We found 1 example of genetic regulation of SP-B expression: genotypes (CC vs. CT vs. TT) of the nonsynonymous SNP at g. 1580 were significantly associated with presence of the Mr 42-kDa (p = 0.0011) or absence of the Mr 40-kDa pro-SP-B peptide (p = 0.0011), respectively, as predicted by the loss of a potential N-linked, amino-terminus glycosylation site of pro-SP-B if isoleucine (T allele) is substituted for threonine (C allele) at codon 131 (table 4) [22]. We also observed that the CC genotype at g. 1580 was associated with the absence of 1 pro-SP-B peptide (P15) (table 4). This observation suggests that release of pro-SP-B proteolytic cleavage products may be influenced by the SNP at g. 1580. Although univariate analysis demonstrated that alleles at g. 1013 (p = 0.05, rs3024798) and g. 5796 (p = 0.1, rs2040349) were weakly associated with the expression of the Mr 24-kDa pro-SP-B peptide, logistic regression analyses did not detect association between any other common SFTPB genotype and any common SP-B peptide (table 4).
Table 3.
Location, race-specific MAF, and Hardy-Weinberg frequency comparisons for each SFTPB polymorphic site in the neonatal RDS cohort
| Rs number | Genomic location | Hardy-Weinberg p value |
MAF |
||||
|---|---|---|---|---|---|---|---|
| total | ED | AD | total | ED | AD | ||
| Rs2077079 | −463 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs45443200 | −429 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs3024791 | −384 | 0.9574 | 0.9827 | 0.4002 | 0.165 | 0.142 | 0.263 |
| Rs35955910 | −375 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs2077079 | −18 | 1 | 0.7301 | 0.3382 | 0.391 | 0.396 | 0.368 |
| Rs3024795 | 600 | 1 | 1 | 1 | 0.020 | 0 | 0.111 |
| Rs3024797 | 750 | 1 | 1 | 1 | 0.101 | 0.098 | 0.118 |
| Rs36210374 | 781 | 1 | 1 | 1 | 0.01 | 0 | 0.053 |
| Rs3024798 | 1013 | 0.4942 | 0.9936 | 0.5174 | 0.317 | 0.36 | 0.132 |
| Rs34463745 | 1392 | 0.0302 | 1 | 0.1714 | 0.015 | 0 | 0.083 |
| Rs3024799 | 1436 | 1 | 1 | 1 | 0.01 | 0 | 0.056 |
| Rs45478492 | 1461 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rsll30866 | 1580 | 0.5414 | 0.6148 | 1 | 0.464 | 0.494 | 0.333 |
| Rs45615839 | 2147 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs34263487 | 2213 | 1 | 1 | 1 | 0.01 | 0 | 0.053 |
| Rs35373464 | 2413 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs34520179 | 2688 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs35049407 | 2870 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs45532532 | 2997 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs3024803 | 3744 | 1 | 1 | 1 | 0.04 | 0.006 | 0.184 |
| Rs3024804 | 3942 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs45456197 | 4238 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs3024809 | 4493 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs3024810 | 4521 | 1 | 1 | 0.7913 | 0.045 | 0.006 | 0.211 |
| Rs36210375 | 4525 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs762548 | 4568 | 0.3396 | 0.512 | 0.418 | 0.105 | 0.068 | 0.263 |
| Rs45520240 | 4671 | 1 | 1 | 1 | 0 | 0 | 0 |
| Rs45504597 | 4719 | 1 | 1 | 1 | 0.005 | 0 | 0.026 |
| Rs2118177 | 5028 | 0.3659 | 0.9332 | 1 | 0.434 | 0.35 | 0.194 |
| Rs3024811 | 5173 | 1 | 1 | 0.7404 | 0.045 | 0.006 | 0.222 |
| Rs35369826 | 5406 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs2040349 | 5796 | 0.5192 | 0.7492 | 1 | 0.443 | 0.361 | 0.194 |
| Rs35297133 | 6051 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs35588863 | 6220 | 1 | 1 | 1 | 0.005 | 0.006 | 0 |
| Rs3024814 | 6299 | 1 | 1 | 1 | 0.01 | 0.006 | 0.028 |
| Rs3024832 | 6537 | 1 | 1 | 1 | 0.035 | 0.043 | 0 |
| Rs3024833 | 6934 | 1 | 1 | 1 | 0.005 | 0.007 | 0 |
MAF = Minor allele frequency; total = total cohort (n = 101).
