Summary
OBJECTIVE
To determine the relationship between mutations in dhfr and dhps and SP treatment failure in Plasmodium falciparum malaria in the Democratic Republic of the Congo (DRC)
METHODS
Between June and September 2002, a therapeutic efficacy trial was conducted in Rutshuru, Eastern DRC, comparing sulfadoxine-pyrimethamine (SP), SP plus amodiaquine (AQSP), and artesunate plus SP (ASSP) regimens for treating malaria in children under 5 years old. We genotyped 212 samples for mutations associated with SP resistance and investigated their association with treatment failure.
RESULTS
In the SP arm, 61% of the subjects experienced treatment failure after 14 days. The failure rate was lower in the combination arms (AQSP: 32%, ASSP: 21%). The dhfr-108 and dhfr-51 mutations were nearly universal while 89% of the samples had at least one additional mutation at dhfr-59, dhps-437, or dhps-540. Dhps mutations had a bigger impact on treatment failure in children with high parasite density: for children with a parasite density less than 45,000 parasites/μl, the risk of treatment failure was 37% for mutations at dhps-437 and dhps-540 mutation and 21% for neither mutation (risk difference (RD) = 17%, 95%CI: −3%, 36%). In children with a parasite density greater than 45,000 parasites/μl, the treatment failure risk was 58% and 8% for children with both mutations or neither mutation, respectively (RD = 51%, 95%CI: 34%, 67%).
CONCLUSIONS
Dhps-437 and dhps-540 are strongly associated with SP treatment failure and should be evaluated further as a method for surveillance of SP-based therapy in the DRC.
Keywords: Plasmodium falciparum, dihydrofolate reductase, dihydropteroate synthase, malaria, drug resistance
Introduction
A major obstacle in the control of Plasmodium falciparum malaria is drug resistance. Drug resistance has increased malaria-related morbidity and mortality (Bjorkman & Bhattarai 2005; Trape 2001). It has also amplified the cost of malaria control, as second line drugs and combination therapy are often more expensive.
Sulfadoxine-pyrimethamine (SP) was the first line agent for uncomplicated malaria in much of Africa until recently due to the development of resistance (EANMAT 2003; Wongsrichanalai et al. 2002). Most African countries have since adopted artemisinin-based combination therapy (ACT) as their official first line drug for uncomplicated malaria. However, in reality, SP is still used widely in Africa due to the limited distribution of ACT (Nosten & White 2007). In addition, SP is the only recommended drug for Intermittent Preventive Treatment in pregnant women (IPTp) (World Health Organization 2004).
SP resistance is conferred by mutations in the dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) genes, which encode for the drugs’ targets. The presence of mutations at dhfr-51, dhfr-59, dhfr-108, dhps-437, and dhps-540 has been previously associated with SP treatment failure (Kublin et al. 2002; Talisuna et al. 2004). The role of mutations at dhps-436, dhps-581, and dhps-613 in treatment failure is less clear, as these mutations have received less attention due to their low prevalence in Africa.
Molecular markers of drug resistance, such as dhfr and dhps, can be used for surveillance (Laufer et al. 2007). For example, the prevalence of dhfr and dhps mutations were used to predict drug efficacy in regions where in vivo efficacy studies were unfeasible (Anderson et al. 2003; Djimde et al. 2004). In addition, molecular markers are used to monitor resistance in areas where the drug is no longer used (Laufer et al. 2006).
In the Democratic Republic of Congo (DRC), SP replaced chloroquine as the national first line drug in 2001 (Kazadi et al. 2003). However, SP has been in use in the Eastern region of the country since 1995 (unpublished data). In vivo efficacy studies have shown that parasites in the East are more resistant to SP than the rest of the country (Kazadi et al. 2003). Methods to extend the current in vivo surveillance is greatly needed in DRC because of its large geographic size, limited health infrastructure, and political instability. We report here the prevalence of dhfr and dhps mutations in Eastern DRC and their relationship to 14 day risk of SP treatment failure in order to evaluate the use of these markers for surveillance of SP resistance.
