Abstract
Patients with biliary tract carcinoma have a poor prognosis. Early detection efforts are urgently needed to ameliorate the dismal prognosis for these patients. Mutations of the KRAS2 gene are one of the most common genetic aberrations in this cancer. In this study, we used LigAmp, an ultrasensitive technology for detecting point mutations, to analyze KRAS2 mutations in patients with a variety of neoplastic and non-neoplastic pancreatobiliary diseases. DNA was isolated from 64 samples, including 44 bile samples and 20 serum samples. Oligonucleotides specific for KRAS2 G35A (GAT, G12D), G35T (GTT, G12V), and G34A (AGT, G12S) mutations were used. KRAS2 mutations were detected in 14 of 16 (87.5%) neoplastic bile samples and in 9 of 28 (32.1%) non-neoplastic bile samples. However, the mutation levels were significantly lower in the non-neoplastic bile (median = 0.4%) compared with those in the neoplastic bile (median = 5.1%). KRAS2 mutations were also detected in 9 of 11 (81.8%) serum samples from patients with biliary tract carcinoma, which was further confirmed by cloning BstN1-refractory PCR products and DNA sequencing. However, KRAS2 mutations were not present in the sera from eight patients with benign pancreatobiliary diseases. These data demonstrate that KRAS2 mutations are detectable in both bile and serum using LigAmp. This technology has the potential for early biliary tract carcinoma detection and possibly for residual disease monitoring post-therapy.
Biliary tract carcinoma arises within the bile duct system, and is anatomically divided into carcinoma of the gallbladder, as well as intrahepatic and extrahepatic cholangiocarcinoma. It carries a poor prognosis, with an overall 5 year-survival of less than 5%.1 In the United States, intrahepatic cholangiocarcinoma is the second most common primary hepatic cancer, and cholangiocarcinoma as a whole accounts for >4500 cancer-related deaths each year. The dismal outcome of biliary tract carcinoma is largely because the majority of cancers are diagnosed at an unresectable advanced stage. Patients with early biliary tract carcinoma have a survival rate as high as 40% following a curative surgical resection,2,3 whereas those diagnosed late have a survival rate of only 5%. Early detection therefore could have a significant impact on overall survival. Although recent progress has been made in various imaging modalities such as endoscopic retrograde cholangiopancreatography, endoscopic ultrasonography, and magnetic resonance imaging, diagnosis of biliary tract carcinoma at an early stage is still difficult in many cases.4,5 In addition, it is technically challenging to perform endoscopic biopsy because of the presence of biliary strictures associated with small lesions. Although biliary tract carcinoma is derived from the bile duct epithelium, endobiliary brush cytology typically yields a low sensitivity of detection for malignancy.6 Furthermore, reactive biliary epithelial atypia associated with inflammatory biliary stricture mimics cancerous epithelium, which makes it problematic to diagnose biliary tract carcinoma cytologically and histologically. On the other hand, since this type of tumor tends to grow less coherently and the neoplastic cells are easily shed into bile, molecular analysis of tumor DNA in bile could be an important ancillary testing for definitive diagnosis of early biliary tract carcinoma.
Patients with biliary tract carcinoma who harbor locally advanced or distant metastatic disease are not candidates for surgery, and receive either systemic chemotherapy or local irradiation. However, there are few avenues available to monitor the effectiveness of therapy, or to assess for minimal residual disease in the face of therapeutic response. To ensure that these patients receive effective treatment and avoid unnecessary toxicities, it is mandatory to have a sensitive and specific test to monitor the therapeutic response in vivo using a relatively non-invasive assay.
Activation of oncogenes and inactivation of tumor suppressor genes have been identified as important contributors in the development of biliary tract carcinoma.7,8,9 The incidence of KRAS2 mutations that have been reported in biliary tract carcinoma varies widely ranging from 0% to 100%, depending on what techniques have been used. KRAS2 mutations most commonly occur at codon 12, including GGT to AGT, GAT, and GTT.9,10 Detection of KRAS2 mutations in bile has been explored as a potential diagnostic test by several groups. However, in these studies, although KRAS mutations were detected in the primary tumors of more than 80% cases, a much lower frequency of KRAS2 mutations was observed in bile from the same population,6,11,12 which probably resulted from the presence of excessive wild-type DNA in the bile. An identical technological pitfall exists for detecting mutant DNA in serum. A sensitive and specific assay could improve the detection of mutant KRAS2 to a great extent.