Table 4.
SFTPB genotype association with SP-B peptides
| Rs number | Genomic location | Minor allele | P401 | P421 | M211 | P241 | P151 | P171 |
|---|---|---|---|---|---|---|---|---|
| Rs2077079 | −18 | C | 0.39 | 0.85 | 0.47 | 0.45 | 0.85 | 0.26 |
| Rs3024798 | 1013 | A | 0.31 | 0.88 | 0.36 | 0.28 | 0.74 | 0.78 |
| Rs1130866 | 1580 | C | 0.00232 | 0.00343 | 0.84 | 0.47 | 0.00034 | 0.0169 |
| Rs2118177 | 5028 | G | 0.74 | 0.10 | 0.88 | 0.12 | 0.70 | 0.46 |
| Rs2040349 | 5796 | G | 0.81 | 0.14 | 0.88 | 0.20 | 0.81 | 0.45 |
We used a logistic approach that corrected for gestational age, gender and race to assess the association between each SFTPB genotype and the absence of each peptide band. A modified Bonferroni and Yekutieli false-discovery rate significance criterion was p value <0.0125 after correcting for 30 tests (5 polymorphic markers for each of 6 peptides).
P40 = Mr 40-kDa pro-SP-B peptide; P42 = Mr 42-kDa pro-SP-B peptide; M21 = mature Mr 21-kDa SP-B peptide; P24 = Mr 24-kDa pro-SP-B peptide; P15 = Mr 15-kDa pro-SP-B peptide; P17 = Mr 17-kDa pro-SP-B peptide.
Probability test p values.
For g. 1580 (Rs1130866), P40, CC (TT as reference, p = 0.0052, ß = −1.17) was associated with absence of the Mr 40-kDa pro-SP-B peptide.
For g. 1580 (Rs1130866), P42, CT (TT as reference, p = 0.04, ß = 0.71) was associated with presence of the Mr 42-kDa pro-SP-B peptide.
For g. 1580 (Rs1130866), P15, CC (TT as reference, p < 0.0001, ß = −1.63) was associated with absence of Mr 15-kDa pro-SP-B peptide.
We found no differences in frequencies of common SFTPB genotypes between the RDS and control groups or between the RDS and control infants in the replication cohort (table 5). Using logistic regression models that included the risk of RDS with respect to genotype, expression of the Mr 21- or 24-kDa SP-B peptides, race, and gender, we found that the Mr 24-kDa SP-B peptide was consistently an independent contributor to the risk of RDS irrespective of genotype (p = 0.02–0.04). These observations suggest that while SFTPB genotype at g. 1580 regulates amino-terminus glycosylation of pro-SP-B, SFTPB genotypes do not contribute to risk of RDS.
Table 5.
SFTPB genotype association with RDS
| Rs number | Genomic location | Minor allele | Case-control cohort p value |
Replication cohort p value | |
|---|---|---|---|---|---|
| AD | ED | ||||
| Rs2077079 | −18 | C | 0.04 | 0.43 | 0.16 |
| Rs3024798 | 1013 | A | 0.14 | 0.39 | 0.54 |
| Rs1130866 | 1580 | C | 0.12 | 0.49 | 0.48 |
| Rs2118177 | 5028 | G | 0.67 | 0.48 | 0.47 |
| Rs2040349 | 5796 | G | 0.67 | 0.32 | 0.77 |
p value for RDS vs. control or replication cohort.
Discussion
Our data suggest 5 possible differences in developmental regulation of pro-SP-B proteolytic processing in vivo between fetuses or newborn infants and adults. First, proteases required for pro-SP-B processing may be developmentally regulated in fetuses and newborn infants. Secondly, pro-SP-B may be released without routing through the multivesicular/composite body/lamellar body pathway in fetuses and newborn infants [9, 11]. Thirdly, fetal or neonatal alveolar epithelial cells may differ from adult cells in their secretory selectivity. Fourthly, pro-SP-B may be misprocessed by proteases not normally involved in pro-SP-B processing. Finally, detection of pro-SP-B peptides in bronchoalveolar lavage fluid from older children and adults with lung injury and from LPS-induced chorioamnionitis in animal models suggests that these SP-B peptides may also result from alveolar epithelial cell injury, the influence of inflammatory mediators, or proteolytic activity in airway effluent [11, 16, 27, 28]. We also cannot exclude the possibility that the pro-SP-B peptides we observed were released after alveolar type II cell death or by Clara cells. However, because we found these peptides in amniotic fluid, it is unlikely that they result from oxygen toxicity or barotrauma alone.