Methods
Study design
A therapeutic efficacy trial was conducted in Rutshuru, a district near the Rwandan border in Eastern DRC, to compare the efficacy of 3 regiments: 1. amodiaquine plus SP (AQSP); 2. artesunate plus SP (ASSP); and 3. SP alone (SP). This trial was a randomized, open label study following the WHO 1996 protocol with the modifications suggested in the 2002 report (Kazadi et al. 2003; World Health Organization 2002; World Health Organization 1996). The study took place between June and September 2002 at four different clinics. Children were included in the study if they met the following criteria: 1. age 6-59 months; 2. they did not suffer from severe malnutrition; 3. they had a P. falciparum infection with no other Plasmodium species detected; 4. their parasitemia was between 2,000 to 200,000 parasites / μl on initial counting of blood smear; 5. there were no signs of severe disease; 6. axillary temperature ≥ 37.5 °C; 7. there was no evidence of illnesses other than malaria; 8. there was no history of allergic reactions to sulfa-containing drugs; 9. they were able to return to clinic for follow-up visits; and 10. the parent or guardian provided consent.
249 children were initially enrolled and randomized to a treatment arm (AQSP: 75, ASSP: 82, SP: 92). At enrollment, children received either SP alone (25 mg/kg sulfadoxine and 1.25 mg/kg pyrimethamine), or amodiaquine (10 mg/kg/day for 3 days) plus SP, or artesunate (4 mg/kg/day/ for 3 days) plus SP. All treatments were directly observed and the treatment was repeated if vomiting occurred within 30 minutes. The initial blood smears were later re-examined by two lab technicians and rechecked by a senior lab technician. If the parasitemia was found to be >200,000 parasites/ul on the second reading, the patients were withdrawn from the efficacy study. However, data was still collected on their clinical course (as they were encouraged to attend the same appointments) and therefore they were retained in the analysis of the effect of mutations on treatment failure. Follow-up visits occurred 1, 2, 3, 7, and 14 days after enrollment, though the subjects were encouraged to return sooner if they experienced any danger sign or fever. Subjects were considered lost to follow up if they failed to return for follow-up appointments despite genuine efforts to find them. They were excluded after enrollment if they reported using an antimalarial drug other than those given in the study, they developed another disease, or their parent or guardian withdrew consent. If any child exhibited any signs of severe malaria, they were sent to the local health center for further evaulation and treatment. In the event of treatment failure, the children were treated with quinine. If any child exhibited any signs of severe malaria, they were sent to the local health center for further evaulation and treatment.
Sample processing and genotyping
Approximately 50 μl of blood was collected from 212 patients at enrollment. The blood was placed on IsoCode Stix filter paper (Schleicher & Schuell, Keene, N.H., USA), desiccated, and then transported to UNC Chapel Hill for processing. DNA was extracted according to IsoCode Stix's protocol.
All samples were genotyped at 3 dhfr (51, 59, 108) and 4 dhps (437, 540, 581, 613) codons using real-time PCR and MGB probes, as described previously (Alker et al. 2004). Genotyping of dhps-437 does not work in the presence of a mutation at dhps-436. Therefore, all samples that were negative in the dhps-437 assay were amplified and sequenced, as previously described (Alker et al. 2004). A random selection of 20 samples were genotyped for dhfr-164 (Alker et al. 2005).
Analysis
All statistical analyses were performed in Stata 8.2 (College Station, TX). The main outcome being investigated was 14 day treatment failure as defined by the 2002 World Health Organization report.
The main exposure of this analysis was the dhfr and dhps genotype. For each individual codon, a binary variable was created that reflected whether the mutant genotype was present. The following genotypes are considered mutant: dhfr-51-Ile dhfr-59-Arg, dhfr-108-Asn/Thr, dhps-437-Gly, dhps-540-Glu, dhps-581-Gly, and dhps-613-Ser/Thr.
For all analyses, risk differences were estimated using linear risk regression (general linear model with the identity link and a binomial error distribution). Linear risk regression is more appropriate than log risk or logistic regression in this study because risk differences more accurately reflect the magnitude of the effect when the outcome is common. In addition, linear risk regression is more appropriate for evaluating effect modification because it is on the additive scale (Prentice & Mason 1986; Rothman et al. 1980). However, since most other studies use ratio measures of effect, RR will also be calculated to aid in the comparison to other studies.
To evaluate the independent effect of each mutation on treatment failure, dhfr-59, dhps-437, and dhps-540 were all included in the model a priori. Interactions between codons were then evaluated using a forward selection technique based on a p-value of 0.05. dhfr-108, dhfr-51, dhps-581, and dhps-613 were not included in this analysis because they are homogeneous in this population.