Recently, we developed an ultra-sensitive technology, LigAmp, for detection of single base mutations in the presence of large amounts of wild-type DNA.13 We have demonstrated that LigAmp is able to detect one mutant in the presence of ∼10,000 wild-type molecules or cells. In the present study, we used LigAmp to analyze KRAS2 mutations in the bile and serum from patients with a variety of non-neoplastic diseases and those with biliary tract carcinoma.
Materials and Methods
We obtained appropriate institutional approval for all experiments involving human subjects.
Sample Collection
Bile was collected during endoscopic retrograde cholangiopancreatography from 16 patients with biliary tract carcinoma (as confirmed by subsequent histology on surgical resection, diagnostic biopsy, or imaging), and from 28 patients with benign pancreatobiliary conditions (as confirmed by serial imaging or clinical follow up). The 16 biliary tract carcinomas included 12 extrahepatic cholangiocarcinomas, 3 intrahepatic cholangiocarcinomas and 1 carcinoma of the gallbladder. Bile samples were kept at −70°C until further use. Blood (10 ml) from patients with biliary tract carcinoma (n = 11, including 9 extrahepatic cholangiocarcinomas, 1 intrahepatic cholangiocarcinoma and 1 carcinoma of the gallbladder) and those with benign pancreatobiliary diseases (n = 9) was drawn into Becton-Dickinson SSAT tubes (Franklin Lakes, NJ) and placed immediately on ice. Then, these samples were centrifuged at 1500 × g for 30 minutes at 4°C. Serum was removed and stored at −20°C for further use.
DNA Extraction
Genomic DNA from a KRAS2 wild-type cell line (HeLa), KRAS2 mutant cell lines LS513 (GAT), and SW480 (GTT) was extracted using the DNeasy Tissue Kit (Qiagen, Valencia, CA). QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) was used to extract DNA from serum (200 μl) and bile (200 μl). DNA was eluted in a final volume of 30 μl.
LigAmp Oligonucleotides and Probes
Ligation oligonucleotides for wild-type and mutant KRAS2 and modified M13 forward and reverse primers (Table 1) were purchased from Invitrogen, Corp. (Carlsbad, CA). The downstream common oligonucleotides were phosphorylated at the 5′ end. The LacZ and 16S rDNA Taqman probes containing different fluorophores and quenchers (Table 1) were purchased from Integrated DNA Technology (Coralville, IA).
Table 1.
Oligonucleotides and Probes
| KRAS2 upstream | GAT | 5′-ACTGTAAAACGACGGCCAGTGT-TCCCCTCAAACTGGCAGATGCACG-C-TTGTGGTAGTTGGAGCCGA*-3′ |
| GTT | 5′-ACTGTAAAACGACGGCCAGTGT-TCCCCTCAAACTGGCAGATGCACG-C-TTGTGGTAGTTGGAGCCGT*-3′ | |
| AGT | 5′-ACTGTAAAACGACGGCCAGTGT-TCCCCTCAAACTGGCAGATGCACG-C-TTGTGGTAGTTGGAGTTA*-3′ | |
| GGT (Wild-type) | 5′-ACTGTAAAACGACGGCCAGTGT-CGTATTACCGCGGCTGCTGGCACC-TTGTGGTAGTTGGAGCTGG*-3′ | |
| KRAS2 common (downstream) | 12b | 5′-PO4-TGGCGTAGGCAAGAGTGCC-TGGTCATAGCTGTTTCCTGCA-3′ |
| M13 primers | Forward | 5′-CTGTAAAACGACGGCCAGTG-3′ |
| Reverse | 5′-TGCAGGAAACAGCTATGACCA-3′ | |
| TaqMan probes | lacZ | FAM-5′-TCCCCTCAAACTGGCAGATGCACG-3′-BHQ-1 |
| 16S rDNA | ROX-5′-CGTATTACCGCGGCTGCTGGCAC-3′-BHQ-2 | |
Underlined, M13 primer binding regions; Italics, probe binding regions (lacZ or 16S rDNA); Bold, target-specific regions; Asterisk, terminal bases with perfect homology to either the wild-type or mutant sequences; FAM, 6-carboxyflurorescein; ROX, 6-carboxy-X-rhodamine; BHQ, black hole quenche; Boxed base, an additional mis-pair in the upstream mutant oligonucleotides was introduced at the third base from the 3' end to improve the specificity of the assay.