Our study extends previous observations of regulation of human SP-B in fetal lung explant cultures, amniotic fluid, tracheal aspirate, and bronchoalveolar lavage [10, 11,13,14,15,16]. In vitro studies of the subcellular processing, localization, and distribution of SP-B in human fetal alveolar type II cells suggest 3 steps in the proteolytic processing of pro-SP-B that include 2 distinct cleavages of the amino-terminus pro-peptide (from 42- or 40-kDa pro-SP-B to 23 kDa and from 9-kDa to the 8-kDa mature SP-B) and 1 cleavage of the carboxy-terminus pro-peptide (from 23 to 9 kDa) [11, 19]. Immunolocalization studies suggest that these proteolytic processing steps likely involving cathepsin H, napsin A, and pepsinogen C occur en route from the Golgi complex through the multivesicular and composite bodies to the lamellar bodies in alveolar type II cells [11, 12,29,30,31]. In vitro studies have also suggested that the amino-terminus, co-translational glycosylation of pro-SP-B is regulated by the genotype of a common, nonsynonymous SNP at g. 1580 [22]. While our study is limited by statistical power to detect associations between rare SFTPB genotypes and SP-B peptides, it confirms the association between genotype at g. 1580 and the presence of pro-SP-B peptides that suggests in vivo genetic regulation of amino terminus glycosylation and developmental regulation of SP-B proteolytic processing. Because of the multiple pro- and mature SP-B peptides we observed in tracheal aspirate samples, our study also demonstrates the importance of comprehensive proteomic resolution in evaluation of regulation of surfactant associated proteins. These observations suggest that pharmacologic strategies that increase SP-B proteolytic processing to produce mature SP-B may improve alveolar type II cell metabolism and pulmonary surfactant function without increased SFTPB transcription.
Acknowledgements
We wish to thank Katherine K. Madden, BSN, for subject recruiting, Timothy E. Weaver, PhD, University of Cincinnati, for providing purified human SP-B, Ann Gronowski, MD, Department of Pathology, Washington University School of Medicine, for providing amniotic fluid samples, and Tomoko Betsuyaku, MD, Hokkaido University School of Medicine, Kita-ku, Japan, for providing adult bronchoalveolar lavage samples.
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL065174 and HL082747 to F.S.C., HL065385 to A.H., and HL059959 to S.H.G.), from the Children's Discovery Institute of St. Louis Children's Hospital (F.S.C. and A.H.), and from the Saigh Foundation (F.S.C. and A.H.).
References
- 1.Hills B. Surface-active phospholipid: a Pandora's box of clinical applications. 1. The lung and air spaces. Intern Med J. 2002;32:170–178. doi: 10.1046/j.1444-0903.2001.00200.x. [DOI] [PubMed] [Google Scholar]
- 2.Mazela J, Merritt TA, Gadzinowski J, Sinha S. Evolution of pulmonary surfactants for the treatment of neonatal respiratory distress syndrome and paediatric lung diseases. Acta Paediatr. 2006;95:1036–1048. doi: 10.1080/08035250600615168. [DOI] [PubMed] [Google Scholar]
- 3.Kramer B. The respiratory distress syndrome (RDS) in preterm infants: physiology, prophylaxis, and new therapeutic approaches. Intensivmed. 2007;44:403–408. [Google Scholar]
- 4.Angus D, Linde-Zwirble WT, Clermont G, Griffin MF, Clark RH. Epidemiology of neonatal respiratory failure in the united states: projections from California and New York. Am J Respir Crit Care Med. 2001;164:1154–1160. doi: 10.1164/ajrccm.164.7.2012126. [DOI] [PubMed] [Google Scholar]
