Abstract
We have previously reported that a quantitative reverse transcription (QRT)-PCR assay accurately analyzes sentinel lymph nodes (SLNs) from breast cancer patients. The aim of this study was to assess a completely automated, cartridge-based version of the assay for accuracy, predictive value, and reproducibility. The triplex (two markers + control) QRT-PCR assay was incorporated into a single-use cartridge for point-of-care use on the GeneXpert system. Three academic centers participated equally. Twenty-nine positive lymph nodes and 30 negative lymph nodes were analyzed to establish classification rules. SLNs from 120 patients were subsequently analyzed by QRT-PCR and histology (including immunohistochemistry), and the predetermined decision rules were used to classify the SLNs; 112 SLN specimens produced an informative result by both QRT-PCR and histology. By histological analysis, 21 SLNs were positive and 91 SLNs were negative for metastasis. QRT-PCR characterization produced a classification with 100% sensitivity, 97.8% specificity, and 98.2% accuracy compared with histology (91.3% positive predictive value and 100% negative predictive value). Interlaboratory reproducibility analyses demonstrated that a 95% prediction interval for a new measurement (ΔCt) ranged between 0.403 and 0.956. This fully automated QRT-PCR assay accurately characterizes breast cancer SLNs for the presence of metastasis. Furthermore, the assay is not dependent on subjective interpretation, is reproducible across three clinical environments, and is rapid enough to allow intraoperative decision making.
Assessment of metastasis to regional lymph nodes (LNs) remains germane to the staging of epithelial malignancies.1,2,3 For breast cancer patients, sentinel lymph node (SLN) mapping and analysis are now routinely performed, improving detection rates of LN metastasis through a combination of increased sampling and improved sensitivity using immunohistochemical (IHC) staining techniques.4,5 The prognostic relevance of previously occult, micrometastatic disease is becoming better understood but remains an area of active investigation and controversy.6,7,8,9
Current methods of SLN analysis are time and labor intensive and require subjective interpretation. Many centers have ceased to perform intraoperative frozen section analysis as this rapid assessment is only ∼60% sensitive compared with final pathological analysis.10,11,12 Furthermore, differences in SLN analysis protocols exist across the pathology community,13 and significant discordance in interpretation of specimens has been reported.14,15,16 Thus, the current standard of care for analysis of SLNs is unable to provide timely information, lacks uniformity, and is dependent on subjective interpretation.
The reverse transcription (RT)-PCR has been explored as a potentially superior tool for characterizing LNs for the presence of metastatic disease.17,18,19,20 Although this technique offers theoretical advantages of improved sampling and objective analysis, the methodology requires considerable technical expertise for accurate results and prevention of contamination, and results have not been particularly reproducible.21,22,23 Thus, RT-PCR analyses for metastasis detection have yet to have been proven reliable or superior to current histology-based methods of analysis and remain experimental in nature.
We have demonstrated previously that a two-marker, quantitative RT (QRT)-PCR assay accurately and rapidly characterizes LNs for the presence of metastatic breast cancer.20 Thus, we aimed to assess a fully automated, GeneXpert (Cepheid, Sunnyvale, CA)-based version of the assay that is well controlled (one-tube triplex assay). The intent was to assess the ability of this assay as a point-of-care tool used by minimally trained personnel, using the current “gold standard” as a concordance comparison.
Materials and Methods
Role of Participating Entities
This project was conducted at four sites: Mount Sinai School of Medicine (MSSM), the University of Pittsburgh Medical Center (UPMC), the Medical University of South Carolina (MUSC), and Cepheid. Cepheid assisted in assay development,20,24,25 manufactured cartridges, and calibrated and supplied the GeneXpert instruments (one per institution). All tissue processing and histological analysis was performed at MSSM or UPMC, and tissue lysates were subsequently distributed for analysis. All raw data were sent to UPMC for analysis by the project statistician (W.E.G.). Cepheid had no role in tissue processing, data collection, or analysis and interpretation.
Source of Tissues
All tissues were collected and the research project was performed under institutional review board-approved protocols. Positive LNs from independent patients with breast cancer and negative LN controls were obtained from tissue banks at UPMC and MSSM. SLNs were obtained from patients enrolled in the Minimally Invasive Molecular Staging of Breast Cancer trial and banked at MUSC.26
Specimen Handling and Cutting Protocol
Tissues were snap-frozen in liquid nitrogen and stored at −80°C until use and then were embedded in OCT for cryostat sectioning. After facing the LN, a pattern of serial, 5-μm sections (2:10:2:10:2) was distributed to slides alternating with RNA lysis buffer (1.6 ml), and this process was repeated based on the LN size (LNs >1 cm = 40 RNA sections; LN <1 cm = 50 RNA sections). The lysis buffer was briefly vortexed and then stored at −80°C until needed. Tissue slides were labeled (including exact position in the cutting sequence), acetone fixed, and stored at −20°C until needed.