LigAmp Analysis of KRAS2 Mutations
LigAmp analysis of KRAS2 mutations has been previously described in detail.13,14 First, a region of KRAS2 including KRAS2 codon 12 (hot spot) was PCR amplified. For ligation, 1 pmol upstream mutant oligonucleotide, 1 fmol wild-type oligonucleotide, and l pmol downstream oligonucleotide were incubated with approximately 200 pg PCR amplified KRAS2 DNA and 4 U Pfu DNA ligase in 1× Pfu Ligase buffer (Stratagene, La Jolla, CA). The ligation condition included denaturation at 95°C for 3 minutes, followed by 90 two-step cycles of 95°C for 30 seconds, alternating with 65°C for 4 minutes. For quantification of both wild-type and mutant KRAS2 DNA in the serum samples, mutant KRAS2 DNA was serially diluted into wild-type DNA (from 200 pg to 0.2 pg into 200 pg Hela DNA), the wild-type DNA was also serially diluted (2000 pg, 200 pg, and 20 pg). LigAmp analyses of mutant DNA mixtures and serial diluted wild-type DNA were used as standard quantitative controls.
Q-PCR amplification of ligated products was performed in a SmartCycler (Cepheid, Sunnyvale, CA). Each reaction (25 μl) contained 5 pmol forward and 5 pmol reverse M13 primers, 2 μl of the unpurified ligation reaction, 12.5 μl platinum Quantitative PCR SuperMix-UDG (Invitrogen), and 2.5 pmol lacZ and 16S rDNA probes. To simultaneously quantify mutant and wild-type KRAS2, we included both lacZ (for mutated KRAS2) and 16S rDNA (for wild-type KRAS2) probes in the reaction. The PCR reaction included pre-incubation at 50°C for 2 minutes and 95°C for 2 minutes, followed by 50 two-step cycles of 95°C for 10 seconds alternating with 64°C for 20 seconds. For each experiment, a cycle threshold (Ct) was manually set to the middle of the linear range (log scale) of the growth curves.
BstN1 Restrict Digestion-PCR, TA Cloning, and DNA Sequencing
To eliminate wild-type and enrich mutant KRAS2, KRAS2 DNA was first amplified using a forward mutant primer to produce a BstN1 site in only wild-type KRAS2 PCR amplified products. The forward and reverse primers are 5′-AATATAAACTTGTGGTAGTTGGACCT-3′ and 5′-TCAAAGACAAGGCGATATGCT-3′ respectively, where an underlined base in the forward primer is the mutant base required to create a BstN1 site in wild-type KRAS2 DNA. A 30-cycle PCR using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) was performed, yielding a 1031 bp PCR product. A second BstN1 restriction site for both wild-type and mutant KRAS2 PCR products was located 136 bases upstream of the reverse primer. Three fragments (136, 888, and 18 bp) for wild-type KRAS2 and two (136 and 906 bp) for mutated KRAS2 were produced after BstN1 enzyme digestion.
Following purification using a Qiagen PCR purification Kit, the PCR product (5 μl) was digested with BstN1 (20 units, New England Biolabs, Beverly, MA) at 65°C for 2 hours. The digestion reaction was analyzed by 2% agarose gel. The restriction digest product (1 μl) was then re-amplified for 35 cycles using the same forward primer and a second reverse primer (R: 5′-CCCTGACATACTCCCAAGGA-3′) located upstream of the second restriction site, and produced a 304 bp PCR product. Wild-type DNA from HeLa cells was used as a negative control. The BstN1-refractory PCR products were saved for TA cloning.
TA Cloning and Sequencing
Purified BstN1 refractory PCR products were cloned into pCRII-TOPO cloning vectors using the TOPO TA Cloning Kit, Version N (Invitrogen). PCR was performed on 20 white colonies using M13 forward and reverse primers. These PCR products were then subjected to BstN1 digestion. The BstN1-refractory PCR products were sequenced using a M13 forward primer, the BigDye Terminator 3.1 Cycle Sequencing Kit and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Results
LigAmp Analyses of KRAS2 Mutations in Bile Samples Obtained from Patients with Biliary Tract Carcinoma Versus Those with Benign Pancreatobiliary Diseases
We first analyzed KRAS2 mutations in bile from 16 patients with biliary tract carcinoma, including 12 extrahepatic cholangiocarcinomas, three intrahepatic cholangiocarcinomas, and one carcinoma of the gallbladder. We had no obvious difficulty amplifying DNA isolated from bile, although bile is known to contain PCR inhibitors.15 The amounts of mutant KRAS2 DNA relative to wild-type KRAS2 are shown in Figure 1A. We detected KRAS2 mutations in 14 of 16 samples (87.5%), and multiple KRAS2 mutations were present in 9 of these 14 positive samples. GAT mutation was the predominant variant in all these samples. The amounts of KRAS2 mutations varied over a wide range, from less than 1% to over 90%. However, majority of the positive samples contained more than 2% of mutant DNA. Preliminary analysis comparing level of mutant KRAS2 with the size and stage of the tumor shows no obvious correlation, although a larger sample size would need to be analyzed for proper analysis. Initial analysis of the cases with multiple mutations versus those with single mutations revealed no obvious correlation to histological grade, size, or stage.
Figure 1.
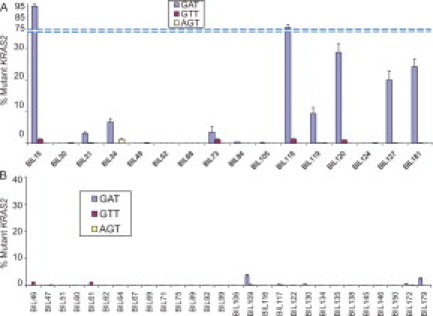
LigAmp quantification of KRAS2 mutations in bile from patients with and without biliary tract carcinoma. A: Percentage of mutant KRAS2 relative to wild-type KRAS2 detected in bile from 16 patients with biliary tract carcinoma. LigAmp analysis of GAT (G12D), GTT (G12V), and AGT (G12S) KRAS2 mutations shown as blue, red, and yellow bars, respectively. The mean data were obtained from four individual experiments. Percent mutant KRAS2 = mutant KRAS2/(mutant KRAS2+wild-type KRAS2). The data are presented as mean ± SEM. B: Percentage of mutant KRAS2 relative to wild-type KRAS2 detected in serum from 28 patients with benign pancreatobiliary diseases. The data are presented as mean ± SEM from four independent experiments.
KRAS2 mutations were also evaluated in bile from patients with benign pancreatobiliary diseases (n = 28). KRAS2 mutations were detected in nine cases (31.4%). However, the levels of the mutations in these positive samples were significantly lower than those from the cancer group, ranging from 0.01% to 3.6% (Figure 1B). Of these positive samples, only two cases had a mutation level of more than 2%. These two cases were obtained from one patient with serous cystadenoma and another with primary sclerosing cholangitis (PSC), respectively. Of note, PSC is the most common precursor lesion for cholangiocarcinoma in the United States,16,17,18,19 so the presence of mutant KRAS2 in this condition is somewhat expected.
We compared KRAS2 mutation levels between the benign and malignant groups. At a threshold of 2% mutational levels, 57% of the samples from the patients with biliary tract carcinomas were positive for KRAS2 mutations, while only 7% of the benign samples are positive (χ2 = 7.333, P < 0.01). The KRAS2 mutation levels in the cancer group was significantly higher than that in the benign group (16.61 ± 7.11% versus 0.24 ± 0.15%, P < 0.01). The median value was 5.1% in the cancer group versus 0.4% in the benign group.
LigAmp Analyses of KRAS2 Mutation in Serum from Patients with Biliary Tract Carcinoma Versus Those with Non-Neoplastic Diseases
We next investigated circulating KRAS2 DNA status in serum from 11 patients with biliary tract carcinoma. LigAmp analysis revealed detectable mutant KRAS2 DNA in 9 of 11 cases (81.8%) (Figure 2A). In three of these positive cases, multiple KRAS2 mutations were detected. Additionally, as we observed in the bile samples, GAT KRAS2 mutation was predominant in most cases. The relative amount of KRAS2 mutations detected in the patients' serum ranged from 0.1% to 24% with a median value of 1.5%.
Figure 2.
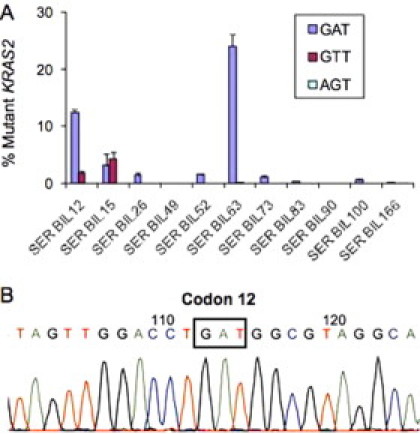
Detection of KRAS2 mutations in serum from 11 patients with biliary tract carcinoma. A: The relative amount of mutant to wild-type KRAS2 DNA detected by LigAmp in serum (n = 11). LigAmp analysis of GAT (G12D), GTT (G12V), and AGT (G12S) KRAS2 mutations shown as blue, red and yellow bars, respectively. The mean data were obtained from four individual experiments. The date is presented as mean ± SEM. B: A representative KRAS2 DNA sequence of a cloned BstN1 refractory PCR product from serum of Bile166. A GGT to GAT mutation at codon 12 is shown in the box.
To rule out whether the detection of low levels of KRAS2 mutations in the serum is caused by a nonspecific reaction, we further confirmed its existence by T-A cloning BstN1-refractory PCR products and DNA sequencing in two cases, designated as Bil100 and Bil166. Bil166 contained approximately 0.1% mutant KRAS2, which was the lowest level of mutant DNA detected among the KRAS2 positive samples. Sequencing of cloned BstN1-refractory PCR products confirmed GAT KRAS2 mutation in both Bil100 (data not shown) and Bil166 (Figure 2B).
Serum samples from nine patients with benign pancreatobiliary disorders were also analyzed as negative controls. Eight of nine samples were amplified by PCR. LigAmp analyses of these eight samples demonstrated wild-type KRAS2 except one sample, which showed a minimal level of GAT mutation (0.03%). This experiment was repeated, however, in the second experiment, no GAT or other KRAS2 mutations were detected.
Correlation of KRAS2 Mutation Status between Bile and Serum of Same Patients with Biliary Tract Carcinoma
Four patients with cholangiocarcinoma had matched bile and serum samples available (Table 2). For case Bil15, GTT and GAT KRAS2 were detected in bile, and both of these mutant variants were also present in serum. Bile from case Bil49 contained neither GAT nor GTT KRAS2, but a minimal level of AGT KRAS2 (<0.1%). No mutant KRAS2 DNA was detected in the corresponding serum. Bile KRAS2 mutation was not identified in case Bil52, but the corresponding serum contained a GAT mutant (1.2%). In case Bil73, we identified two mutants, GAT and GTT in bile, whereas only the GAT KRAS2 mutant was present in serum.
Table 2.
Correlation of KRAS2 Mutation Status between Bile and Serum of Four Patients with Biliary Tract Carcinoma
| Location | Bile | Serum | |
|---|---|---|---|
| Bil 15 | Extrahepatic | GAT (93.4%) | GAT (3.2%) |
| GTT (1.2%) | GTT (4.3%) | ||
| Bil 49 | Extrahepatic | AGT (<0.1%) | Wild type |
| Bil 52 | Gallbladder | Wild type | GAT (1.2%) |
| Bil 73 | Intrahepatic | GAT (3.5%) | GAT (1.0%) |
| GTT (1.2%) |
Discussion
In the present study, we analyzed mutant KRAS2 genes by LigAmp assay in bile or serum from patients with biliary tract carcinoma, and compared this to corresponding clinical samples obtained from individuals with benign pancreatobiliary disorders. Our data showed the presence of mutant KRAS2 in bile from majority of the cancer patients, and in a minor subset of benign pancreatobiliary diseases. However, the levels of KRAS2 mutations in bile of the cancer group as compared with the benign group were significantly higher, consistent with our previous findings in pancreatic cancer.14 As noted previously, one of the higher mutant KRAS2 levels in the “benign” category was obtained from a patient with PSC, an established cholangiocarcinoma precursor lesion. In addition, we demonstrated that we were able to detect KRAS2 mutations in serum from majority of the cancer patients. Detection of KRAS2 mutations in bile and/or serum might serve as, at least, an important ancillary test for diagnosis of biliary tract carcinoma. In addition, this data indicates the possibility of being able to monitor therapeutic responses in patients with biliary tract carcinoma by quantitative analysis of KRAS2 mutations in serum using LigAmp.