- 5.Hamvas A, Devine T, Cole FS. Surfactant therapy failure identifies infants at risk for pulmonary mortality. Am J Dis Child. 1993;147:665–668. doi: 10.1001/archpedi.1993.02160300071026. [DOI] [PubMed] [Google Scholar]
- 6.Richardson D, Torday JS. Racial differences in predictive value of the lecithin/sphingomyelin ratio. Am J Obstet Gynecol. 1994;170:1273–1278. doi: 10.1016/s0002-9378(94)70141-5. [DOI] [PubMed] [Google Scholar]
- 7.Nogee LM de Mello DE, Dehner LP, Colten HR. Brief report: deficiency of pulmonary surfactant protein B in congenital alveolar proteinosis. N Engl J Med. 1993;328:406–410. doi: 10.1056/NEJM199302113280606. [DOI] [PubMed] [Google Scholar]
- 8.Clark R. The epidemiology of respiratory failure in neonates born at an estimated gestational age of 34 weeks or more. J Perinatol. 2005;25:251–257. doi: 10.1038/sj.jp.7211242. [DOI] [PubMed] [Google Scholar]
- 9.Weaver T, Conkright JJ. Function of surfactant proteins B and C. Annu Rev Physiol. 2001;63:555–578. doi: 10.1146/annurev.physiol.63.1.555. [DOI] [PubMed] [Google Scholar]
- 10.Guttentag S, Beers MF, Bieler BM, Ballard PL. Surfactant protein B processing in human fetal lung. Am J Physiol. 1998;275:L559–L566. doi: 10.1152/ajplung.1998.275.3.L559. [DOI] [PubMed] [Google Scholar]
- 11.Brasch F, Johnen G, Winn-Brasch A, Guttentag SH, Schmiedl A, Kapp N, Suzuki Y, Muller KM, Richter J, Hawgood S, Ochs M. Surfactant protein B in type II pneumocytes and intra-alveolar surfactant forms of human lungs. Am J Respir Cell Mol Biol. 2004;30:449–458. doi: 10.1165/rcmb.2003-0262OC. [DOI] [PubMed] [Google Scholar]
- 12.Ueno T, Linder S, Na CL, Rice WR, Johansson J, Weaver TE. Processing of pulmonary surfactant protein B by napsin and cathepsin H. J Biol Chem. 2004;279:16178–16184. doi: 10.1074/jbc.M312029200. [DOI] [PubMed] [Google Scholar]
- 13.Dilger I, Schwedler G, Dudenhausen JW. Determination of the pulmonary surfactant-associated protein SP-B in amniotic fluid with a competition ELISA. Gynecol Obstet Invest. 1994;38:24–27. doi: 10.1159/000292439. [DOI] [PubMed] [Google Scholar]
- 14.Mori K, Ikeda K, Tanaka M. Different expression of surfactant protein B mature peptide and proprotein at 21 weeks’ gestation in human fetal pulmonary epithelial cells. Pediatr Int. 2002;44:500–504. doi: 10.1046/j.1442-200x.2002.01605.x. [DOI] [PubMed] [Google Scholar]
- 15.Stahlman M, Gray ME, Whitsett JA. The ontogeny and distribution of surfactant protein B in human fetuses and newborns. J Histochem Cytochem. 1992;40:1471–1480. doi: 10.1177/40.10.1527371. [DOI] [PubMed] [Google Scholar]
- 16.Griese M, Schumacher S, Tredano M, Steinecker M, Braun A, Guttentag S, Beers MF, Bahuau M. Expression profiles of hydrophobic surfactant proteins in children with diffuse chronic lung disease. Respir Res. 2005;6:80. doi: 10.1186/1465-9921-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S, Akinbi HT, Breslin JS, Weaver TE. Structural requirements for targeting of surfactant protein B (SP-B) to secretory granules in vitro and in vivo. J Biol Chem. 1996;271:19689–19695. doi: 10.1074/jbc.271.33.19689. [DOI] [PubMed] [Google Scholar]
- 18.Akinbi H, Breslin JS, Ikegami M, Iwamoto HS, Clark JC, Whitsett JA, Jobe AH, Weaver TE. Rescue of SP-B knockout mice with a truncated SP-B proprotein. Function of the c-terminal propeptide. J Biol Chem. 1997;272:9640–9647. doi: 10.1074/jbc.272.15.9640. [DOI] [PubMed] [Google Scholar]
- 19.Korimilli A, Gonzales LW, Guttentag SH. Intracellular localization of processing events in human surfactant protein B biosynthesis. J Biol Chem. 2000;275:8672–8679. doi: 10.1074/jbc.275.12.8672. [DOI] [PubMed] [Google Scholar]
