Abstract
Evidence that activating mutations of the KRAS oncogene abolish the response to anti-epidermal growth factor receptor therapy has revolutionized the treatment of advanced colorectal cancer. This has resulted in the urgent demand for KRAS mutation testing in the clinical setting to aid choice of therapy. The aim of this study was to evaluate six different KRAS mutation detection methodologies on two series of primary colorectal cancer samples. Two series of 80 frozen and 74 formalin-fixed paraffin-embedded tissue samples were sourced and DNA was extracted at a central site before distribution to seven different testing sites. KRAS mutations in codons 12 and 13 were assessed by using single strand conformation polymorphism analysis, pyrosequencing, high resolution melting analysis, dideoxy sequencing, or the commercially available TIB Molbiol (Berlin, Germany) or DxS Diagnostic Innovations (Manchester, UK) kits. In frozen tissue samples, concordance in KRAS status (defined as consensus in at least five assays) was observed in 66/80 (83%) cases. In paraffin tissue, concordance was 46/74 (63%) if all assays were considered or 71/74 (96%) using the five best performing assays. These results demonstrate that a variety of detection methodologies are suitable and provide comparable results for KRAS mutation analysis of clinical samples.
The epidermal growth factor receptor (EGFR) mediates molecular events critical to cellular growth and survival. This receptor is up-regulated in the majority of colorectal cancers (CRCs) and contributes to cancer progression by modulation of events such as proliferation, adhesion, and angiogenesis.1,2 EGFR therefore presents an excellent candidate for targeted therapy.3 Monoclonal antibody treatments such as cetuximab have been developed that target the extracellular binding domain of EGFR, thus competitively inhibiting the binding of ligand to its receptor. However, a significant proportion of EGFR positive CRCs remain resistant to this therapy. One reason for resistance is that the pathway may also be activated downstream of EGFR due to mutation of the KRAS oncogene, which occurs in approximately 30 to 40% of CRCs.4 The overall survival benefit in patients with stage IV CRC following treatment with anti-EGFR therapies has been modest; however, when patients are stratified for KRAS mutation status, overall and progression-free survival rates are significantly improved for patients with wild-type KRAS.5,6,7,8,9,10
The recent and consistent demonstrations of efficacy for anti-EGFR therapies in KRAS wild-type CRC has resulted in a rapid increase in demand for mutation testing in the clinical setting. Although a provisional clinical opinion statement has recently been released by the American Society of Clinical Oncology,7 no published studies have evaluated the different testing methodologies that are currently available. The sensitivity and specificity of each methodology is influenced by the proportion of cancer cells in the sample, the quality of DNA obtained, and inherent limitations of the technology. CRCs are infiltrated by varying amounts of stromal epithelium and normal cells such as fibroblasts and lymphocytes, with the result that some clinical samples may contain only a small proportion of mutated DNA. The sensitivity of most assays for KRAS mutation detection is therefore reliant on careful selection of tissue samples that are enriched for tumor cells. This can be further improved by microdissection or by core biopsy sampling. Another major consideration for KRAS mutation screening is the tissue source. The majority of assays will necessarily be performed on archival formalin-fixed paraffin embedded (FFPE) samples from the patient's primary lesion, although biopsies from metastatic cancer sites may be available in some instances.
The vast majority of KRAS mutations occur in codon 12 or 13 of the gene, with a small minority of mutations occurring in codon 61.11 A number of different KRAS mutation detection methods are in routine laboratory use, including automated dideoxy sequencing, pyrosequencing, single strand conformation polymorphism (SSCP) analysis, and high resolution melting (HRM) analysis. Sequence information is provided by the former two methods, whereas a binary result is provided by the latter two. Although dideoxy sequencing has long been considered the “gold standard” for mutation detection, there is considerable evidence to suggest that it may not be the most sensitive of these four methods.12 The current study was designed to evaluate four different KRAS mutation detection assays (dideoxy sequencing, pyrosequencing, SSCP, and HRM) currently in use across five clinical testing sites, as well as two commercially available kits (TIB Molbiol [Berlin, Germany] and DxS Diagnostic Innovations [Manchester, UK]).
Materials and Methods
CRC Tissue Samples
CRC tissue samples were obtained from the CRC tissue bank in the School of Surgery, University of Western Australia. The first cohort was a consecutive series of 80 surgically resected, primary CRC specimens that were snap frozen in liquid nitrogen and stored at −70°C before extraction of genomic DNA by using standard phenol chloroform methods. The second cohort comprised 74 FFPE CRC samples. H&E-stained sections from this cohort were evaluated by a pathologist (K.T.) to estimate the percentage of tumor cells. For FFPE samples, 2 to 3 sections (10-μm thickness) were incubated for 10 minutes at 94°C in 300 μl of digestion buffer (10 mmol/L Tris-hydrochloric acid, pH 8.3; 1 mmol/L EDTA; 0.5% Tween 20) followed by centrifugation at 13,000 rpm for 10 minutes.13 The surface paraffin layer was removed with a tip and the tissue pellet was placed in a new tube containing 200 μl of digestion buffer. Proteinase K solution was added (20 μl of 20 mg/ml, Promega, Madison, WI) and the samples were incubated for 72 hours at 55°C with rotation. The enzyme was then inactivated by heating for 10 minutes at 94°C and the samples were centrifuged at 13,000 rpm for 10 minutes. The upper 100 μl of solution beneath the residual paraffin layer and containing the extracted DNA was removed and stored at 4°C for PCR. All patients gave informed consent for the use of stored tissues for molecular analysis.
Testing Sites and Methodologies Assessed
DNA from the two CRC cohorts was distributed to four testing sites in Australia, two sites in Singapore, and one site in the United Kingdom (Table 1). To limit variability between testing sites, DNA was extracted from FFPE samples at one site (Perth, Australia) before distribution. Additional sections of FFPE blocks were sent to each site for re-extraction by individual testing centers as needed. KRAS mutation assays were performed without knowledge of the percentage of tumor cells estimated for each sample.
Table 1.
Mutation Detection Frequencies at Each Site
| Frozen samples (n = 80) |
FFPE samples (n = 75) |
||||
|---|---|---|---|---|---|
| Testing site | Method* | No. mutations detected (%) | % of consensus† | No. mutations detected (%) | % of consensus† |
| A | SSCP | 30/80 (37.5) | 111.1 | 27/74 (36.5) | 100.0 |
| B | Pyrosequencing | 22/80 (27.5) | 81.5 | 26/74 (35.1) | 96.1 |
| C | HRM 128 bp | 27/80 (33.8) | 100.0 | 28/74 (37.8) | 103.6 |
| D | HRM 80 bp | 27/80 (33.8) | 100.0 | 28/74 (37.8) | 103.6 |
| E | Sequencing | 28/80 (35.0) | 104.5 | 23/70 (32.9) | 90.1 |
| F | TIB Molbiol Kit | 31/80 (38.8) | 114.8 | 44/74 (59.5) | 163.0 |
| G | DxS Kit | 28/75 (37.3) | 110.4 | 28/71 (39.4) | 107.9 |
| Consensus‡ | 27/80 (33.8) | 27/74 (36.5) | |||
A, Perth, Australia; B, Newcastle, Australia; C, Melbourne, Australia; D, Brisbane, Australia; E, Department of Pathology, National University of Singapore; F, Cancer Science Institute, National University of Singapore; G, Manchester, UK.
Note that for the HRM 128 bp assay the target sequence is 92 bp, whilst the amplicon size is 128 bp due to the presence of M13 tags.
The percentage of consensus value for each method is defined as the number of mutations detected by the method divided by the consensus number of mutations (27 for both frozen and FFPE samples).
Consensus mutations were defined as those detected at five or more of the seven test sites.
SSCP Analysis
Fluorescent single SSCP analysis was performed in Perth, Australia. Primers used for the KRAS assay (forward, 5′-GACTGAATATAAACTTGTGG-3′; reverse, 5′-CTATTGTTGGATCATATTCG-3′) were labeled at the 5′ end with fluorescent HEX dye (GeneWorks, Adelaide, Australia). PCR conditions were 5 minutes of denaturation at 94°C during which time 1 μl of DNA solution was added, followed by 32 cycles of denaturation at 94°C, annealing at 54°C, and extension at 72°C (40 seconds each). The program was terminated by 7 minutes extension at 72°C. Three μl of PCR product was mixed with 6 μl of deionized formamide loading buffer containing 0.05% dextran blue and heated to 94°C for 5 minutes. One microliter of this mixture was then loaded onto a nondenaturing 10% polyacrylamide/2% glycerol gel (100-μm thickness, 18-cm length) and mounted in a DNA fragment analyzer (GS-3000, Corbett Life Sciences, New South Wales, Australia). Samples were pulse-loaded for 20 seconds at 1400V before rinsing the wells and then running the gel for 1 hour at 1400V in 0.8× Tris-Borate-EDTA buffer. Gel temperature was maintained at a constant 24°C throughout the run by an inbuilt cooling unit. The electrophoretogram was analyzed by using ONE-Dscan 1.3 software (Scanalytics, Billerica, MA). DNA samples that displayed bands additional to the wild-type bands were classified as showing a mutation. Positive controls comprising of DNA with known KRAS mutations were run with each gel.
Pyrosequencing
The Pyrosequencing assay (Newcastle, Australia) was performed according to the method of Ogino et al.12 A 5% threshold value was chosen arbitrarily to define the presence of mutation.
HRM Analysis
HRM analysis was performed in Melbourne, Australia by using a LightCycler 480 (Roche Diagnostics, Penzberg, Germany) in a modification of the previously published method of Krypuy et al.14 The reaction mixture contained 1× PCR buffer, 2.5 mmol/L MgCl2, 200 nmol/L of each primer (5′-TGTAAAACGACGGCCAGTTTATAAGGCCTGCTGAAAATGACTGAA-3′ and 5′-CAGGAAACAGCTATGACCTGAATTAGCTGTATCGTCAAGGCACT-3′, tagged with M13 sequences [in bold] to allow direct sequencing of the HRM product in the clinical setting), DNA template of various concentrations, 200 μmol/L dNTPs (Fisher Biotec Australia, Wembley, Western Australia), 5 μmol/L SYTO 9 (Invitrogen, Carlsbad, CA), and 0.5U HotStarTaq polymerase (Qiagen, Hilden, Germany) in a 10-μl final reaction volume. The cycling and melting conditions were as follows: one cycle of 95°C for 10 minutes; 56 cycles of 95°C for 10 seconds; 60°C for 10 seconds with an initial 20 cycles of touchdown from 70°C (0.5°C/cycle); 72°C for 20 seconds; one cycle of 97°C for 1 minute; and a melt from 65°C to 95°C rising at 4.4°C/second. All samples were tested in triplicate. Mutations were visualized using the LightCycler 480 software version 1.5 (Roche Diagnostics).
HRM analysis was also performed (Brisbane, Australia) on a Corbett Research RotorGene 6000 real-time PCR machine (Qiagen, Hilden, Germany). The reaction consisted of 25 ng of DNA template, 1× PCR buffer, 2.5 mmol/L MgCl2, 5 μm of SYTO 9 (Invitrogen), 200 nmol/L each of forward 5′-TGAAAATGACTGAATATAAACTTGTGG-3′ and reverse 5′-CTGAATTAGCTGTATCGTCAAGG-3′ primers, 0.5U of AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA), and water to a final volume of 16 μl. The polymerase was activated at 95°C for 12 minutes before 40 amplification cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The product was denatured at 95°C for 5 minutes, heteroduplexes were allowed to form at 50°C for 2 minutes and then the 80 base pair (bp) product was melted over a gradient from 72 to 86°C rising by 0.2°C/second. Mutations were visualized by using the normalized graph and melt curve analysis software (Corbett Research).
Re-extraction of DNA as necessary from paraffin sections was performed by using the QIAamp DNA FFPE tissue kit (Qiagen) according to the manufacturer's instructions.
Dideoxy Sequencing
Automated dideoxy sequencing was performed in the Department of Pathology, National University of Singapore, according to the methods of Krypuy et al.14 Following PCR, products were purified with the ExoSap-IT reagent (USB, Cleveland, Ohio). Purified PCR product was sequenced by using the BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) and the same primers were used for PCR. The 3100 Genetic Analyzer (Applied Biosystems) was used for capillary electrophoresis and sequence analysis. All samples were sequenced in both directions.
TIB Molbiol Kit
The TIB Molbiol KRAS mutation detection kit (LightMix Kit, TIB Molbiol) was trialed at the Cancer Science Institute, National University of Singapore and performed according to the kit instructions. Briefly, 16 μl of LightCycler FastStart DNA Master Hyprobe (Roche Diagnostics) was added to 113.8 μl of TIB Molbiol reaction mixes and 15 μl of this solution was aliquoted into a capillary. Five microliters (∼100 ng) of DNA was then added and PCR amplification and melting curve analyses were performed on a LightCycler 2.0 (Roche Diagnostics).
DxS Kit
Mutation analysis for all samples was also performed by using the DxS KRAS mutation detection kit by the manufacturers (DxS Diagnostic Innovations). This technology combines allele specific PCR (ARMS) with real-time PCR (Scorpions).
Statistical Analysis
Data from individual testing sites was sent to a central site (Brisbane, Australia) for collation and analysis. Data were analyzed by using GraphPad Prism 5 (GraphPad Software, Inc, San Diego, CA). Direct estimates of sensitivity and specificity were not possible as there was no reliable predetermined gold standard method for comparison.
Results
The frequency of mutation detection across the different testing centers is shown in Figure 1. A clear bimodal distribution was observed for both frozen and FFPE samples. The raw data for frozen and FFPE samples is shown in Tables 2 and 3, respectively. A “consensus mutation” was called if a mutation was detected at five or more sites (Table 1). Frequencies of the consensus mutations for both frozen (27/80, 33.8%) and FFPE (27/74, 36.5%) samples were consistent with reports in the literature. A mutation was detected at just one or two testing sites in 9/80 (11.3%) frozen tissue samples compared with 21/74 (28.4%) FFPE samples. The poor concordance for FFPE samples was largely due to the abnormally high mutation frequency detected with the TIB Molbiol kit (44/74, 59.5%) compared with the consensus frequency of 36.5% (Table 3). For both frozen and FFPE samples, 100% concordance was observed between the two sites that used HRM, despite the use of different primers, reagents, assay conditions, and instruments.
Figure 1.
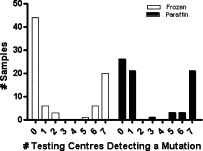
Frequency of KRAS mutation detection for frozen and FFPE primary CRC samples among the different test sites. Samples were deemed wild-type for KRAS when none of the centers or one center only detected a mutation. The consensus for presence of KRAS mutation was when five or more centers detected a mutation in that sample.
Table 2.
Mutation Detection in Frozen Tissue Samples
| Sample no. | SSCP | Pyrosequencing | HRM 92 bp | HRM 80 bp | Tib MolBiol | Dideoxy sequencing | DxS | % Mutant |
|---|---|---|---|---|---|---|---|---|
| F1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F2 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F3 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F5 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F6 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F7 | ○ | ○ | ○ | ○ | ○ | ○ | ND | 0.0 |
| F8 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F9 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F10 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F11 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F12 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F13 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F14 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F15 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F16 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F17 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F18 | ○ | ○ | ○ | ○ | ○ | ○ | ND | 0.0 |
| F19 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F20 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F21 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F22 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F23 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F24 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F25 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F26 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F27 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F28 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F29 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F31 | ○ | ○ | ○ | ○ | ○ | ○ | ND | 0.0 |
| F32 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F33 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F34 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F35 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F36 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F37 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F38 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F39 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F40 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F41 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F42 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F43 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F44 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| F45 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| F46 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| F47 | ○ | ○ | ○ | ○ | ○ | ○ | • | 14.3 |
| F48 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| F49 | ○ | ○ | ○ | ○ | ○ | ○ | • | 14.3 |
| F50 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| F51 | • | ○ | ○ | ○ | • | ○ | ○ | 28.6 |
| F52 | • | ○ | ○ | ○ | ○ | ○ | • | 28.6 |
| F53 | • | ○ | ○ | ○ | ○ | • | ○ | 28.6 |
| F54 | • | ○ | • | • | ○ | • | • | 71.4 |
| F55 | • | ○ | • | • | • | • | • | 85.7 |
| F56 | • | ○ | • | • | • | • | • | 85.7 |
| F57 | • | ○ | • | • | • | • | • | 85.7 |
| F58 | • | ○ | • | • | • | • | • | 85.7 |
| F59 | • | • | • | • | • | • | • | 100.0 |
| F60 | • | • | • | • | • | • | • | 100.0 |
| F61 | • | • | • | • | • | • | • | 100.0 |
| F62 | • | • | • | • | • | • | ND | 100.0 |
| F63 | • | • | • | • | • | • | • | 100.0 |
| F64 | • | • | • | • | • | • | • | 100.0 |
| F65 | • | • | • | • | • | • | • | 100.0 |
| F66 | • | • | • | • | • | • | • | 100.0 |
| F67 | • | • | • | • | • | • | • | 100.0 |
| F68 | • | • | • | • | • | • | • | 100.0 |
| Sample no. | SSCP | Pyrosequencing | HRM 92 bp | HRM 80 bp | Tib MolBiol | Dideoxy sequencing | DxS | % Mutant |
|---|---|---|---|---|---|---|---|---|
| F69 | • | • | • | • | • | • | • | 100.0 |
| F70 | • | • | • | • | • | • | • | 100.0 |
| F71 | • | • | • | • | • | • | • | 100.0 |
| F72 | • | • | • | • | • | • | • | 100.0 |
| F73 | • | • | • | • | • | • | ND | 100.0 |
| F74 | • | • | • | • | • | • | • | 100.0 |
| F75 | • | • | • | • | • | • | • | 100.0 |
| F76 | • | • | • | • | • | • | • | 100.0 |
| F77 | • | • | • | • | • | • | • | 100.0 |
| F78 | • | • | • | • | • | • | • | 100.0 |
| F79 | • | • | • | • | • | • | • | 100.0 |
| F80 | • | • | • | • | • | • | • | 100.0 |
Closed circles indicate that a mutation was detected, whereas open circles indicate no mutation was detected. ND indicates no data were provided by the testing site. Percent of samples with a KRAS mutation was calculated based on the number of sites where data were available.
Table 3.
Mutation Detection in FFPE Samples
| Sample no. | % Tumor cells | SSCP | Pyrosequencing | HRM 92 bp | HRM 80 bp | Tib MolBiol | Dideoxy sequencing | DxS | % Mutant |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 80 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P2 | 30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P3 | 60 | ○ | ○ | ○ | ○ | ○ | ○ | ND | 0.0 |
| P4 | 30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P5 | 50 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P6 | 60 | ○ | ○ | ○ | ○ | ○ | ○ | ND | 0.0 |
| P7 | 40 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P8 | 15 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P9 | 45 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P10 | 70 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P11 | 15 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P12 | 50 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P13 | 70 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P14 | 30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P15 | 50 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P16 | 80 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P17 | 50 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P18 | 30 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P19 | 70 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P20 | 40 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P21 | 40 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P22 | 60 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P23 | 50 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P24 | 80 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P25 | 80 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | 0.0 |
| P26 | 20 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P27 | 40 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P28 | 10 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P29 | 30 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P30 | 30 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P31 | 50 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P32 | 50 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P33 | 40 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P34 | 80 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P35 | 20 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P36 | 30 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P37 | 60 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P38 | 30 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P39 | 40 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P40 | 50 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P41 | 70 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P42 | 60 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P43 | 70 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P44 | 80 | ○ | ○ | ○ | ○ | • | ○ | ○ | 14.3 |
| P45 | 20 | ○ | ○ | ○ | ○ | • | ○ | ND | 16.7 |
| P46 | 20 | ○ | ○ | ○ | ○ | • | ND | ○ | 16.7 |
| P47 | 60 | ○ | ○ | • | • | ○ | ○ | • | 42.9 |
| P48 | 40 | • | • | • | • | ○ | ND | • | 83.3 |
| P49 | 30 | • | • | • | • | ○ | ND | • | 83.3 |
| P50 | 50 | • | • | • | • | ○ | ND | • | 83.3 |
| P51 | 10 | • | ○ | • | • | • | • | • | 85.7 |
| P52 | 30 | • | • | • | • | ○ | • | • | 85.7 |
| P53 | 40 | • | • | • | • | • | ○ | • | 85.7 |
| P54 | 80 | • | • | • | • | • | • | • | 100.0 |
| P55 | 60 | • | • | • | • | • | • | • | 100.0 |
| P56 | 30 | • | • | • | • | • | • | • | 100.0 |
| P57 | 30 | • | • | • | • | • | • | • | 100.0 |
| P58 | 30 | • | • | • | • | • | • | • | 100.0 |
| P59 | 60 | • | • | • | • | • | • | • | 100.0 |
| P60 | 70 | • | • | • | • | • | • | • | 100.0 |
| P61 | 80 | • | • | • | • | • | • | • | 100.0 |
| P62 | 20 | • | • | • | • | • | • | • | 100.0 |
| P63 | 25 | • | • | • | • | • | • | • | 100.0 |
| P64 | 40 | • | • | • | • | • | • | • | 100.0 |
| P65 | 50 | • | • | • | • | • | • | • | 100.0 |
| P66 | 30 | • | • | • | • | • | • | • | 100.0 |
| P67 | 30 | • | • | • | • | • | • | • | 100.0 |
| (table continues) |
| Sample no. | % Tumor cells | SSCP | Pyrosequencing | HRM 92 bp | HRM 80 bp | Tib MolBiol | Dideoxy sequencing | DxS | % Mutant |
|---|---|---|---|---|---|---|---|---|---|
| P68 | 60 | • | • | • | • | • | • | • | 100.0 |
| P69 | 50 | • | • | • | • | • | • | • | 100.0 |
| P70 | 30 | • | • | • | • | • | • | • | 100.0 |
| P71 | 20 | • | • | • | • | • | • | • | 100.0 |
| P72 | 30 | • | • | • | • | • | • | • | 100.0 |
| P73 | 60 | • | • | • | • | • | • | • | 100.0 |
| P74 | 40 | • | • | • | • | • | • | • | 100.0 |
Closed circles indicate that a mutation was detected, whereas open circles indicate no mutation was detected. ND indicates no data were provided by the testing site. Percent of samples with a KRAS mutation was calculated based on the number of sites where data were available.
Mutation Detection in Frozen Tissue Samples
In the series of frozen CRC tissues, full concordance (all wild-type or all mutant) was observed for 66/80 (82.5%) samples (Table 2). Mutations were detected in four samples (F55 to F58) by all methods except pyrosequencing, and in one sample (F54) by all methods except pyrosequencing and the TIB Molbiol kit. KRAS mutations were not detected in five samples (F54 to F58) by pyrosequencing, possibly reflecting low tumor cell content in these samples. This was suggested by the low intensity or magnitude of the aberrant bands observed for sample F55 by using SSCP, HRM, and dideoxy sequencing (Figure 2, A–F). Mutations were detected in an additional nine samples (F45 to F53) by two or fewer sites. Mutations were detected in three samples by SSCP and one additional site. One of these was detected by SSCP and sequencing (F53), and proved to be a bona fide mutation, albeit outside of the codons 12 and 13 hotspots (codon 20, ACG to ATG). One mutation was detected by SSCP and DxS only and one by SSCP and the TIB Molbiol kit only. Of the six mutations detected by a single site only, four were by the TIB Molbiol kit alone and two were by the DxS kit alone.
Figure 2.
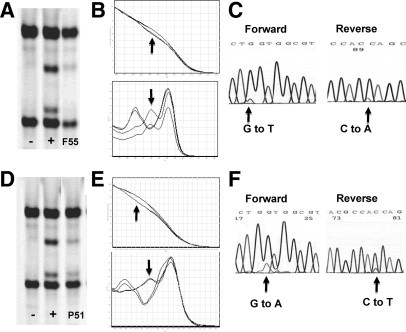
Mutation detection by using SSCP, HRM, and dideoxy sequencing. Raw data were shown for frozen sample 55 (F55). Where a faint band was detected by SSCP (A; −, negative control; +, positive control), a clear difference in melt temperature was detected by HRM 82 bp assay (B, mutant indicated by arrows) and subtle variant peak identified by dideoxy sequencing (C). Similarly, a mutation in FFPE sample 51 (P51) estimated to have a 10% tumor cell content is shown by using SSCP (D), HRM 82 bp assay (E, mutant indicated by arrows), and dideoxy sequencing (F).
Mutation Detection in FFPE Tissue Samples
Concordance was seen across all testing sites in 46/74 (62.2%) FFPE samples. Compared with the overall mutation frequency detected with other methodologies (range, 32.9 to 39.4%), a very high mutation frequency of 59.5% was seen by using the TIB Molbiol kit. If data from this assay were excluded from the paraffin series, an excellent concordance between sites was observed (100% concordance in 71/74 samples). Excluding data from the TIB Molbiol kit, discordance was seen in only three samples. A mutation in sample P47 was not detected by SSCP, pyrosequencing, or dideoxy sequencing (60% tumor cell content), in P53 by dideoxy sequencing alone (40% tumor cell content) or in P51 by pyrosequencing alone (10% tumor cell content). No result was reported in four samples by dideoxy sequencing (P46, P48, P49, and P50) and three samples by DxS (P3, P6, and P45) because they did not meet the quality assurance conditions imposed by these testing sites.
Detection of Different Types of KRAS Mutations
An advantage of automated dideoxy sequencing, pyrosequencing, and the TIB Molbiol and DxS mutation detection kits is that the type of KRAS mutation is determined. We therefore compared the specificity of detection of each mutation type across these four methodologies in both the frozen and FFPE series (Table 4). Excellent concordance was seen for all but the TIB Molbiol kit where the incorrect mutation type was reported in 7/26 (26.9%) frozen samples and 2/22 (9.1%) FFPE samples. Consensus for mutation type was not reached in one other FFPE sample (P73), with dideoxy sequencing and DxS reporting a 12V mutation, whereas pyrosequencing and the TIB Molbiol kit reported a 12A mutation.
Table 4.
Types of Mutations Detected
| Frozen Tissue |
Paraffin Tissue |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample no. | Dideoxy sequencing | Pyrosequencing | DxS | TIB-Molbiol | Sample no. | Dideoxy sequencing | Pyrosequencing | DxS | TIB-Molbiol |
| F65 | 12A | 12A | 12A | 12D | P60 | 12A | 12A | 12A | 12D |
| F55 | 12C | wt | 12C | 12C | P54 | 12A | 12A | 12A | 12D |
| F77 | 12C | 12C | 12C | 12C | P49 | ND | 12A | 12A | wt |
| F79 | 12C | 12C | 12C | 12C | P73 | 12V | 12A | 12V | 12A |
| F54 | 12C | wt | 12C | wt | P59 | 12C | 12C | 12C | 12C |
| F66 | 12D | 12D | 12D | 12D | P68 | 12C | 12C | 12C | 12C |
| F69 | 12D | 12D | 12D | 12D | P52 | 12C | 12C | 12C | wt |
| F74 | 12D | 12D | 12D | 12D | P48 | ND | 12C | 12C | wt |
| F76 | 12D | 12D | 12D | 12D | P61 | 12D | 12D | 12D | 12D |
| F62 | 12D | 12D | ND | 12D | P63 | 12D | 12D | 12D | 12D |
| F60 | 12D | 12D | 12D | 12D | P70 | 12D | 12D | 12D | 12D |
| F61 | 12D | 12D | 12D | 12D | P72 | 12D | 12D | 12D | 12D |
| F56 | 12D | wt | 12D | 12D | P57 | 12S | 12S | 12S | 12S |
| F58 | 12D | wt | 12D | 12D | P64 | 12S | 12S | 12S | 12S |
| F78 | 12S | 12S | 12S | 12S | P55 | 12V | 12V | 12V | 12V |
| F63 | 12V | 12V | 12V | 12A | P58 | 12V | 12V | 12V | 12V |
| F64 | 12V | 12V | 12V | 12A | P67 | 12V | 12V | 12V | 12V |
| F71 | 12V | 12V | 12V | 12A | P56 | 12V | 12V | 12V | 12V |
| F72 | 12V | 12V | 12V | 12A | P53 | wt | 12V | 12V | 12V |
| F59 | 12V | 12V | 12V | 12V | P69 | 12V | 12V | 12V | 12V |
| F67 | 12V | 12V | 12V | 12V | P50 | ND | 12V | 12V | wt |
| F75 | 12V | 12V | 12V | 12V | P62 | 13D | 13D | 13D | 13D |
| F73 | 12V | 12V | ND | 12V | P65 | 13D | 13D | 13D | 13D |
| F57 | 12V | wt | 12V | 12A | P66 | 13D | 13D | 13D | 13D |
| F68 | 13D | 13D | 13D | 13D | P71 | 13D | 13D | 13D | 13D |
| F70 | 13D | 13D | 13D | 13D | P74 | 13D | 13D | 13D | 13D |
| F80 | 12A | 12A | 12A | 12D | P51 | 12D | wt | 12D | 12D |
wt, wild-type; ND, no data; discordant data shown in bold.
Data are shown only if a mutation was detected at two or more sites.
Discussion
This study evaluated various KRAS mutation screening techniques in two series of primary CRC tissues totaling 154 samples. The first series comprised high quality DNA extracted from unfixed, frozen tumor specimens, while the second series comprised archival FFPE samples. The latter are more representative of the source material used in KRAS testing centers. DNA was extracted centrally and distributed to seven sites that independently screened for KRAS mutations by using different methodologies, including two commercially available mutation detection kits. This is the first study to assess the concordance of different methods for the detection of KRAS mutation across multiple testing sites and using clinical specimens. Although excellent agreement between methods was observed, it was less than 100%. This raises issues about what the gold standard for KRAS mutation detection should be for routine clinical testing.
DNA quality is a potentially important factor affecting the performance of KRAS mutation assays. In the clinical setting the tissue source will usually be archival FFPE blocks from surgically resected primary cancers, although there are also a significant proportion of biopsy samples from the primary or metastatic lesion. Formalin fixation dramatically reduces DNA quality; however, this did not appear to have a negative impact on assay performance in this study, as judged by the high degree of concordance between different techniques. The exception to this was the TIB Molbiol kit. Although this kit performed reasonably well with frozen tissue DNA, it gave an unacceptably high frequency of false positive results with FFPE samples. The DNA extraction procedure used here for FFPE tissues was a simple Proteinase K-based digestion method. It worked particularly well for the SSCP and pyrosequencing assays; however, other purification methods may have increased the data concordance for some of the other methods, especially the TIB Molbiol kit. No result could be obtained for five frozen tissue and three FFPE tissue samples by the DxS method and for four FFPE series samples by dideoxy sequencing.
The use of a small amplicon or target DNA fragment size is important for assay success when using FFPE samples. Assay design is critical for methods such as dideoxy sequencing where the amplicon should be of sufficient size to allow high quality sequence information across the region of interest, and that region should be centrally placed in the amplicon to allow high quality forward and reverse sequence.
Another key determinant of assay performance is the relative tumor cell content of tissue samples. Four of the FFPE samples in this study were estimated to have a tumor cell content of less than 20% (P8, P11, P28, and P51). One of these (P51, estimated 10% tumor cells) was found to contain a mutation by using all methods except pyrosequencing. On the other hand, three FFPE samples with 20 to 25% tumor cell content (P62, P63, and P71) were found to contain a mutation at all seven test sites, indicating that a 20% threshold for minimum tumor cell content is appropriate. Significant enrichment for tumor cells can be achieved by routine microdisection or core biopsy of FFPE blocks following identification of suitable areas by a histopathologist. Case review by a histopathologist is essential in solid tumor testing and further involvement in block selection and tumor cell enrichment by a histopathologist with an understanding of the associated molecular test will enhance this process.15 It should be noted that, in the clinical setting, tissue microdissection is routinely conducted by the Melbourne (HRM) and Singapore (dideoxy sequencing) sites and core sampling is performed by the Newcastle site (pyrosequencing), thus minimizing the impact of a low tumor to normal cell ratio. In addition to samples with a high degree of stromal involvement, histological evaluation will be particularly important for cancers treated with chemoradiation where few tumor cells may remain. In this instance it may be necessary to access the original diagnostic biopsy rather than surgical resection specimen.
A further consideration regarding tissue source is whether the sample is obtained from the primary cancer or a metastatic site. It could be argued that therapy should be directed against the metastatic disease. However, in the large majority of cases only the primary tumor specimen will be available for analysis. KRAS is believed to be mutated early in the adenoma-carcinoma progression and hence the metastatic tissues should be clonally derived from the primary tumor. This is supported by a recent study showing a high level of concordance for KRAS mutation status in 95/99 (96.0%) matched primary and metastatic CRC samples.16
The utility of sequence information should also be considered when choosing the assay. Although uncommon, mutations do exist outside the major codon 12/13 hotspots.17,18 These may not activate MAPK signaling and this should be considered before denial of therapy. A single example of such a mutation (codon 20, ACG to ATG) was detected by SSCP and dideoxy sequencing (F53). A database of sequence variants may be useful for future determination of the clinical relevance of uncommon mutations. Such a database should include reports of uncommon mutations, details of clinical management and patient outcome as well as any available evidence for impact on MAPK pathway signaling.
Cost and turnaround time of assays are also important factors for routine clinical testing (Table 5). SSCP, HRM, and pyrosequencing are rapid, high-throughput and inexpensive methods that performed well in this study for both frozen and FFPE samples. The dideoxy sequencing assay was more expensive than each of these methods. However, one advantage of this method is that DNA sequencing services are widely available in many centers. In addition, sequencing is a necessary quality control step for the SSCP and HRM assays.
Table 5.
Estimates of Assay Cost and Turnover Time
| Method | Reagent costs | Labor time | Turnover time, hours* |
|---|---|---|---|
| SSCP | + | ++ | 5 |
| HRM | + | + | 2.5 |
| Pyrosequencing | ++ | ++ | 3.5 |
| Sequencing | ++ | +++ | 5 |
| TIB Molbiol | +++++ | +++++ | 2.5 |
| DxS | +++++ | ++ | 2.5 |
Exclusive of DNA extraction.
The DxS kit performed well in this study and offers a rapid, standardized and high-throughput platform. However, the cost of this assay means that it could be prohibitive for many clinical testing facilities. In addition, patients with rarer codon 13 mutations will not be detected. The poor performance of the TIB Molbiol kit for FFPE samples indicates that it may not be suitable for use in the clinical setting.
A further important consideration in the method of choice for KRAS mutation screening is the equipment and technical expertise that are available at each testing site. These tests must be performed in conjunction with an anatomical pathologist to review the tumor cell content of samples and with an experienced molecular geneticist to review the mutation results.
Other molecular events may account for the resistance to anti-EGFR therapies seen in some patients with wild-type KRAS. There is clinical evidence that the V600E hotspot mutation in the BRAF oncogene also confers resistance to cetuximab.19 BRAF functions downstream of KRAS in the MAPK pathway and mutations of BRAF and KRAS are mutually exclusive. Clinical testing for BRAF mutation will therefore also play an important role in predicting response to anti-EGFR therapies and will need to be implemented with consideration of the same issues discussed here for KRAS testing. Other evidence is accumulating for a possible role of disrupted PIK3CA, PTEN, amphiregulin, and epiregulin in determining the response to anti-EGFR therapies.20,21,22,23,24
This study has demonstrated a high degree of concordance in KRAS mutation detection for routine testing methodologies across different testing sites. Discordant results for pyrosequencing in the frozen tumor series may have been due to lower tumor cell content in these samples. Based on the results of our study, the SSCP, HRM, dideoxy sequencing, and pyrosequencing methods appear to be suitable and cost-effective assays for KRAS testing of clinical specimens. It is important, however, that suitably qualified personnel are used to establish these techniques and to perform ongoing quality control testing. The sensitivity of some methods may be increased by prior tumor cell enrichment by using microdissection or core biopsy, and by the use of column-based DNA extraction procedures. Because metastatic CRC has a high frequency of KRAS mutation and the time required to obtain a result is often dependent on the speed of retrieval of archival FFPE blocks, routine KRAS mutation testing at the time of surgical resection may also be worth considering.
The discovery that response to anti-EGFR therapy is modulated by the mutation status of the KRAS oncogene represents a major advance in clinical oncology and is an exciting step toward personalized medicine for advanced CRC. This has prompted the rapid introduction of molecular testing in molecular pathology laboratories worldwide. The data presented here indicate that a variety of techniques are suitable for KRAS mutation analysis in the clinical setting. As further molecular targets are shown to have a role in predicting response to anti-EGFR and other targeted therapies, there will be a rapid increase in the need for mutation testing in the clinical setting. The choice of method used by different laboratories is likely to depend on the equipment and technical expertise available, as well as the importance of cost and throughput of each method to those laboratories.
Footnotes
Supported by Infrastructure from the Hunter Medical Research Institute, the Peter MacCallum Centre, Queensland Health Clinical and Statewide Services, Pathology Queensland, and Singapore Cancer Syndicate (SCS BU51).
Australian KRAS Mutation Testing Advisory Board Members (V.W., S.F., R.J.S., A.D., and B.I.) were paid honoraria by Merck Serono to attend advisory meetings. No funds were received to perform this study. Merck Serono paid for the costs associated with the publication of this article. M.S.T.'s laboratory provides KRAS analysis for schemes and clinical trials generated by Merck Serono, but did not receive funding to assist with the current study.
References
- 1.Hemming AW, Davis NL, Kluftinger A, Robinson B, Quenville NF, Liseman B, LeRiche J. Prognostic markers of colorectal cancer: an evaluation of DNA content, epidermal growth factor receptor, and Ki-67. J Surg Oncol. 1992;51:147–152. doi: 10.1002/jso.2930510304. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart AC, Berlin JD. The epidermal growth factor receptor as a target for colorectal cancer therapy. Semin Oncol. 2005;32:52–60. doi: 10.1053/j.seminoncol.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 3.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 4.Andreyev JJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 5.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 6.Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 7.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 8.De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 9.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 10.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 11.Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, Hunter C, Smith R, Stephens P, Goldstraw P, Nicholson A, Chan TL, Velculescu VE, Yuen ST, Leung SY, Stratton MR, Futreal PA. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soong R, Iacopetta BJ. A rapid and nonisotopic method for the screening and sequencing of p53 gene mutations in formalin-fixed, paraffin-embedded tumors. Mod Pathol. 1997;10:252–258. [PubMed] [Google Scholar]
- 14.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salto-Tellez M. A case for integrated morphomolecular diagnostic pathologists. Clin Chem. 2007;53:1188–1190. doi: 10.1373/clinchem.2007.088088. [DOI] [PubMed] [Google Scholar]
- 16.Santini D, Loupakis F, Vincenzi B, Floriani I, Stasi I, Canestrari E, Rulli E, Maltese PE, Andreoni F, Masi G, Graziano F, Baldi GG, Salvatore L, Russo A, Perrone G, Tommasino MR, Magnani M, Falcone A, Tonini G, Ruzzo A. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13:1270–1275. doi: 10.1634/theoncologist.2008-0181. [DOI] [PubMed] [Google Scholar]
- 17.Rouleau E, Spyratos F, Dieumegard B, Guinebretiere JM, Lidereau R, Bieche I. KRAS mutation status in colorectal cancer to predict response to EGFR targeted therapies: the need for a more precise definition. Br J Cancer. 2008;99:2100. doi: 10.1038/sj.bjc.6604815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchetti A, Gasparini G. K-ras mutations and cetuximab in colorectal cancer. N Engl J Med. 2009;360:833–834. doi: 10.1056/NEJMc082346. author reply 835–836. [DOI] [PubMed] [Google Scholar]
- 19.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 20.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, Nasser S, Arango D, Shin J, Klampfer L, Augenlicht LH, Perez-Soler R, Mariadason JM. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA, III, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 23.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 24.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]