We detected multiple KRAS2 mutations in bile and serum of most cases. Multiple mutations in a single tumor have been reported previously by several groups, including our own.14,20,21 This could be attributed to either multiple independent clones existing in the cancer,22 or in theory due to the presence of precursor lesion(s) with different KRAS2 mutation(s).
When comparing the results from bile to those from serum of the same patients, we observed, in one case, a KRAS2 mutation is present in the serum, but not in the bile. This can be explained by following mechanisms: 1) little if any tumor DNA shed into the bile because of its extremely scirrhous nature; 2) extraluminal development of cancer; or 3) obstruction of the bile duct or cystic duct in the case of gallbladder carcinoma. Indeed, this is a case with a gallbladder carcinoma, one would imagine, with a completely obstructed cystic duct, no tumor cells from the gallbladder could access to the bile duct. In another case, two mutations were detected in the bile, while only one mutation was present in the serum. The mutation present in both bile and serum could be derived from the cancer, whereas the mutation only present in bile could be in theory from a coexisting precursor lesion which harbored a different mutant from the cancer.
Several candidate molecular markers in serum have been described for detection of biliary tract carcinoma.23,24 However, the role of these markers in early detection or diagnosis is uncertain. Currently, the most commonly used serum marker for biliary tract carcinoma is CA19-9. Serum CA19-9 has been shown to be increased in 77.9% of patients with biliary tract carcinoma, but it is also raised in patients with colorectal, gastric, and gynecological malignancies, as well as in other benign conditions such as cholangitis and cholestasis.24,25 In patients with PSC, serum CA19-9 has an even lower sensitivity and specificity for diagnosis of cholangiocarcinoma. In addition, only a small portion of cholangiocarcinoma patients with increased serum CA19-9 are surgically resectable. However, there are some promising serum markers currently being studied, including tumor-associated antigen receptor-binding cancer antigen expressed on SiSo cells, the cytokeratin 19 fragment CYFRA 21-1, and MUC5AC.24
Detecting tumor markers in bile might be an alternative approach to finding biliary tract carcinoma at an early stage. Proteomic analyses of biliary tract carcinomas have identified overexpressed proteins in tumor cells,24,26 and these can be shed from tumor tissues into bile during tumor cell death. Secreted proteins derived from biliary tract carcinoma should be at higher local concentrations in bile. Kristiansen et al have identified several malignancy-related proteins in bile from patients with biliary tract carcinoma using a liquid chromatography and tandem mass spectrometric approach.27 The same group further demonstrated that one of the identified proteins, Mac-2-binding protein, is a promising diagnostic marker for patients with biliary carcinoma.28
Detection of mutated genes in bile might be another approach for early detection of biliary tract carcinoma. In the current study, KRAS2 mutations were detected in bile from 87.5% patients with biliary tract carcinoma. Therefore, analysis of KRAS2 mutations in bile could be a potential early detection technique. Sensitivity might be further increased by including additional lower frequency KRAS2 mutations (G12C, G12R, G12A, and G13 mutations), which our assay is currently not designed to detect. However, KRAS2 mutations have been reported to be present in benign pancreatobiliary conditions too, such as chronic pancreatitis and primary biliary cirrhosis.5,29,30 The mutant cells in these lesions could be shed into the fluid, and mutant DNA could be detected in pancreatic juice or bile. This is supported by our previous and current studies.14 However, the mutation levels in these conditions were significantly lower than those detected in the patients with biliary tract carcinoma. In 28 controls of this study, only two bile samples contained more than 2% mutant KRAS2, one of them was from a patient with PSC. PSC is a major risk factor for biliary tract carcinoma (specifically, cholangiocarcinoma) in the west. An estimated 8% to 40% of patients with PSC develop cholangiocarcinoma. Kubicka and colleagues analyzed KRAS2 mutations in bile from patients with PSC, and found that 30% of patients had KRAS2 mutant DNA in their bile fluid.31 In addition, the presence of KRAS2 mutations in bile of these patients was associated with an increased risk for development of cholangiocarcinoma. Therefore, dynamic monitoring of KRAS2 mutant DNA in bile fluid might be helpful for early detection of malignant transformation in patients with PSC.