- 20.Nogee L, Garnier G, Dietz HC, Singer L, Murphy AM, deMello DE, Colten HR. A mutation in the surfactant protein B gene responsible for fatal neonatal respiratory disease in multiple kindreds. J Clin Invest. 1994;93:1860–1863. doi: 10.1172/JCI117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melton K, Nesslein LL, Ikegami M, Tichelaar JW, Clark JC, Whitsett JA, Weaver TE. Sp-B deficiency causes respiratory failure in adult mice. Am J Physiol Lung Cell Mol Physiol. 2003;285:L543–L549. doi: 10.1152/ajplung.00011.2003. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Christensen ND, Wigdahl B, Guttentag SH, Floros J. Differences in n-linked glycosylation between human surfactant protein-B variants of the C or T allele at the single-nucleotide polymorphism at position 1580: implications for disease. Biochem J. 2003;369:179–184. doi: 10.1042/BJ20021376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballard P, Merrill JD, Godinez RI, Godinez MH, Truog WE, Ballard RA. Surfactant protein profile of pulmonary surfactant in premature infants. Am J Respir Crit Care Med. 2003;168:1123–1128. doi: 10.1164/rccm.200304-479OC. [DOI] [PubMed] [Google Scholar]
- 24.Merrill J, Ballard RA, Cnaan A, Hibbs AM, Godinez RI, Godinez MH, Truog WE, Ballard PL. Dysfunction of pulmonary surfactant in chronically ventilated premature infants. Pediatr Res. 2004;56:918–926. doi: 10.1203/01.PDR.0000145565.45490.D9. [DOI] [PubMed] [Google Scholar]
- 25.Hamvas A, Wegner DJ, Carlson CS, Bergmann KR, Trusgnich MA, Fulton L, Kasai Y, An P, Mardis ER, Wilson RK, Cole FS. Comprehensive genetic variant discovery in the surfactant protein B gene. Pediatr Res. 2007;62:170–175. doi: 10.1203/PDR.0b013e3180a03232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamvas A, Wegner DJ, Trusgnich MA, Madden K, Heins H, Liu Y, Rice T, An P, Watkins-Torry J, Cole FS. Genetic variant characterization in intron 4 of the surfactant protein B gene. Hum Mutat. 2005;26:494–495. doi: 10.1002/humu.9378. [DOI] [PubMed] [Google Scholar]
- 27.Bachurski C, Ross GF, Ikegami M, Kramer BW, Jobe AH. Intra-amniotic endotoxin increases pulmonary surfactant proteins and induces SP-B processing in fetal sheep. Am J Physiol Lung Cell Mol Physiol. 2001;280:L279–L285. doi: 10.1152/ajplung.2001.280.2.L279. [DOI] [PubMed] [Google Scholar]
- 28.Kramer B, Kramer S, Ikegami M, Jobe AH. Injury, inflammation, and remodeling in fetal sheep lung after intra-amniotic endotoxin. Am J Physiol Lung Cell Mol Physiol. 2002;283:L452–L459. doi: 10.1152/ajplung.00407.2001. [DOI] [PubMed] [Google Scholar]
- 29.Brasch F, Ochs M, Kahne T, Guttentag S, Schauer-Vukasinovic V, Derrick M, Johnen G, Kapp N, Muller KM, Richter J, Giller T, Hawgood S, Buhling F. Involvement of napsin A in the C- and N-terminal processing of surfactant protein B in type-II pneumocytes of the human lung. J Biol Chem. 2003;278:49006–49014. doi: 10.1074/jbc.M306844200. [DOI] [PubMed] [Google Scholar]
- 30.Foster C, Aktar A, Kopf D, Zhang P, Guttentag S. Pepsinogen C: a type 2 cell-specific protease. Am J Physiol Lung Cell Mol Physiol. 2004;286:L382–L387. doi: 10.1152/ajplung.00310.2003. [DOI] [PubMed] [Google Scholar]
- 31.Guttentag S, Robinson L, Zhang P, Brasch F, Buhling F, Beers M. Cysteine protease activity is required for surfactant protein B processing and lamellar body genesis. Am J Respir Cell Mol Biol. 2003;28:69–79. doi: 10.1165/rcmb.2002-0111OC. [DOI] [PubMed] [Google Scholar]