Adjusted effects of these mutations were estimated by including confounders and effect measure modifiers (EMM) in the linear risk model. Potential covariates included: initial parasite density (coded as a binary variable with a cut point at 45,000 parasites/μl), age (in years), hemoglobin (coded as a binary variable with the cut point at 10), fever, z-score of weight for age (coded as a binary variable with the cut point of –2), clinic, and treatment arm. For the parasitemia, there was no cut-off that maximally modeled the relationship between treatment failure and parasitemia. Therefore the median value was used.Covariates were first evaluated as potential EMMs using forward selection. The interaction term was retained in the model and the covariate was considered an EMM when p ≤ 0.05 in the Wald test. All non-EMM were evaluated in a directed acyclic diagram to select a sufficient set of covariates that should be included in the model to control for confounding (Greenland et al. 1999).
Sensitivity analyses were performed to quantify the uncertainty caused by the 20 exclusions (including the 3 deaths). The data were re-analyzed under the following scenarios: 1. all the excluded patients failed treatment and 2. all the excluded patients did not fail treatment.
Ethics
The original study was reviewed by the Ministry of Health Technical Panel on behalf of the ethics committee. The Institutional Review Board at UNC Chapel Hill School of Public Health approved the genotyping and data analysis.
Results
Efficacy study
For study participants whose samples were genotyped, the average age was 28.7 months (range: 6-59). The mean parasitemia at enrollment was 74,173 parasites per μl (range: 1,043-454,664) while the mean hemoglobin was 10.0 g/dl (range: 5.6-14.5 g/dl). A quarter of the subjects were underweight (weight for age z-scores ≤ −2 in reference to the NCHS/WHO international dataset) (Dibley et al. 1987). The genotyped sub-cohort was similar in these clinical characteristics to the entire cohort (results not shown).
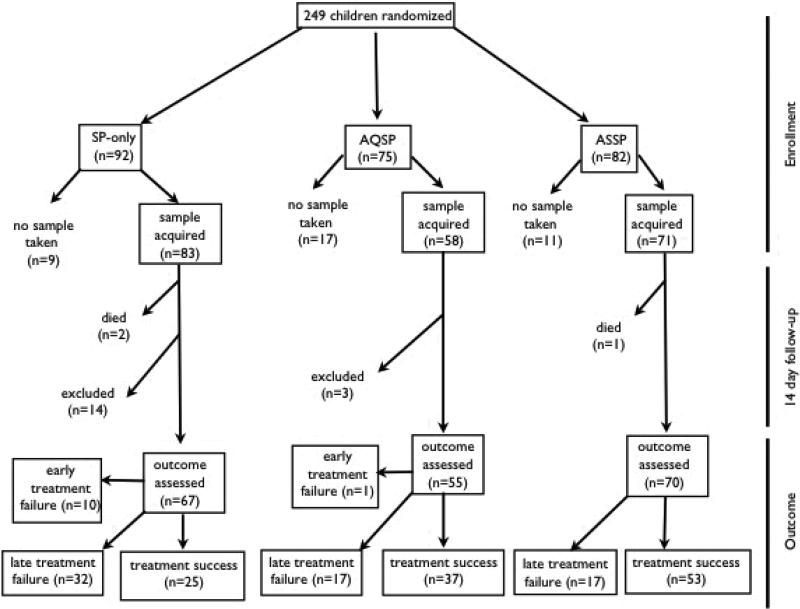
For the 212 subjects that were genotyped, 17 patients were excluded, mostly due to loss to follow-up (AQSP: 3, ASSP: 1, SP: 13). In addition, three children died during the course of the study (AQSP: 0, ASSP: 1, SP: 2). These children all had an initial parasitemia of less than 200,000 parasites/μl and showed no danger signs during the study appointments they attended. All these children subsequently did not show up for follow-up appointments and their deaths were discovered later by the survey team. Therefore, the exact cause of death was not determined, though in one case a home remedy for malaria was suspected. In the remaining 192 subjects, the treatment failure was high (AQSP: 33%, ASSP: 24%, SP: 63%). The majority of the treatment failures occurred after day 3 (late clinical failures & late parasitological failures), except for the SP arm in which 10 out of the 42 failures were classified as early treatment failures (Figure 1). The treatment failure rate was slightly higher in the genotyped subjects compared to the entire cohort (Entire cohort failure rate: AQSP: 32% ASSP: 22% SP: 61%). These results were not PCR corrected to differentiate recrudescence from new infections.
Figure 1.
Flowchart for the in vivo efficacy trial in Rutshuru, DRC. Clinical outcome was classified according to the WHO in vivo efficacy protocol for intense transmission areas (World Health Organization 2002). ACPR = adequate clinical and parasitological response.
Genotyping
All real-time PCR genotyping was successful except for 10 samples for dhps-437. Sequencing revealed one sample contained 436-Ser 437-Gly, eight samples were 436-Ala 437-Ala, and one sample had a 436-Cys 437-Ala dhps genotype. The 436-Cys mutation, which is a two nucleotide difference from the wild-type sequence, has only been reported once before (Basco et al. 2000).
The prevalence of mutant and mixed (mutant and wild-type) genotypes in dhfr, and dhps are presented in Figure 2. Most (99%) samples had a mutant dhfr-108 and dhfr-51 component. Mutations at dhfr-59, dhps-437 and dhps-540 were very common (dhfr-59: 66%, dhps-437: 72% dhps-540: 67%). No mutations at dhps-613 and dhfr-164 were observed. Only one sample (1%) was wild-type at all dhfr and dhps codons while 92 samples (43%) contained the quintuple mutant (mutation at dhfr-108, -51, -59, dhps-437 and –540).
Figure 2.
Prevalence of mutant (black) and mixed (gray) genotypes at codons associated with drug resistance in dhfr, and dhps in children ages 6-59 months in Rutshuru, DRC in 2002. Sample size is 212 except for dhfr-164 (n=20).
Dhfr and dhps mutations and treatment failure
Dhps-437 and –540 mutations were strongly associated with increased risk of treatment failure (Table 1). Interestingly, the presence of both these mutations was associated with a slightly smaller increase treatment failure risk than with either of these mutations alone (interaction term between 437 and 540: χ2 = 4.84, p=0.023). This finding suggests that these two mutations are antagonistic. Dhfr-59 was only weakly associated with treatment failure. These results did not substantially change when only subjects in the SP arm were analyzed.
Table 1.
Risk differences (RD) and 95% confidence intervals (CI) for the independent effect of dhfr and dhps mutations on 14 day treatment failure in Rutshuru, DRC. The referent group for all comparisons is parasites with the wild-type genotype at dhfr-59, dhps-437, -540, and –581.
| Mutation | RD (%) | CI (%) |
|---|---|---|
| dhfr-59 | 9 | −4, 22 |
| dhps-437 | 44 | 15, 73 |
| dhps-540 | 50 | 9, 91 |
| dhps-437 and -540 | 30 | 15, 45 |
Because dhps-437 and dhps-540 had the biggest impact on treatment failure, they were evaluated further in multivariable modeling. The purpose of this analysis was to test for EMMs and to obtain an unconfounded estimate effect of dhps mutations on treatment failure.
In the EMM screening, initial parasite density was the only covariate identified (χ2 = 10.70, p=0.005). This result implies that the effect of dhps-437 and dhps-540 differs by parasitemia. Of particular interest, neither treatment arm nor age were significant EMMs (treatment arm: χ2 = 2.03, p=0.567; age: χ2 = 0.12, p=0.940), which implies that the effect of the dhps mutations does not differ by regimen or by age in this study population.
In subjects with parasitemia less than 45,000 parasites/μl, the dhps-437 and –540 mutations were associated with a small increase in risk of treatment failure compared to subjects with neither mutation (for one mutation: RD = 19%, 95%CI: 14%, 53%; for both mutations: RD = 17%, 95%CI: −4%, 36%). The corresponding RR is 1.9 (95%CI: 0.7, 5.7) for one mutation and 1.8 (95%CI: 0.8, 4.2) for both mutations. In subjects with parasitemia equal or greater than 45,000 parasites/μl, the risk difference was 80% (95% CI: 54%, 105%) for one mutation and 51% (95%CI: 34% , 67%) for both mutations. The corresponding RR for one mutation is 10.9 (95%CI: 2.8, 42.4) and for both mutations is 7.3 (95%CI: 1.9, 28.2). The adjusted risk differences were similar to the crude estimates (Table 2). The sensitivity analyses revealed that the risk differences changed little by assuming either all the excluded children failed or all were successfully treated (Table 3).
Table 2.
Risk differences (RD) and 95% confidence intervals (CI) for the association between dhps mutations and 14 day risk of treatment failure by parasitemia in Rutshuru, DRC
| mutation at dhps-437 + 540 | Failed (n) | Total (n) | Risk (%) | Crude | Adjusted† | |||
|---|---|---|---|---|---|---|---|---|
| Parasitemia | RD (%) | 95%CI | RD (%) | 95%CI | ||||
| < 45,000 | none | 5 | 24 | 20.8 | 0.* | 0. | ||
| 1 mutation | 4 | 10 | 40.0 | 19 | −15, 53 | 19 | −14, 53 | |
| both mutations | 25 | 67 | 37.3 | 17 | −4 , 36 | 17 | −3, 36 | |
| ≥ 45,000 | none | 2 | 25 | 8.0 | 0. | 0. | ||
| 1 mutation | 7 | 8 | 87.5 | 80 | 54, 105 | 85 | 71, 98 | |
| both mutations | 34 | 58 | 58.6 | 51 | 34, 67 | 49 | 35, 64 | |
Referent level
Adjusted for clinic, age, and z-score of weight for age
Table 3.
Sensitivity analysis of the effect of dhps mutations on treatment failure by parasitemia in Rutshuru, DRC as measured by unadjusted risk differences (RD) and 95% confidence intervals (CI).
| Scenario | Parasitemia | dhps-437/540 | Risk (%) | RD | CI |
|---|---|---|---|---|---|
| All excluded failed treatment | < 45,000 | none | 24.0 | 0.* | |
| 1 mutation | 40.0 | 16 | −19, 51 | ||
| both mutations | 40.8 | 17 | −3, 37 | ||
| ≥ 45,000 | none | 23.3 | 0. | ||
| 1 mutation | 90.0 | 67 | 43, 91 | ||
| |
|
both mutations |
63.6 |
40 |
21, 59 |
| All excluded were successfully treated | < 45,000 | none | 20.0 | 0. | |
| 1 mutation | 40.0 | 20 | −14, 54 | ||
| both mutations | 35.2 | 15 | −4, 34 | ||
| ≥ 45,000 | none | 6.7 | 0. | ||
| 1 mutation | 70.0 | 63 | 34, 93 | ||
| both mutations | 51.5 | 45 | 30, 60 |
Referent level
Discussion
In this study investigating the molecular determinants of SP treatment failure, mutations at dhps-437 and dhps-540 were strongly associated with treatment failure. However, this relationship differed by parasitemia. In the low parasitemic group, subjects with either dhps-437/540 mutation had a 19% greater absolute risk of 14 day treatment failure than subjects with neither mutation. In contrast, among those with high parasitemia, the absolute risk of treatment failure was 80% greater in subjects with either dhps-437/540 mutation.
The differential effect of genetic markers of resistance by parasitemia has been reported before: higher parasitemia was related to decreased ability to clear infections with pfcrt-76 and pfmdr1-86 mutations after chloroquine treatment (Khalil et al. 2005). High parasitemia may be a sign of low partial immunity. Therefore, the larger effect of these mutations at high parasitemias might be caused by the inability of the immune system to clear resistant parasites. These results suggest that resistance might have a greater impact in less immue individuals. However, these results need to be replicated in areas of lower resistance to determine their generalizability.
In the evaluation of the independent effects of dhfr and dhps mutations, both dhps-437 and dhps-540 were strongly related to risk of treatment failure. Dhfr-59 was only weakly associated. This is in contrast to a previous study that found these three mutations have similar independent effects (Staedke et al. 2004). However, the effect of mutations has been shown to vary by region (Alifrangis 2003; Omar et al. 2001), which is likely caused by variation in the prevalence of treatment failure, the presence of other dhfr/dhps mutations, and the prevalence of other risk factors for treatment failure across sites.
In contrast to previous reports of the quintuple mutation being found in 0.9% to 27% of samples in DRC, the prevalence in Rutshuru was 42% (Swarthout et al. 2006; Cohuet et al. 2006). While the difference in prevalence might be due to methodological differences, it is more likely reflective of a high level of geographic variation of drug resistance in this country.
The main strength of this study is analytical technique: effect measure modifiers were systematically screened, potential confounders were controlled for, and the uncertainty caused by exclusions was quantified. The main limitation of this study is the limited follow-up. SP failures have occurred up to 28 days after treatment (Checchi et al. 2004) and therefore it is possible that some subjects failed after the 14 day follow-up ended. A review of in vivo efficacy studies found that, in general, the 14 day follow-up period has limited sensitivity in detecting treatment failures (Stepniewska et al. 2004). However, the 14 day follow-up had the best sensitivity (~80%) in areas of high resistance and high levels of transmission, such as Rutshuru (Figures 3C and 4 in Stepniewska et al. 2004). Therefore, in the Rutshuru study, it is likely that most treatment failures were captured.
Another limitation is the lack of PCR correction to distinguish true recrudescence from re-infections. However, the risk of re-infection is low during the first two weeks after treatment: Stephniewska et al. (2004) found that 73% of the time, all instances of treatment failure in the first two weeks were due to recrudescence. Since outcome misclassification caused by the lack of PCR correction and the limited follow-up is nondifferential with respect to the exposure (dhfr/dhps genotype), these potential biases would likely bias the effect estimate towards the null (Rothman & Greenland 1998). Another limitation of this study is that not all determinants of treatment failure were measured, such as drug pharmacokinetics and HIV status.
The high failure rate of SP and SP-combination therapy indicates that neither SP alone nor SP-combination should be used in this area for the treatment of falciparum malaria. It was based on this in vivo study and evidence from other sentinel sites that the DRC decided to shift from SP to amodiaquine plus artesunate as the new first line antimalarial. However, DRC policy continues to recommend using two doses of SP for IPTp. dhfr and dhps could provide an alternative technique to efficacy trials to explore the geographic and temporal changes in resistance. In addition, dhfr and dhps can help identify areas where SP efficacy should be investigated further (such as by IPTp efficacy trials) so that interventions can be targeted where they are needed the most. This study suggests that the collection of clinical information, such as parasite density, might be needed to use molecular markers for the monitoring of drug resistance.
Acknowledgements
We thank the children and their guardians for their participation. We acknowledge the assistance provided by Jesse Kwiek, Sarah Landis, William Miller, Annelies Van Rie, Melissa Miller, Phuc Nguyen-Dinh, Robert Ryder and Paul Wilson. The sample genotyping was funded by Global Network for Women's and Children's Health Research, NIH-NICHD 1U01 HD043475. Competing Interests: none declared
References
- Alker AP, Mwapasa V, Meshnick SR. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrobial Agents and Chemotherapy. 2004;48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alker AP, Mwapasa V, Purfield A, et al. Mutations associated with sulfadoxine-pyrimethamine and chlorproguanil resistance in Plasmodium falciparum isolates from Blantyre, Malawi. Antimicrobial Agents and Chemotherapy. 2005;49:3919–3921. doi: 10.1128/AAC.49.9.3919-3921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Jacobzone C, Zavai A, Balkan S. Molecular assessment of drug resistance in Plasmodium falciparum from Bahr El Gazal province, Sudan. Tropical Medicine and International Health. 2003;8:1068–1073. doi: 10.1046/j.1360-2276.2003.01144.x. [DOI] [PubMed] [Google Scholar]
- Basco LK, Tahar R, Keundjian A, Ringwald P. Sequence variations in the genes encoding dihydropteroate synthase and dihydrofolate reductase and clinical response to sulfadoxine-pyrimethamine in patients with acute uncomplicated falciparum malaria. Journal of Infectious Diseases. 2000;182:624–628. doi: 10.1086/315731. [DOI] [PubMed] [Google Scholar]
- Bjorkman A, Bhattarai A. Public health impact of drug resistant Plasmodium falciparum malaria. Acta Tropica. 2005;94:163–169. doi: 10.1016/j.actatropica.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Checchi F, Piola P, Kosack C, et al. Antimalarial efficacy of sulfadoxine-pyrimethamine, amodiaquine and a combination of chloroquine plus sulfadoxine-pyrimethamine in Bundi Bugyo, western Uganda. Tropical Medicine and International Health. 2004;9:445–450. doi: 10.1111/j.1365-3156.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- Cohuet S, Bonnet M, Van Herp M, Van Overmeir C, D'Alessandro U, Guthmann JP. Short report: molecular markers associated with Plasmodium falciparum resistance to sulfadoxine-pyrimethamine in the Democratic Republic of Congo. American Journal of Tropical Medicine and Hygiene. 2006;75:152–154. [PubMed] [Google Scholar]
- Dibley MJ, Goldsby JB, Staehling NW, Towbridge FL. Development of normalized curves for the international growth reference: historical and technical considerations. American Journal of Clinical Nutrition. 1987;46:736–748. doi: 10.1093/ajcn/46.5.736. [DOI] [PubMed] [Google Scholar]
- Djimde A, Dolo A, Ouattara A, Diakite S, Plowe CV, Doumbo O. Molecular diagnosis of resistance to antimalarial drugs during epidemics and in war zones. Journal of Infectious Diseases. 2004;190:853–855. doi: 10.1086/422758. [DOI] [PubMed] [Google Scholar]
- EANMAT The efficacy of antimalarial monotherapies, sulphadoxine-pyrimethamine and amodiaquine in East Africa: implications for sub-regional policy. Tropical Medicine and International Health. 2003;8:860–867. doi: 10.1046/j.1360-2276.2003.01114.x. [DOI] [PubMed] [Google Scholar]
- Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999;10:37–48. [PubMed] [Google Scholar]
- Kazadi WM, Vong S, Makina BN, et al. Assessing the efficacy of chloroquine and sulfadoxine-pyrimethamine for treatment of uncomplicated Plasmodium falciparum malaria in the Democratic Republic of Congo. Tropical Medicine and International Health. 2003;8:868–875. doi: 10.1046/j.1365-3156.2003.01098.x. [DOI] [PubMed] [Google Scholar]
- Khalil IF, Alifrangis M, Tarimo DS, et al. The roles of the pfcrt 76T and pfmdr1 86Y mutations, immunity and the initial level of parasitaemia, in predicting the outcome of chloroquine treatment in two areas with different transmission intensities. Annals of Tropical Medicine and Parasitology. 2005;99:441–448. doi: 10.1179/136485905X46441. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. Journal of Infectious Diseases. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Laufer MK, Djimde AA, Plowe CV. Monitoring and deterring drug-resistant malaria in the era of combination therapy. American Journal of Tropical Medicine and Hygiene. 2007;77:160–169. [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, et al. Return of chloroquine antimalarial efficacy in Malawi. New England Journal of Medicine. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. American Journal of Tropical Medicine and Hygiene. 2007;77:181–192. [PubMed] [Google Scholar]
- Prentice RL, Mason MW. On the application of linear relative risk regression models. Biometrics. 1986;42:109–120. [PubMed] [Google Scholar]
- Rothman KJ, Greenland S. Modern Epidemiology. Lippincott Williams & Wilkins; Philadelphia, PA: 1998. [Google Scholar]
- Rothman KJ, Greenland S, Walker AM. Concepts of interaction. American Journal of Epidemiology. 1980;112:467–470. doi: 10.1093/oxfordjournals.aje.a113015. [DOI] [PubMed] [Google Scholar]
- Stepniewska K, Taylor WR, Mayxay M, et al. In vivo assessment of drug efficacy against Plasmodium falciparum malaria: duration of follow-up. Antimicrobial Agents and Chemotherapy. 2004;48:4271–4280. doi: 10.1128/AAC.48.11.4271-4280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout TD, van den Broek IV, Kayembe G, Montgomery J, Pota H, Roper C. Artesunate + amodiaquine and artesunate + sulphadoxine-pyrimethamine for treatment of uncomplicated malaria in Democratic Republic of Congo: a clinical trial with determination of sulphadoxine and pyrimethamine-resistant haplotypes. Tropical Medicine and International Health. 2006;11:1503–1511. doi: 10.1111/j.1365-3156.2006.01710.x. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Nalunkuma-Kazibwe A, Langi P, et al. Two mutations in dihydrofolate reductase combined with one in the dihydropteroate synthase gene predict sulphadoxine-pyrimethamine parasitological failure in Ugandan children with uncomplicated falciparum malaria. Infection, genetics and evolution. 2004;4:321–327. doi: 10.1016/j.meegid.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Trape J. The public health impact of chloroquine resistance in Africa. American Journal of Tropical Medicine and Hygiene. 2001;64:12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- Wongsrichanalai C, Pickard A, Wernsdorfer W, Meshnick S. Epidemiology of drug-resistant malaria. The Lancet Infectious Diseases. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- World Health Organization Assessment and monitoring of antimalarial drug efficacy for uncomplicated falciparum malaria in areas with intense transmission. 1996. WHO/MAL/96.1077.
- World Health Organization Monitoring Antimalarial Drug Resistance, Report of a WHO consultation. 2002. WHO/CDS/RBM/2002.39.
- World Health Organization A strategic framework for malaria prevention and control during pregnancy in the Africa Region. Brazzaville: WHO Regional Office for Africa. 2004. AFR/MAL/04/01.