Histological Analysis
Histological examination was performed in several stages. Initially, the first and last sections from each LN were stained with H&E and evaluated by two pathologists (R.B. and J.M.). Next, the second and second to last sections were used for IHC evaluation with AE1/AE3 antibody (DAKO, Carpinteria, CA) as described previously,20 and these slides were evaluated by the pathologists. Finally, for selected SLNs all intervening pairs of slides were stained (1 H&E and 1 IHC) and evaluated.
GeneXpert Breast Cancer Assays
RNA isolation and QRT-PCR were performed on the GeneXpert system. This instrument utilizes single-use disposable cartridges to perform automated sample preparation and QRT-PCR-based gene expression analysis in approximately 30 minutes.25 The triplex QRT-PCR assay comprises two target genes (TACSTD1 and PIP) and an endogenous control gene (GUSB). Sample lysis buffer was loaded into the cartridge and all subsequent steps of the assay, including analysis, were automated.
For the current study, the cartridges were hand-assembled by Cepheid and contained lyophilized reagent beads for all primers, probes, reaction buffers, and enzymes (reverse transcriptase and thermostable polymerase). Cartridges were bar coded and vacuum packed for storage at 4°C until needed. In addition to the cartridges, the GeneXpert breast cancer assay kit provided by Cepheid included pre-aliquoted tubes of RNA binding buffer, wash buffer, and RNA elution buffer, all in squeezable plastic containers for easy addition to the cartridge. Assays were performed by first filtering 800 μl of specimen lysate through a 0.22-μm syringe filter (Osmonics Inc., West Borough, MA), mixing the filtered lysate with the pre-aliquoted RNA binding buffer (∼470 μl), and loading the mixture into the cartridge through a designated opening. Next, the wash and elution buffers were added to the cartridge through separate, labeled openings, and the cartridge lid was closed. The cartridge bar code was then scanned into the GeneXpert to identify the appropriate assay protocol, the sample name was typed in, and the assay was started. At this point the GeneXpert performed a pressure check (during fluidic transportation between chambers) and a probe fluorescence check before initiating RNA isolation. If either of these checks was not passed, the assay was automatically aborted.
The assays were performed by individuals of varying experience. At UPMC, all assays were performed by a pathology associate who had no previous experience in RT-PCR or other RNA-based experimental techniques. The assays performed at MUSC were run by graduate students as part of an introductory class taught by one of the authors with extensive experience in QRT-PCR, and those performed at MSSM were completed in essentially equal numbers by two individuals: a research technician who routinely performs QRT-PCR assays and a high school student laboratory volunteer with no previous experience.
Quantitative PCR Assay Design
In the current breast cancer study, we designed a triplex QRT-PCR assay comprising two target genes (TACSTD1 and PIP) and an endogenous control gene (GUSB). The primer and probe sequences used for reverse transcription and for quantitative PCR were modified from previous reports20 to optimize the triplex assay and to obtain quantitative data regardless of whether the target genes or the endogenous control gene are most abundant in any single sample. Primer and probe sequences are provided in Table 1 and Supplemental Figures 1–3 (see http://jmd.amjpathol.org). In the triplex assay, the amplification of GUSB can be stopped at any point using our previously reported temperature controlled primer limiting method,24 thus allowing quantitative detection of the target genes if GUSB expression is higher than target gene expression (the majority of cases). In addition, the target gene PCR amplicons are designed to have higher melting temperatures than the GUSB amplicon (86°C versus 80°C). Thus, by reducing the denaturation temperature during PCR cycling, this modification can be used to stop target gene amplification while allowing amplification of GUSB to proceed. Reduced denaturation temperature is used in samples in which target gene expression is higher than GUSB expression and allows quantitative expression data to be obtained in such cases. The ability to perform this bidirectional multiplexing strategy is facilitated by the independent site control on the GeneXpert and assay-specific software that changes cycling parameters in real time, depending on the observed fluorescence values in different fluorescence channels at each cycle.
Table 1.