LigAmp analysis of serum KRAS2 mutations appears promising for diagnosis of biliary tract carcinoma. We were able to detect KRAS2 mutations in serum of more than 80% of cancer patients. In addition, serum KRAS2 mutations should be, in theory, more specific for cancerous diseases than that of other body fluids including bile because of a unique vascular invasive characteristic of malignant cells. Use of KRAS2 testing in practice will ultimately require correlating KRAS2 cutoff values below the size of cancers that we want to detect and above premalignant lesions that can sometimes harbor these mutations (eg, benign PanIN1 lesions in the pancreas). The patient will then need to be scanned to discover the source of mutant KRAS2, and if that is unsuccessful, may require endoscopic retrograde cholangiopancreatography sampling or even angiographic sampling of venous return from organs most likely to be the source. In addition, periodic analysis of serum KRAS2 mutations in these patients could help monitor therapeutic responses to non-invasive modalities like systemic chemotherapy or radiation, where direct access to tissues is not available. Currently, the majority of patients with biliary tract carcinomas are diagnosed at an advanced stage and they are not considered as candidates for curative surgery. An effective monitoring system would greatly facilitate the measurement of patients' responses to existing, as well emerging, therapies. LigAmp may be ideal for this purpose because of its high sensitivity and accurate quantification.
In conclusion, while LigAmp analysis of KRAS2 mutations in bile might be a sensitive assay for an early detection of biliary tract carcinoma, analysis of mutant KRAS2 in serum could be more specific for cancer. By dynamic monitoring KRAS2 mutations in serum of cancer patients, we may be able to use LigAmp to determine patients' response to anticancer treatment and detect minimal residual disease.
Footnotes
Supported by R01 CA130938 (J.R.E.) and R01CA120432 (M.G.) from the National Cancer Institute, the Maryland Cigarette Restitution Fund (J.R.E.) and by a Fellowship from the Canadian Institute of Health Research (C.S.). We are grateful to the Breakstone Foundation for Gallbladder Cancer Research, Inc. for their support of the Maitra laboratory. A.M. and J.R.E. are also supported by the Sol Goldman Pancreatic Cancer Research Center and the Michael Rolfe Foundation for Pancreatic Cancer Research.
Some of the data in this manuscript has been presented at United States and Canadian Academy of Pathology in 2005.
References
- 1.Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353–1357. doi: 10.1053/jhep.2001.25087. [DOI] [PubMed] [Google Scholar]
- 2.Jarnagin WR, Fong Y, DeMatteo RP, Gonen M, Burke EC, Bodniewicz BJ, Youssef BM, Klimstra D, Blumgart LH. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507–517. doi: 10.1097/00000658-200110000-00010. discussion 517–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gores GJ. Cholangiocarcinoma: current concepts and insights. Hepatology. 2003;37:961–969. doi: 10.1053/jhep.2003.50200. [DOI] [PubMed] [Google Scholar]
- 4.Freeny PC. Computed tomography in the diagnosis and staging of cholangiocarcinoma and pancreatic carcinoma. Ann Oncol. 1999;(10 Suppl 4):12–17. [PubMed] [Google Scholar]
- 5.van Leeuwen DJ, Reeders JW. Primary sclerosing cholangitis and cholangiocarcinoma as a diagnostic and therapeutic dilemma. Ann Oncol. 1999;(10 Suppl 4):89–93. [PubMed] [Google Scholar]
- 6.Sturm PD, Rauws EA, Hruban RH, Caspers E, Ramsoekh TB, Huibregtse K, Noorduyn LA, Offerhaus GJ. Clinical value of K-ras codon 12 analysis and endobiliary brush cytology for the diagnosis of malignant extrahepatic bile duct stenosis. Clin Cancer Res. 1999;5:629–635. [PubMed] [Google Scholar]
- 7.Wistuba II, Miquel JF, Gazdar AF, Albores-Saavedra J. Gallbladder adenomas have molecular abnormalities different from those present in gallbladder carcinomas. Hum Pathol. 1999;30:21–25. doi: 10.1016/s0046-8177(99)90295-2. [DOI] [PubMed] [Google Scholar]
- 8.Wistuba II, Albores-Saavedra J. Genetic abnormalities involved in the pathogenesis of gallbladder carcinoma. J Hepatobiliary Pancreat Surg. 1999;6:237–244. doi: 10.1007/s005340050113. [DOI] [PubMed] [Google Scholar]
- 9.Sasatomi E, Tokunaga O, Miyazaki K. Precancerous conditions of gallbladder carcinoma: overview of histopathologic characteristics and molecular genetic findings. J Hepatobiliary Pancreat Surg. 2000;7:556–567. doi: 10.1007/s005340070004. [DOI] [PubMed] [Google Scholar]
- 10.Nakanuma Y, Harada K, Ishikawa A, Zen Y, Sasaki M. Anatomic and molecular pathology of intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Surg. 2003;10:265–281. doi: 10.1007/s00534-002-0729-3. [DOI] [PubMed] [Google Scholar]
- 11.Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin N Am. 2002;11:995–1009. doi: 10.1016/s1055-3207(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Yamaguchi Y, Watanabe H, Ohtsubo K, Wakabayashi T, Sawabu N. Usefulness of p53 gene mutations in the supernatant of bile for diagnosis of biliary tract carcinoma: comparison with K- ras mutation. J Gastroenterol. 2002;37:831–839. doi: 10.1007/s005350200137. [DOI] [PubMed] [Google Scholar]
- 13.Shi CES, Jones D, Fukushima N, Hua L, Parker AR, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. LigAmp: sensitive detection of single nucleotide differences. Nature Meth. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 14.Shi C, Fukushima N, Abe T, Bian Y, Hua L, Wendelburg BJ, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. Sensitive and quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chronic pancreatitis, potential for early detection. Cancer Biol Ther. 2008;7:353–360. doi: 10.4161/cbt.7.3.5362. [DOI] [PubMed] [Google Scholar]
- 15.Al-Soud WA, Ouis IS, Li DQ, Ljungh S, Wadstrom T. Characterization of the PCR inhibitory effect of bile to optimize real-time PCR detection of Helicobacter species. FEMS Immunol Med Microbiol. 2005;44:177–182. doi: 10.1016/j.femsim.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Yachimski P, Pratt DS. Cholangiocarcinoma: natural history, treatment, and strategies for surveillance in high-risk patients. J Clin Gastroenterol. 2008;42:178–190. doi: 10.1097/MCG.0b013e31806daf89. [DOI] [PubMed] [Google Scholar]
- 17.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 18.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 19.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366:1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 20.Mariyama M, Kishi K, Nakamura K, Obata H, Nishimura S. Frequency and types of point mutation at the 12th codon of the c-Ki-ras gene found in pancreatic cancers from Japanese patients. Jpn J Cancer Res. 1989;80:622–626. doi: 10.1111/j.1349-7006.1989.tb01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levi S, Urbano-Ispizua A, Gill R, Thomas DM, Gilbertson J, Foster C, Marshall CJ. Multiple K-ras codon 12 mutations in cholangiocarcinomas demonstrated with a sensitive polymerase chain reaction technique. Cancer Res. 1991;51:3497–3502. [PubMed] [Google Scholar]
- 22.Laghi L, Orbetegli O, Bianchi P, Zerbi A, Di Carlo V, Boland CR, Malesci A. Common occurrence of multiple K-RAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers. Oncogene. 2002;21:4301–4306. doi: 10.1038/sj.onc.1205533. [DOI] [PubMed] [Google Scholar]
- 23.Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin Liver Dis. 2004;24:139–154. doi: 10.1055/s-2004-828891. [DOI] [PubMed] [Google Scholar]
- 24.Bonney GK, Craven RA, Prasad R, Melcher AF, Selby PJ, