Abstract
An accurate molecular diagnosis for viral pathogens is highly dependent on pre-analytical procedures. The efficiencies of two viral RNA extraction methods (liquid phase partition and silica-based adsorption chromatography) and the effects of handling and storage on the stability of RNA isolated from dengue virus (DENV) were studied. Viral RNA extracted from spiked sera or clinical samples characterized with DENV infection were quantified by TaqMan real-time PCR. The presence of high serum proteins severely affected the recovery of DENV RNA by the liquid phase partition, but not the silica-based method. The recovery with Trizol liquid phase partition method was significantly improved by a concomitant addition of a co-precipitant and the reduction of sera proteins, resulting in recoveries similar to that of the silica-based methods. Repeated freeze-thaw cycles did not affect the recovery of viral RNA. While intact DENV was found to be stable in serum for up to 2 hour at 25°C, recovery of viral RNA from sera stored in the lysis/binding buffer was stable for up to 5 days. These data indicate that the choice of viral RNA extraction methods, the conditions for handling, and storing of clinical sera critically affect the quantification of viral nucleic acid from clinical samples. This will impact the accuracy and reproducibility of DENV diagnosis by PCR-based assays.
Dengue virus (DENV) has emerged as an important vector-borne viral disease,1 usually afflicting rural areas of endemic countries and posing tremendous health problems in these regions.2 Hence, to prevent and control the progression of dengue disease, the World Health Organization has recommended the augmentation of active and accurate laboratory-based surveillance for early reporting of dengue virus infections to the public health authorities.3
Pre-analytical variables, including the storage and transport of patient samples, the stability of viral RNA in the samples and methods of isolating viral RNA of high yield and quality, have major impacts on the development and performance of any successful molecular diagnostics.4,5 Hence, the technical challenges associated with these pre-analytical issues must first be identified and optimized. Various commercially available viral RNA extraction methods are currently in use for clinical diagnostics of viral diseases. These extraction kits are generally based on two methods—liquid phase partition (eg, TRIzol LS) and silica-based nucleic acid adsorption chromatography (eg, High Pure Viral RNA and QIAamp Viral RNA kits). The recoveries of viral RNA by these two methods differ substantially with different viruses,6,7,8,9,10,11,12 thus making the choice of any particular method for the isolation of viral RNA uncertain. While there were several studies on the isolation and stability of other viruses, few reports have focused on the systematic evaluation of the stability and recovery of DENV RNA from sera.13,14,15 This study examined the contributions of various pre-analytical variables to the sensitivity of DENV RNA detection in serum. Using reverse transcription quantitative real-time PCR (RT-qPCR), critical pre-analytical variables, including the performances of commercially available liquid phase partition and silica-based viral RNA extraction methods and the effects of storage, handling and freeze/thaw on the stability of the DENV RNA in serum were investigated.
Materials and Methods
Virus, Cell Lines, and Clinical Samples
DENV-1 (strain Singapore 8114/93), DENV-2 (strain NGC), DENV-3 (strain Singapore 8120/95), and DENV-4 (strain Singapore 8976/95) were propagated in C6/36 mosquito cells, and titered in BHK-21 cells as described.16 Except for DENV-2 all other dengue virus were obtained from the Singapore General Hospital, Singapore. All cell lines used in the study were obtained from the American Type Culture Collection (ATCC, VA). The clinical serum samples used in this study were obtained from de-identified confirmed dengue cases during a dengue outbreak in Malaysia, 2005.17 In brief, confirmed dengue cases were identified both serologically by IgM-capture enzyme-linked immunosorbent assay (Panbio, Australia), and by virus isolation and subsequent molecular detection of DENV genome by reverse transcription-PCR,18 using the acute serum samples. The acute serum samples were stored at −80°C until use. Only serum specimens from patients with laboratory-confirmed dengue infection were used in this study. Normal human serum from clotted human whole blood (H1388) and human serum albumin (hSA, A9511) were obtained from Sigma- Aldrich (St. Louis, MO). All studies were approved by the National University of Singapore Institutional Review Board.
Viral RNA Extraction
RNA extraction and stability studies were performed with both DENV-2 and DENV-3. Viral RNA was extracted with TRIzol LS (Invitrogen, CA) in the presence or absence of linear acrylamide (20 μg/ml, Ambion, Tx) as a co-precipitant,19 High Pure Viral RNA Isolation Kit (Roche Applied Sciences, Germany) or QIAamp Viral RNA Kit (Qiagen, Germany). Briefly, physiologically relevant titrated concentrations of DENV virions (1 × 104 plaque forming units [PFUs]/ml) or genomic RNA (105 to 107 copies/ml)20 were added to commercially available normal human sera or PBS that were premixed with the lysis/binding buffer to prevent viral RNA degradation before extraction. All extractions were performed according to the respective manufacturer's protocols. As controls for total viral RNA, the same amounts of DENV were directly lysed in the presence of RNasin Ribonuclease Inhibitor (Promega, WI) by heating at 95°C for 5 minutes before RT-qPCR.
For the study on RNA stability in human serum, DENV-2 genomic RNA or virions were spiked into human serum and incubated at 25°C. For studies on the stability of viral RNA in the lysis/binding buffer, DENV-spiked normal human sera were added to the Roche High Pure Viral RNA isolation lysis/binding buffer and kept at 25°C. For freeze-thaw studies, normal human sera spiked with DENV or patient sera were frozen at −70°C, thawed in a temperature-controlled water bath at 25°C, and the freeze-thaw cycles repeated. The recovery of viral RNA extraction was calculated using the formula 10−ΔCt/ε, where ΔCt = Ct (recovered RNA) − Ct (control RNA) and ε is the amplification efficiency. The efficiency of extraction of variable-sized single-stranded RNA using the silica-based extraction method was evaluated using the Experion Automated Electrophoresis System (BioRad, CA) according to manufacturer's instructions.
Real-Time PCR (TaqMan) Assays
Serotype-specific DENV-2 and DENV-3 primers and fluorogenic probes were designed using the Beacon Designer version 2.0.1 (PREMIER Biosoft International, CA) and Vector NTI version 8.0 (Invitrogen, CA) softwares. The serotype specificities of primers were evaluated by comparisons of available published sequences of the complete genome of each DENV-2 and DENV-3, followed by comparisons of specific regions using a combination of pan-DENV alignment analyses and BLAST searches. The DENV-2 probe was labeled at the 5′ end with the HEX reporter dye and at the 3′ end with black hole quencher 1 (BHQ-1); the DENV-3 probe was labeled with Texas Red and BHQ-2 (Table 1). The performances of the primer-probe sets were determined using in vitro transcribed genomic viral RNA. In both DENV-2 and DENV-3 assays, the dynamic ranges of detection were greater than 106 fold, with detection limits of 50 copies/reaction. The serotype specificity of the each assay was greater than 106-fold over the detection of the other heterologous DENV serotypes.
Table 1.
TaqMan Real-Time PCR Primers and Probes
| Primers/Probes* | Nucleotide sequence | Genome position† | Virus |
|---|---|---|---|
| 8800F | 5′-GCCATATTCACTGATGAGAACA-3′ | 8800–8821 | DENV-2 |
| 8868R | 5′-CCTTTCCTTGTCAACCAGC-3′ | 8868–8886 | |
| 8836P | 5′-AACCTACTATCTTCAACAGCCTCACG-3′ | 8861–8836 | |
| 1403F | 5′-GGAGTCACGG(C/T)TGAGATAAC-3′ | 1403–1422 | DENV-3 |
| 1495R | 5′-AATCATTTCATTGAAATCCAAACCT-3′ | 1495–1519 | |
| 1472P | 5′-CCGTGGTGAGCATTCTAGCCC-3′ | 1472–1492 |
RT-qPCR
RT-qPCR was performed with the one-step TaqMan RNA Amplification kit (Roche Molecular Systems, NJ) using 2.0 mmol/L Mn2+, 1 μmol/L of each primer and 0.2 μmol/L TaqMan probe, on the Bio-Rad iQ4 Multicolor Real-Time iCycler (Bio-Rad Laboratories, CA) with the following conditions: 5 minutes AmpErase step at 50°C, 1 minute denaturation at 95°C and 30 minutes reverse transcription at 60°C, followed by 40 cycles of 20 seconds denaturation at 95°C, 20 seconds annealing at 60°C, and 20 seconds extension at 72°C. Viral RNA copy numbers were interpolated from standard curves generated using in vitro transcribed DENV-2 or DENV-3 genomic RNA using the MEGAscript Transcription kit (Ambion, Tx) according to the manufacturer's instructions. The sequences of the primers and probes used for the DENV-2 and DENV-3 TaqMan assays are shown in Table 1. All experiments were done in quadruplicates and assayed in duplicates. All studies were performed by two independent operators and the cumulative results were analyzed. All statistical analyses were performed using the GraphPad Prism 4.0 software (GraphPad Prism Software Inc., CA).
Assay Precision
The intra-assay imprecision was performed to determine the “within-run” and “total” standard deviations,21 and was calculated from the Ct values of 12 replicate samples with low (104 copies/50 μl reaction volume) and high (106 copies/50 μl reaction volume) concentrations of DENV-2 viral RNA. Interassay imprecision was similarly calculated from the Ct values collected over a period of 3 consecutive days with the same lot of reagents. The intra-assay values of Ct ± SD (CV) were determined to be 29.47 ± 0.55 (1.11%) and 22.92 ± 0.25 (1.88%) for the low and high concentrations of the viral RNA respectively. Similarly, the interassay values of Ct ± SD (CV) were determined to be 29.35 ± 0.1513 (0.52%) and 22.74 ± 0.2095 (0.92%) for low and high concentrations of the viral RNA respectively. The DENV-2 and DENV-3 TaqMan assays showed similar performances.
Results
DENV RNA Extraction by Liquid Phase Partition and Silica-Based Adsorption Methods
The extractions of RNA from sera containing different amount of viral genomic RNA or titers of DENV with TRIzol LS, in the absence of linear acrylamide, were found to be inefficient (<1% of control RNA) when compared with the recoveries from virus-spiked PBS samples (11% to 16% of control RNA) (Figure 1A). The inclusion of linear acrylamide into the sera however significantly improved the recoveries of viral RNA (Figure 1B). To test the hypothesis that serum proteins contribute to the poor recovery of RNA, DENV, or viral genomic RNA were spiked into varying dilutions of serum or different amounts of hSA, and the extractions were performed using TRIzol LS with linear acrylamide. The results indicated an inverse linear relationship between the recoveries of RNA and the increase in sera protein concentration for both DENV (r2 = 0.993 and 0.963 for sera and hSA respectively; Figure 1C) and viral genomic RNA (r2 = 0.990). Consistent with previous reports, the PFUs were significantly lower than RNA copy numbers.22,23
Figure 1.
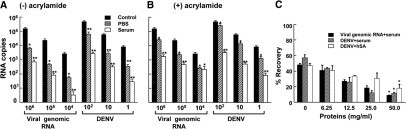
Recovery of DENV viral RNA from PBS or human sera samples using the liquid phase partition method. Different amounts of DENV-2 (100, 10 or 1 PFU/ml) or viral genomic RNA (106, 105, or 104 copies/ml) were added to either PBS (hatched bars) or human serum (open bars) and extracted using TRIzol LS in the absence (A) or presence (B) of 20 μg/ml linear acrylamide carrier. The asterisks denote statistical significance (Tukey's test, *P < 0.01, **P < 0.005) in the amount of viral RNA recovered compared with the control RNA. C: Viral RNA was isolated from either virus (DENV-2, 104 PFU/ml) or DENV-2 genomic RNA (106 copies/ml) using TRIzol LS reagent with the linear acrylamide additive, in the presence of varying dilutions of normal human serum (50 mg/ml total protein) or human serum albumin (hSA). The asterisks denote statistical significance (paired Student's t-test, *P < 0.01) in the amount of viral RNA recovered from samples with high protein content (50 mg/ml) as compared with the control RNA. The isolated viral RNA was assayed using the Roche TaqMan RNA amplification kit in parallel with the genomic viral RNA standards. The control reference is the amount of viral RNA obtained from the direct lysis method. The experiments were performed in quadruplicates and assayed in duplicates. The data are expressed as mean ± SEM.
We next explored the use of silica filter-based adsorption of nucleic acids for the extraction of viral RNA from biological fluids. The recoveries of RNA from DENV and viral genomic RNA samples were comparable between the two silica-based kits assessed, and were efficient as compared with the controls (Figure 2, A and B). Remarkably, the silica-based method did not show preferential binding for higher molecular mass RNA and was also efficient in recovering small RNAs (0.28 kb) (Figure 3, A–C). The recovery of RNA from serum was however found to be statistically lower (Figure 3D) as compared with extraction from PBS, consistent with the results on the effects of sera proteins (Figures 1C and 2B). The extraction studies described above were also performed with DENV-3, and similar results were obtained (data not shown). As PCR may be inhibited by chemicals co-purified during extractions, control studies using exogenous transcripts in the presence of serum or PBS were performed. RT-qPCR of green fluorescent protein RNA (106, 105, and 104 copies/ml) were not affected by the eluates produced from the various extraction procedures and reagents used (data not shown).
Figure 2.
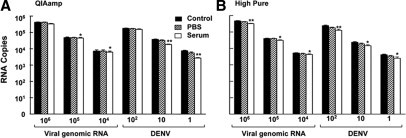
Recovery of DENV-2 viral RNA from PBS or human sera samples using various silica-based extraction methods. PBS (hatched bars) or human sera (open bars) were spiked with relevant titrated amounts of DENV-2 genomic RNA or virus, and the viral RNA isolated using silica-based methods of a QIAamp Viral RNA Kit (A) and a High Pure Viral RNA Isolation Kit (B). The control reference is the amount of viral RNA obtained from the direct lysis method. The experiments were performed in quadruplicates and assayed in duplicates. The data are expressed as mean ± SEM. The asterisks denote statistical significance (Tukey's test, *P < 0.01, **P < 0.005) in the amount of viral RNA recovered compared with the control RNA.
Figure 3.

Quantification of single-stranded RNA using microfluidics. Known amounts of RNA standards were added into PBS (A) or human serum (B), which were premixed with the binding buffe, and isolated using the Roche High Pure Viral RNA isolation kit. The recovered RNAs were then analyzed using the Experion Automated Electrophoresis System, with the unextracted RNA standards acting as controls (C). Three representative electrochromatograms are shown and the amount of RNA recovered for each molecular size was calculated using Experion software version 1.1. The numbers shown in the electrochromatograms represent relative fluorescence units calculated from the area under the peaks. D: Analysis of the composite recoveries of the various molecular-sized RNA from the experiments described above. All experiments were performed in quadruplicates and assayed in duplicates. The data are expressed as mean ± SEM. The asterisks denote statistical significance (Tukey's test, *P < 0.01) in the amount of viral RNA recovered compared with the control RNA.
Clinical Application
A comparison of the liquid phase partition and silica-based method was next performed on thirteen retrospective DENV-3-infected patient sera, and the viral RNA isolated using either TRIzol LS with linear acrylamide or High Pure Viral RNA kit. In the presence of serum, the recovery of the DENV RNA by TRIzol LS was consistently lower and more variable as compared with the silica-based method (Figure 4A; two-tail t-test; P ≤ 0.005 for all patient sera). While the recovery of DENV RNA using the silica-based method was similar with or without the dilution of patient sera with PBS (Figure 4, A and B; two-tail t-test, P > 0.05 for all patients), there were significant improvements in the recovery of viral RNA from the PBS-diluted patient sera using TRIzol LS (Figure 4, A and B; two-tail t-test, P < 0.001 for all sera tested).
Figure 4.
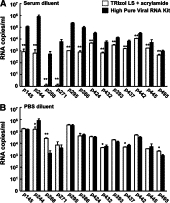
Comparison of Trizol LS and High Pure Viral RNA isolation kits in the isolation of viral RNA from DENV-3 infected patient sera. A volume of 50 μl of serum samples from dengue-infected patients were re-suspended in 150 μl of human serum (A) or PBS (B), and the viral RNA isolated using either TRIzol LS in the presence of linear acrylamide, or the High Pure Viral RNA isolation kit. The viral RNA was then quantified using the DENV-3 TaqMan real-time PCR assay in parallel with in vitro transcribed RNA standards. All experiments were done in quadruplicate and assayed in duplicate. The data are expressed as mean ± SEM. The asterisks denote statistical significance (paired Student's t-test, *P < 0.01; **P < 0.005).
Handling and Storage of DENV Sera Samples
The existence of cell-free circulating cellular and viral RNA in the plasma and serum has been reported.24,25,26 The stability of free DENV RNA in sera was found to be exceedingly poor at 25°C, with extensive loss of detectable RNA (Figure 5A). However, the DENV-2 virus was relatively more stable in serum at 25°C over a longer period of time. To simplify sample handling, while maintaining and prolonging viral RNA integrity before extraction, the feasibility of using the same lysis/binding buffer of the extraction kit, which contain chaotropes,27 to store DENV-containing samples was examined. Viral RNA isolated from DENV-infected sera that was kept in the lysis/binding buffer was found to be stable for at least 5 days at 25°C (Figure 5B; Tukey's test, P > 0.05). Additionally, since it is common for patient sera to undergo cycles of freeze-thaw in retrospective studies, the effects of repeated freeze-thaw cycles on DENV RNA recovery and integrity were also examined. Unexpectedly, the amount of intact DENV RNA was not affected with repeated freeze-thaw cycles (Figure 5C; 1-way analysis of variance, P = 0.84).
Figure 5.

Sample handling and viral RNA stability. A: DENV-2 virus (104 PFU/ml) or genomic DENV-2 RNA (107 copies/ml) was added to human serum and incubated at 25°C for 0, 15, 30, 60, 180, and 360 minutes. B: DENV-2 virus (104 PFU/ml) in serum was added to the binding buffer (Roche High Pure RNA Isolation Kit) and kept for 1, 3, 5, and 7 days at 25°C. C: DENV-2-spiked sera (104 PFU/ml) or DENV-3 positive patient sera (titer range of 105 to 106 copies/ml) were frozen at −70°C and thawed at 25°C. The freeze-thaw cycle was repeated for up to 5 times. The viral RNA was isolated by the High Pure viral RNA kit and quantified using the DENV-2 and DENV-3 TaqMan real-time PCR assay, together with the in vitro transcribed and genomic RNA standards. The percentage recovery was normalized to the non-incubated samples. All experiments were done in quadruplicate and assayed in duplicate. The asterisks denote statistically significant differences (Tukey's test, P < 0.01). The data are expressed as mean ± SEM.
Discussion
The aim of this study was to gain better insights into the performances of two widely used RNA extraction methods in virological and diagnostics laboratories, and to determine the optimal conditions for sample handling so as to minimize the loss of DENV viral RNA integrity.
The liquid phase partition-based extraction is accepted for routine extraction of clinical samples with various viral diseases28,29,30 and has been recommended for the detection of HIV, hepatitis C virus and hepatitis GB virus-C/hepatitis G, where maximum sensitivity was needed.7,10 However, the results from the present study indicated that the recoveries of DENV RNA from sera were poor with TRIzol LS, consistent with some reports.9,13,31 This could be significantly improved by the use of linear acrylamide as a co-precipitant and the reduction of serum protein. Similarly, enhancements in the recoveries of viral RNA were apparent when diluted DENV-infected patient sera were used.
The improved performances of silica-based RNA extraction methods in the presence of sera were likely to be due to the reduced interference of sera proteins, which is predominantly serum albumin. Contrary to the observation that silica preferentially bind high molecular weight RNA,19 which may exclude their use for the isolation of small size RNA resulting from alternative viral transcription, discontinuous extension of subgenomic viral RNA or partial degradation,32,33 we found that small single-stranded RNA did bind efficiently, with no preferential binding of higher molecular mass RNA. These data therefore implied that both the silica-based and liquid-phase partition methods (TRIZOL Reagent product insert: Invitrogen, Carlsbad, CA) can effectively recover various genomic or subgenomic viral RNAs of a wide range of sizes.
The suggested protocol for the storage of DENV-infected patient sera recommended by the World Health Organization-SEARO is at −70°C without thawing.3 Repeated freeze-thaw cycles have been shown to affect the integrity of cell-free RNA25 and viral RNA significantly.34 Unexpectedly, the recovery of DENV RNA was found not to be affected by repeated freeze-thawing of virus in serum. This is likely to be due to the protective effects of serum protein on the stability of viral particles, similar to that reported previously for another member of the Flaviviridae family, Hepatitis C virus.35 However, unlike Hepatitis C virus, which showed a statistically significant increase in RNA recovered with progressive freeze-thaw cycle, no significant changes were observed for the recovery of DENV RNA. The present study also showed that chaotrope-containing lysis/binding buffer could be used to store and maintain viral RNA integrity for at least 5 days.
In conclusion, it is recommended that for DENV RNA isolation using liquid phase partition methods, it is necessary to include co-precipitant and to reduce serum protein before phase partition. However, while diluting sera in PBS may help in the recovery of viral RNA, the sensitivity of the assay at low viral levels may be affected. A distinct advantage of using silica-based adsorption method is that it is easily automated using robotics.36 For cost efficiency and convenience while maintaining optimal viral RNA integrity, sera can be stored in the lysis/binding buffer and transported at room temperature to the diagnostics laboratories for extraction using silica-based adsorption methods, without the need for cold-chain storage.
Acknowledgements
We thank Tan Yin Jie and Yoong Li Foong for their assistance in the cloning and sequencing of the dengue constructs.
Footnotes
Supported by a collaborative grant awarded to the Division of Biomedical Sciences, Johns Hopkins in Singapore, and the National University of Singapore by Roche Diagnostics (Asia Pacific), and by a NIH grant U19 AI 056541.
A.A. and G.W. contributed equally to this paper.
Current address of A.A., Duke-National University of Singapore Graduate Medical School, 2 Jalan Bukit Merah, Singapore 169547; current address of K.-B.C., International Medical University, Sesama Center, Kuala Lumpur, Malaysia.
References
- 1.Halstead SB. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 2.Guzman MG, Kouri G. Dengue diagnosis, advances and challenges. Int J Infect Dis. 2004;8:69–80. doi: 10.1016/j.ijid.2003.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Rafei UM. Prevention and control of Dengue and Dengue haemorrhagic fever—Comprehensive guidelines. World Health Organization-South-East Asia Regional Office; 1999. [Google Scholar]
- 4.Lippi G, Guidi GC, Mattiuzzi C, Plebani M. Preanalytical variability: the dark side of the moon in laboratory testing. Clin Chem Lab Med. 2006;44:358–365. doi: 10.1515/CCLM.2006.073. [DOI] [PubMed] [Google Scholar]
- 5.Kahan LS. Medical devices; Immunology and microbiology devices; Classification of ribonucliec acid preanalytical systems. Federal Register. 2005;70:49862–49864. [PubMed] [Google Scholar]
- 6.de Paula VS, Villar LM, Coimbra Gaspar AM. Comparison of four extraction methods to detect hepatitis A virus RNA in serum and stool samples. Braz J Infect Dis. 2003;7:135–141. doi: 10.1590/s1413-86702003000200007. [DOI] [PubMed] [Google Scholar]
- 7.Fanson BG, Osmack P, Di Bisceglie AM. A comparison between the phenol-chloroform method of RNA extraction and the QIAamp viral RNA kit in the extraction of hepatitis C and GB virus-C/hepatitis G viral RNA from serum. J Virol Methods. 2000;89:23–27. doi: 10.1016/s0166-0934(00)00192-0. [DOI] [PubMed] [Google Scholar]
- 8.Fransen K, Mortier D, Heyndrickx L, Verhofstede C, Janssens W, van der Groen G. Isolation of HIV-1 RNA from plasma: evaluation of seven different methods for extraction (part two) J Virol Methods. 1998;76:153–157. doi: 10.1016/s0166-0934(98)00115-3. [DOI] [PubMed] [Google Scholar]
- 9.Knepp JH, Geahr MA, Forman MS, Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532–3536. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhofstede C, Fransen K, Marissens D, Verhelst R, van der Groen G, Lauwers S, Zissis G, Plum J. Isolation of HIV-1 RNA from plasma: evaluation of eight different extraction methods. J Virol Methods. 1996;60:155–159. doi: 10.1016/0166-0934(96)02062-9. [DOI] [PubMed] [Google Scholar]
- 11.Shafer RW, Levee DJ, Winters MA, Richmond KL, Huang D, Merigan TC. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–522. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolanaki E, Kottaridi C, Dedepsidis E, Kyriakopoulou Z, Pliaka V, Pratti A, Levidiotou-Stefanou S, Markoulatos P. Direct extraction and molecular characterization of enteroviruses genomes from human faecal samples. Mol Cell Probes. 2008;22:156–161. doi: 10.1016/j.mcp.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 13.De Paula SO, Nunes C, Matos R, de Oliveira ZM, Lima DM, da Fonseca BA. Comparison of techniques for extracting viral RNA from isolation-negative serum for dengue diagnosis by the polymerase chain reaction. J Virol Methods. 2001;98:119–125. doi: 10.1016/s0166-0934(01)00371-8. [DOI] [PubMed] [Google Scholar]
- 14.Prado I, Rosario D, Bernardo L, Alvarez M, Rodriguez R, Vazquez S, Guzman MG. PCR detection of dengue virus using dried whole blood spotted on filter paper. J Virol Methods. 2005;125:75–81. doi: 10.1016/j.jviromet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Matheus S, Meynard JB, Lacoste V, Morvan J, Deparis X. Use of capillary blood samples as a new approach for diagnosis of Dengue virus infection. J Clin Microbiol. 2007;45:887–890. doi: 10.1128/JCM.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould EA, Clegg JCS. Growth, titration and purification of alphaviruses and flaviviruses. In: Mahy BWJ, editor. Virology: A Practical Approach. IRL Press; Oxford: 1985. pp. 43–78. [Google Scholar]
- 17.Kumarasamy V, Wahab AH, Chua SK, Hassan Z, Chem YK, Mohamad M, Chua KB. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J Virol Methods. 2007;140:75–79. doi: 10.1016/j.jviromet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haimov-Kochman R, Fisher SJ, Winn VD. Modification of the standard trizol-based technique improves the integrity of RNA isolated from RNase-rich placental tissue. Clin Chem. 2006;52:159–160. doi: 10.1373/clinchem.2005.059758. [DOI] [PubMed] [Google Scholar]
- 20.Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linnet K, Boyd J. Selection and analytical evaluation of methods-with statistical techniques. In: Burtis C, Ashwood E, Bruns D, editors. Tietz Textbook Of Clinical Chemistry And Molecular Diagnostics. Saunders; St. Louis: 2006. pp. 353–407. [Google Scholar]
- 22.Richardson J, Molina-Cruz A, Salazar MI, Black Wt. Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes aegypti. Am J Trop Med Hyg. 2006;74:132–141. [PubMed] [Google Scholar]
- 23.Wang WK, Sung TL, Tsai YC, Kao CL, Chang SM, King CC. Detection of dengue virus replication in peripheral blood mononuclear cells from dengue virus type 2-infected patients by a reverse transcription-real-time PCR assay. J Clin Microbiol. 2002;40:4472–4478. doi: 10.1128/JCM.40.12.4472-4478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo KW, Lo YM, Leung SF, Tsang YS, Chan LY, Johnson PJ, Hjelm NM, Lee JC, Huang DP. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45:1292–1294. [PubMed] [Google Scholar]
- 25.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961–1965. [PubMed] [Google Scholar]
- 26.Tsui NB, Ng EK, Lo YM. Molecular analysis of circulating RNA in plasma. Methods Mol Biol. 2006;336:123–134. doi: 10.1385/1-59745-074-X:123. [DOI] [PubMed] [Google Scholar]
- 27.Castellino FJ, Barker R. The denaturing effectiveness of guanidinium, carbamoylguanidinium, and guanylguanidinium salts. Biochemistry. 1968;7:4135–4138. doi: 10.1021/bi00851a049. [DOI] [PubMed] [Google Scholar]
- 28.El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, Godfrey TE. Characterization of amplifiable. Circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50:564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 29.Kramvis A, Bukofzer S, Kew MC. Comparison of hepatitis B virus DNA extractions from serum by the QIAamp blood kit. GeneReleaser, and the phenol-chloroform method. J Clin Microbiol. 1996;34:2731–2733. doi: 10.1128/jcm.34.11.2731-2733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smalling TW, Sefers SE, Li H, Tang YW. Molecular approaches to detecting herpes simplex virus and enteroviruses in the central nervous system. J Clin Microbiol. 2002;40:2317–2322. doi: 10.1128/JCM.40.7.2317-2322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verheyden B, Thielemans A, Rombaut B, Kronenberger P. RNA extraction for quantitative enterovirus RT-PCR: comparison of three methods. J Pharm Biomed Anal. 2003;33:819–823. doi: 10.1016/s0731-7085(03)00312-1. [DOI] [PubMed] [Google Scholar]
- 32.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 34.Gessoni G, Barin P, Valverde S, Giacomini A, Di Natale C, Orlandini E, Arreghini N, De Fusco G, Frigato A, Fezzi M, Antico F, Marchiori G. Biological qualification of blood units: considerations about the effects of sample's handling and storage on stability of nucleic acids. Transfus Apher Sci. 2004;30:197–203. doi: 10.1016/j.transci.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Krajden M, Minor JM, Rifkin O, Comanor L. Effect of multiple freeze-thaw cycles on hepatitis B virus DNA and hepatitis C virus RNA quantification as measured with branched-DNA technology. J Clin Microbiol. 1999;37:1683–1686. doi: 10.1128/jcm.37.6.1683-1686.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarrazin C, Dragan A, Gartner BC, Forman MS, Traver S, Zeuzem S, Valsamakis A. Evaluation of an automated, highly sensitive, real-time PCR-based assay (COBAS Ampliprep/COBAS TaqMan) for quantification of HCV RNA. J Clin Virol. 2008;43:162–168. doi: 10.1016/j.jcv.2008.06.013. [DOI] [PubMed] [Google Scholar]