Oligonucleotide Primer and Probe Sequences Used in the GeneXpert Assay
| Gene | Oligonucleotide | Sequence |
|---|---|---|
| GUSB | RT primer | 5′-TTTGGT TGTCTCTGCCGAGT-3′ |
| Forward primer | 5′-CTCATTTGGAATTTTGCCGATTTCA-3′ | |
| Reverse primer | 5′-CCGAGTGAAGATCCCCTT-3′ | |
| Probe | 5′-TGACTGAACAGTCACCGACGAGAGTGCTGG-3′ | |
| PIP | RT primer | 5′-GGGAATGTCAAAATTCTTT-3′ |
| Forward primer | 5′-GCGAGCTCCAGCTCCTGTTCAG-3′ | |
| Reverse primer | 5′-GGCGCAATTATGATCTTCCGAGTGTTGTC-3′ | |
| Probe | 5′-CCAGCCCTGCCACCCTGCTCCTG-3′ | |
| TACSTD1 | RT primer | 5′-AGTTTACGGCCAGCTTG-3′ |
| Forward primer | 5′-GACGCGTTCGGGCTTCTGCTT-3′ | |
| Reverse primer | 5′-GGCGAAGTTTTCACAGACACATTCTTCCTGAG-3′ | |
| Probe | 5′-CGGCGACGGCGACTTTTGC-3′ |
Bases indicated in bold are noncomplementary tails added to increase primer melting temperature in later PCR cycles.
Reproducibility Test
Samples for reproducibility analysis were prepared by spiking PIP-positive RNA from the MDA-MB-453 cell line and TACSTD1-positive normal human colon RNA (Stratagene, La Jolla, CA) into benign LN tissue lysate. The spiked sample was serially diluted (1:2) in the negative LN lysate to give seven samples with serial twofold reductions in PIP and TACSTD1 expression. Enough lysate was prepared to allow four GeneXpert analyses for each concentration at each institution (12 measurements per concentration).
Statistical Analysis
To characterize intra- and interlaboratory reproducibility, within-laboratory replicate values were used to estimate the intraclass correlation coefficient, the coefficient of variation, the SE of measurement, and a 95% prediction interval for a new observation. The average of the four replicates was then used to compare the three laboratories with one another. Linear regression models tested whether expression-concentration slopes were parallel and coincident.
To classify LNs by their expression of PIP and TACSTD1, a risk score (risk of metastasis) based on logistic regression with interaction was used. This risk score is a monotonic function of the likelihood ratio test and produces the same receiver operating characteristic curve as the likelihood ratio27 The risk score was developed from a training set of 29 positive and 30 negative LNs. To facilitate classification, equal probability contours were drawn at probabilities 0.01, 0.50, and 0.99. These contours were then used to classify 115 SLNs, with each contour providing a different threshold of certainty. Sensitivity, specificity, positive and negative predictive values, and overall classification accuracy were calculated relative to each contour.
Results
Histological Analysis
Two pathologists independently reviewed all H&E and IHC stained slides. The 29 positive LNs contained 20 to 100% tumor (median 85%). The 30 negative LNs were confirmed to be free of tumor and any contaminating tissues. The 120 SLN specimens were analyzed with both H&E and IHC staining; 4 lacked LN tissue. The remaining SLNs ranged from 0.2 to 4.5 cm in diameter with a median of 0.9 cm. Based on H&E staining of these 116 SLNs, both pathologists determined that 17 were positive and 98 were negative. One SLN was discordantly interpreted, but this SNL was resolved to be positive by subsequent IHC staining. However, 6 SLNs determined to be negative by H&E analysis were discordantly interpreted by the pathologists when examined with IHC staining. After reviewing the slides together, consensus was reached on 3 SLNs whereas 3 specimens remained equivocal. These data are also available in Supplemental Table 1 (see http://jmd.amjpathol.org). Thus, the final SLN tissue set for QRT-PCR analysis comprised 21 positive, 92 negative, and 3 indeterminate SLNs. The 21 positive SLNs were determined to contain 1 to 100% tumor cell representation with a median of 30%.
QRT-PCR Assay Characteristics
The GeneXpert assay was initially tested and optimized for PCR efficiency, linearity of amplification, and dynamic range using laboratory standards (Supplemental Figures 4–6, see http://jmd.amjpathol.org). The optimized assay was then assessed for reproducibility. The three sites (MSSM, UPMC; and MUSC) each received blinded, synthetic LN samples (4 samples each of 7 dilutions for 28 tests per site). The distribution of samples between sites and the overall performance of the GeneXpert breast cancer assay are shown in Table 2. Four reproducibility assays were not run at MUSC; therefore, a total of 256 assays were performed with 7 failures (2.7%): 1 was aborted, 1 for probe-check failure, and 5 for endogenous control gene failure.
Table 2.
Overall Performance of GeneXpert Assays
| GeneXpert run |
||||||
|---|---|---|---|---|---|---|
| Trial site | Sample type | No. of samples | Valid | Error | ||
| Aborted | Probe check failed | GUSB failed | ||||
| UPMC | Positive LN | 10 | 10 | |||
| Negative LN | 10 | 10 | ||||
| SLN | 30 | 30 | ||||
| Reproducibility | 28 | 27 | 1 | |||
| MUSC | Positive LN | 10 | 10 | |||
| Negative LN | 10 | 9 | 1 | |||
| SLN | 30 | 29 | 1 | |||
| Reproducibility | 24 | 22 | 2 | |||
| MSSM | Positive LN | 10 | 10 | |||
| Negative LN | 10 | 10 | ||||
| SLN | 56 | 56 | ||||
| Reproducibility | 28 | 26 | 1 | 1 | ||
| Totals | 256 | 249 | 1 | 1 | 5 | |
Intra- and intersite reproducibility was calculated for each target gene using ΔCt as the comparator (Table 3). Coefficients of variation ranged between 4 and 8%. We tested the equality of intra-assay reliability with a permutation test (2000 permutations). Intra-assay variation was comparable among the three sites with the exception of TACSTD1 measurement at MUSC (P = 0.0215). The 95% prediction interval for a new value using the Cox statistic of homogeneity of variance varied between 0.4 and 1 cycle. For interlaboratory comparisons, linear regression models were fit to the dilution data such that the equality of slope and intercepts among laboratories could be estimated and tested (Figure 1A–D). There were no significant laboratory differences in the measurement of TACSTD1. For PIP, the slopes were not different (P = 0.1854), but the intercepts were (P < 0.0001), with each site differing from the others. The resulting model for estimated PIP expression shows parallel slopes with constant differences in intercept. Thus, we normalized PIP expression to MSSM and applied correction factors to PIP measured subsequently in SLN at MUSC and UPMC.
Table 3.
Intralaboratory Reproducibility
|
PIP |
TACSTD1 |
|||||
|---|---|---|---|---|---|---|
| MSSM | MUSC | UPMC | MSSM | MUSC | UPMC | |
| Within-dilution coefficient of variation (%) | 5.8 | 7.3 | 5.8 | 4.4 | 6.8 | 3.7 |
| Intraclass correlation coefficient | 0.967 | 0.938 | 0.976 | 0.981 | 0.965 | 0.991 |
| SE of measurement (ΔCt) | 0.389 | 0.488 | 0.330 | 0.265 | 0.373 | 0.205 |
| 95% prediction interval for a new measurement (ΔCt) | ± 0.763 | ± 0.956 | ± 0.647 | ± 0.519 | ± 0.732 | ± 0.403 |
Figure 1.
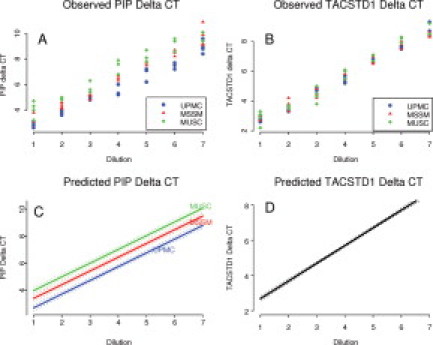
Observed and predicted PIP and TACSTD1 relative expression from intralaboratory reproducibility experiments. Synthetic LN samples representing seven serial dilutions (four samples for each dilution) were sent to three participating laboratories. The objective was to assess assay reproducibility between and within laboratories. A and B: Observed target mRNA concentrations in ΔCt as determined by UPMC, MSSM, and MUSC laboratories. C: The estimated concentration-dilution curve based on a linear regression model. This model supports a separate function for each laboratory that differs by vertical distance and not by slope. D: The best fitting regression model for TACSTD1 permits a single concentration-dilution function for all laboratories. Dotted lines in C and D are 95% confidence bands for the expected ΔCt.
Determination of Molecular Classification Rules
Twenty-nine positive control LNs and 30 negative control LNs were successfully analyzed on the GeneXpert (one assay failed). TACSTD1 and PIP expression data from these 59 valid assays are plotted in Figure 2A and show clear separation of positive and negative LNs. These data were then used to generate a logistic regression model that produced equal probability risk scores based on the probability that a LN is positive, given the observed distribution of dual-marker expression in negative control LNs. Thus, the lines in Figure 2A represent equal probability cutoff contours at which any node below the line has a less than 1, 50, or 99% probability of being positive, respectively. Importantly, measured relative expression levels for both markers were independent of the percentage of metastatic cells within the LN (data not shown).
Figure 2.
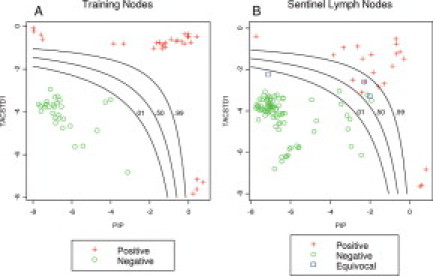
A: A total of 59 training nodes (29 positive and 30 negative) were used to train a risk probability classifier using expression of PIP and TACSTD1. Three equal probability contours were drawn to illustrate low (probability = 0.01), moderate (probability = 0.50), and high (probability = 0.99) probability that a lymph node is positive for micrometastasis. B: The same classifier was then plotted against the observed PIP and TACSTD1 in 115 sentinel lymph nodes. The plotting symbols show the final pathological results as negative (N = 91), positive (N = 21), or equivocal (N = 3).
Sentinel Lymph Node QRT-PCR Analysis
The equal probability risk contours were next applied one at a time to the SLN data to provide a molecular classification (positive or negative) for each SLN (Figure 2B). Molecular classification using each of three probability contours was then compared with the consensus pathology classification to calculate accuracy parameters for each classification rule. Accuracy for all cutoffs was high (minimum accuracy = 97.3%). In addition, Table 4 demonstrates how the use of different probability contours influences sensitivity, specificity, positive predictive value and negative predictive value. Notably, either 100% sensitivity or 100% specificity can be achieved while maintaining >97% overall accuracy. Finally, Figure 2B also shows the three SLNs for which pathological consensus could not be achieved. According to the molecular analysis, one LN with a marginal level of PIP expression had a 0.06 probability of being positive and the other two LNs, with intermediate levels of both PIP and TACSTD1 had probabilities of 0.79 and 0.84 of being positive.
Table 4.
Probability Risk Assessment of QRT-PCR Characterization of SLN Compared with Histology
| Rule | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Low probability (0.01) of LN positive | 100 | 96.7 | 87.5 | 100 | 97.3 |
| Equal probability (0.50) of LN positive | 100 | 97.8 | 91.3 | 100 | 98.2 |
| High probability (0.99) of LN positive | 85.7 | 100 | 100 | 96.8 | 97.3 |
Discussion
We have shown for the first time that a rapid QRT-PCR assay can be fully automated, reducing the technical demands of performing the assay and resulting in exceptional reproducibility across testing environments. The assay accurately characterized SLNs from breast cancer patient specimens compared with conventional pathological analysis including IHC and clearly surpasses current methods of rapid assessment. Similar automated assays for infectious disease applications have received “moderate complexity” certification under the Clinical Laboratory Improvements Amendment from the Food and Drug Administration, expanding the number of clinical environments capable of performing such analyses to include most community hospitals (http://www.genomeweb.com/dxpgx/fda-clears-cepheid-expert-mrsa-test-first-moderate-complexity-diagnostic, last accessed September 10, 2009).
Others have reported the use of RT-PCR assays to assess SLNs from breast cancer patients,28,29,30,31 and two groups have focused on the value of a rapid assessment. Tsujimoto et al32 used a single-marker assay (CK-19) compared with histology including IHC in a concordance study of 325 LNs from 101 patients. They reported a 98.2% rate of concordance between the two techniques using the upper bound of marker expression from histology-negative LNs as the discriminator rule. Likewise an RT-PCR assay for the rapid detection of metastasis to LNs from breast cancer patients has recently become commercially available (Veridex, LLC, Raritan, NJ). This assay uses a two-marker approach (mammaglobin 1 and CK-19) amplified in separate reaction tubes. Veridex reported that in a study of 412 patients, the RT-PCR assay classification of SLNs using linear discriminator methods to establish decision rules resulted in an 86.2% positive predictive value and a 94.9% negative predictive value. Importantly, heterogeneity of results across laboratories in this study almost reached statistical significance (P = 0.066). Thus, the findings of others are in agreement with our conclusion that an RT-PCR analysis can rapidly and accurately characterize SLNs from breast cancer patients, but the reproducibility of other assays is questionable.
In contrast to these reports and in addition to being automated, the GeneXpert assay differs in design because it incorporates an endogenous control gene in a fully multiplexed assay, thus controlling for variations in RNA loading and integrity that will occur during routine clinical use. Determination of target expression relative to this endogenous control may have contributed to the high level of intra- and interlaboratory reproducibility we observed; however, this reproducibility is also probably related to the automated nature of the assay. In addition, this control in combination with the single-use, sealed cartridge minimizes the risk of false-positive results from laboratory contamination with amplification products. Finally, the GeneXpert assay is uniquely controlled in that amplification of all three targets is performed in a single tube, eliminating the potential for differences in reagent availability or quality or cycling parameters between reaction tubes.
We believe the most important aspect of our findings centers on the ability of the assay to achieve 100% negative predictive value with minimal loss of accuracy. Because approximately 80% of SLN biopsies should prove to be negative, implementation of this assay would result in the majority of breast cancer patients being informed of a favorable SLN biopsy result on emergence from anesthesia. Clinical dilemmas such as the appropriateness of immediate breast reconstruction could be resolved. The objective nature of QRT-PCR assay characterization of SLNs may prove to be equally important. Most high-volume breast cancer centers routinely review outside pathology specimens, and differences in pathological methodology and interpretation are a real clinical phenomenon.13,14,15,16 Thus, a fully objective assessment such as that provided by this QRT-PCR assay may have implications in the performance of multicenter trials.
Although the assay is able to produce 100% negative predictive value, this is not in the setting of 100% accuracy. By our risk model, 3 of the 112 SLNs were found to contain intermediate marker expression in the absence of metastasis by histological analysis. Because this assay is quantitative and the decision rules are based on the probability that the SLN is negative, these “false-positive” results cannot be simply dismissed as such. The statistical probabilities that these three nodes are “like” benign nodes from patients without cancer are 0.94, 0.21, and 0.19 (6, 79, and 81% probability that the nodes are positive). Unfortunately, a much larger sample set and long-term patient outcome data are necessary to assess the clinical significance of this level of marker expression. Other studies are currently awaiting adequate follow-up to address whether RT-PCR is capable of identifying clinically significant signals in IHC-negative SLNs from breast cancer patients.26,33
Our application of this statistical method to the QRT-PCR characterization of SLNs is novel. We did investigate several well-known classification methods including linear discriminant, nearest neighbors, and recursive partitioning analyses (data not shown). None of these methods was superior to probability assessment. We believe the risk score method has merit because it permits predicting the actual probability of the presence of LN metastasis for an individual patient, allowing the clinician to interpret these intermediate values relative to the clinical scenario and not be tied to the absolute (realizing the method will produce 100% predictive value in ∼97% of tests). We strongly support an approach in which these rare specimens with an intermediate risk score are subjected to further analysis in the hope of clarifying ambiguity in the status of the LN.
It is important to note that this study is limited by the use of fresh-frozen tissues rather than formalin-fixed, paraffin-embedded tissues. This optimizes the QRT-PCR assay while compromising histological analyses, which may have contributed to the rate of observed discordance between the study pathologists and an equivocal result in three specimens. Although methods of extracting RNA from formalin-fixed, paraffin-embedded tissues are known, these were inappropriate for our purposes as the goal was development of a rapid assay.
In summary, a fully automated QRT-PCR assay is exceptionally accurate compared with conventional analysis of breast cancer SLNs. The assay is reproducible across three laboratory environments.
Footnotes
Supported in part by the National Institutes of Health (grants CA-01958 to S.J.H. and CA-099123 to J.C., T.E.G., and S.J.H.) and the Department of Defense (grants N00014-99-1-0784 to D.J.C., J.M., and M.M.).
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Supplementary data
References
- 1.Fisher B, Bauer M, Wickerham DL, Redmond CK, Fisher ER, Cruz AB, Foster R, Gardner B, Lerner H, Margolese R. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer. an NSABP update. Cancer. 1983;52:1551–1557. doi: 10.1002/1097-0142(19831101)52:9<1551::aid-cncr2820520902>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Fisher ER, Sass R, Fisher B. Pathologic findings from the National Surgical Adjuvant Project for Breast Cancers (protocol no. 4). X. Discriminants for tenth year treatment failure. Cancer. 1984;53:712–723. doi: 10.1002/1097-0142(19840201)53:3+<712::aid-cncr2820531320>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 3.Fisher ER, Costantino J, Fisher B, Redmond C. Pathologic findings from the National Surgical Adjuvant Breast Project (protocol 4). Discriminants for 15-year survival. National Surgical Adjuvant Breast and Bowel Project Investigators. Cancer. 1993;71:2141–2150. doi: 10.1002/1097-0142(19930315)71:6+<2141::aid-cncr2820711603>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 4.Cote RJ, Peterson HF, Chaiwun B, Gelber RD, Goldhirsch A, Castiglione-Gertsch M, Gusterson B, Neville AM. Role of immunohistochemical detection of lymph-node metastases in management of breast cancer. Lancet. 1999;354:896–900. doi: 10.1016/s0140-6736(98)11104-2. [DOI] [PubMed] [Google Scholar]
- 5.Giuliano AE, Dale PS, Turner RR, Morton DL, Evans SW, Krasne DL. Improved axillary staging of breast cancer with sentinel lymphadenectomy. Ann Surg. 1995;222:394–399. doi: 10.1097/00000658-199509000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakorafas GH, Geraghty J, Pavlakis G. The clinical significance of axillary lymph node micrometastases in breast cancer. Eur J Surg Oncol. 2004;30:807–816. doi: 10.1016/j.ejso.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Chen SL, Hoehne FM, Giuliano AE. The prognostic significance of micrometastases in breast cancer: a SEER population-based analysis. Ann Surg Oncol. 2007;14:3378–3384. doi: 10.1245/s10434-007-9513-6. [DOI] [PubMed] [Google Scholar]
- 8.Rutgers EJ. Sentinel node biopsy: interpretation and management of patients with immunohistochemistry-positive sentinel nodes and those with micrometastases. J Clin Oncol. 2008;26:698–702. doi: 10.1200/JCO.2007.14.4667. [DOI] [PubMed] [Google Scholar]
- 9.Kuijt GP, Voogd AC, van de Poll-Franse LV, Scheijmans LJ, van Beek MW, Roumen RM. The prognostic significance of axillary lymph-node micrometastases in breast cancer patients. Eur J Surg Oncol. 2005;31:500–505. doi: 10.1016/j.ejso.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Dabbs DJ, Fung M, Johnson R. Intraoperative cytologic examination of breast sentinel lymph nodes: test utility and patient impact. Breast J. 2004;10:190–194. doi: 10.1111/j.1075-122X.2004.21313.x. [DOI] [PubMed] [Google Scholar]
- 11.Chao C, Wong SL, Ackermann D, Simpson D, Carter MB, Brown CM, Edwards MJ, McMasters KM. Utility of intraoperative frozen section analysis of sentinel lymph nodes in breast cancer. Am J Surg. 2001;182:609–615. doi: 10.1016/s0002-9610(01)00794-2. [DOI] [PubMed] [Google Scholar]
- 12.Gulec SA, Su J, O'Leary JP, Stolier A. Clinical utility of frozen section in sentinel node biopsy in breast cancer. Am Surg. 2001;67:529–532. [PubMed] [Google Scholar]
- 13.Hunt JL, Baloch ZW, LiVolsi VA. Sentinel lymph node evaluation for tumor metastasis. Semin Diagn Pathol. 2002;19:263–277. [PubMed] [Google Scholar]
- 14.Roberts CA, Beitsch PD, Litz CE, Hilton DS, Ewing GE, Clifford E, Taylor W, Hapke MR, Babaian A, Khalid I, Hall JD, Lindberg G, Molberg K, Saboorian H. Interpretive disparity among pathologists in breast sentinel lymph node evaluation. Am J Surg. 2003;186:324–329. doi: 10.1016/s0002-9610(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 15.Turner RR, Weaver DL, Cserni G, Lester SC, Hirsch K, Elashoff DA, Fitzgibbons PL, Viale G, Mazzarol G, Ibarra JA, Schnitt SJ, Giuliano AE. Nodal stage classification for breast carcinoma: improving interobserver reproducibility through standardized histologic criteria and image-based training. J Clin Oncol. 2008;26:258–263. doi: 10.1200/JCO.2007.13.0179. [DOI] [PubMed] [Google Scholar]
- 16.Cserni G, Bianchi S, Boecker W, Decker T, Lacerda M, Rank F, Wells CA. Improving the reproducibility of diagnosing micrometastases and isolated tumor cells. Cancer. 2005;103:358–367. doi: 10.1002/cncr.20760. [DOI] [PubMed] [Google Scholar]
- 17.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, Hermans J, van Krieken JH, Cornelisse CJ, Tollenaar RA. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. doi: 10.1056/NEJM199807233390403. [DOI] [PubMed] [Google Scholar]
- 18.Coello MC, Luketich JD, Litle VR, Godfrey TE. Prognostic significance of micrometastasis in non-small-cell lung cancer. Clin Lung Cancer. 2004;5:214–225. doi: 10.3816/CLC.2004.n.002. [DOI] [PubMed] [Google Scholar]
- 19.Godfrey TE, Raja S, Finkelstein SD, Gooding WE, Kelly LA, Luketich JD. Prognostic value of quantitative reverse transcription-polymerase chain reaction in lymph node-negative esophageal cancer patients. Clin Cancer Res. 2001;7:4041–4048. [PubMed] [Google Scholar]
- 20.Hughes SJ, Xi L, Raja S, Gooding W, Cole DJ, Gillanders WE, Mikhitarian K, McCarty K, Silver S, Ching J, McMillan W, Luketich JD, Godfrey TE. A rapid, fully automated, molecular-based assay accurately analyzes sentinel lymph nodes for the presence of metastatic breast cancer. Ann Surg. 2006;243:389–398. doi: 10.1097/01.sla.0000201541.68577.6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keilholz U, Willhauck M, Rimoldi D, Brasseur F, Dummer W, Rass K, de VT, Blaheta J, Voit C, Lethe B, Burchill S. Reliability of reverse transcription-polymerase chain reaction (RT-PCR)-based assays for the detection of circulating tumour cells: a quality-assurance initiative of the EORTC Melanoma Cooperative Group. Eur J Cancer. 1998;34:750–753. doi: 10.1016/s0959-8049(97)10105-8. [DOI] [PubMed] [Google Scholar]
- 22.Bolufer P, Lo CF, Grimwade D, Barragan E, Diverio D, Cassinat B, Chomienne C, Gonzalez M, Colomer D, Gomez MT, Marugan I, Roman J, Delgado MD, Garcia-Marco JA, Bornstein R, Vizmanos JL, Martinez B, Jansen J, Villegas A, de Blas JM, Cabello P, Sanz MA. Variability in the levels of PML-RARα fusion transcripts detected by the laboratories participating in an external quality control program using several reverse transcription polymerase chain reaction protocols. Haematologica. 2001;86:570–576. [PubMed] [Google Scholar]
- 23.Reinhold U, Berkin C, Bosserhoff AK, Deutschmann A, Garbe C, Glaser R, Hein R, Krahn G, Peter RU, Rappl G, Schittek B, Seiter S, Ugurel S, Volkenandt M, Tilgen W. Interlaboratory evaluation of a new reverse transcriptase polymerase chain reaction-based enzyme-linked immunosorbent assay for the detection of circulating melanoma cells: a multicenter study of the Dermatologic Cooperative Oncology Group. J Clin Oncol. 2001;19:1723–1727. doi: 10.1200/JCO.2001.19.6.1723. [DOI] [PubMed] [Google Scholar]
- 24.Raja S, El Hefnawy T, Kelly LA, Chestney ML, Luketich JD, Godfrey TE. Temperature-controlled primer limit for multiplexing of rapid, quantitative reverse transcription-PCR assays: application to intraoperative cancer diagnostics. Clin Chem. 2002;48:1329–1337. [PubMed] [Google Scholar]
- 25.Raja S, Ching J, Xi L, Hughes SJ, Chang R, Wong W, McMillan W, Gooding WE, McCarty KS, Jr, Chestney M, Luketich JD, Godfrey TE. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin Chem. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- 26.Gillanders WE, Mikhitarian K, Hebert R, Mauldin PD, Palesch Y, Walters C, Urist MM, Mann GB, Doherty G, Herrmann VM, Hill AD, Eremin O, El Sheemy M, Orr RK, Valle AA, Henderson MA, Dewitty RL, Sugg SL, Frykberg E, Yeh K, Bell RM, Metcalf JS, Elliott BM, Brothers T, Robison J, Mitas M, Cole DJ. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Ann Surg. 2004;239:828–837. doi: 10.1097/01.sla.0000128687.59439.d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntosh MW, Pepe MS. Combining several screening tests: optimality of the risk score. Biometrics. 2002;58:657–664. doi: 10.1111/j.0006-341x.2002.00657.x. [DOI] [PubMed] [Google Scholar]
- 28.Bostick PJ, Huynh KT, Sarantou T, Turner RR, Qi K, Giuliano AE, Hoon DS. Detection of metastases in sentinel lymph nodes of breast cancer patients by multiple-marker RT-PCR. Int J Cancer. 1998;79:645–651. doi: 10.1002/(sici)1097-0215(19981218)79:6<645::aid-ijc16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 29.Bostick PJ, Morton DL, Turner RR, Huynh KT, Wang HJ, Elashoff R, Essner R, Hoon DS. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol. 1999;17:3238–3244. doi: 10.1200/JCO.1999.17.10.3238. [DOI] [PubMed] [Google Scholar]
- 30.Mori M, Mimori K, Inoue H, Barnard GF, Tsuji K, Nanbara S, Ueo H, Akiyoshi T. Detection of cancer micrometastases in lymph nodes by reverse transcriptase-polymerase chain reaction. Cancer Res. 1995;55:3417–3420. [PubMed] [Google Scholar]
- 31.Schoenfeld A, Luqmani Y, Smith D, O'Reilly S, Shousha S, Sinnett HD, Coombes RC. Detection of breast cancer micrometastases in axillary lymph nodes by using polymerase chain reaction. Cancer Res. 1994;54:2986–2990. [PubMed] [Google Scholar]
- 32.Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, Tamaki Y, Noguchi S, Kataoka TR, Nakajima H, Komoike Y, Inaji H, Tsugawa K, Suzuki K, Nakamura S, Daitoh M, Otomo Y, Matsuura N. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res. 2007;13:4807–4816. doi: 10.1158/1078-0432.CCR-06-2512. [DOI] [PubMed] [Google Scholar]
- 33.Mikhitarian K, Martin RH, Mitas M, Mauldin PD, Palesch Y, Metcalf JS, Cole DJ, Gillanders WE. Molecular analysis improves sensitivity of breast sentinel lymph node biopsy: results of a multi-institutional prospective cohort study. Surgery. 2005;138:474–481. doi: 10.1016/j.surg.2005.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


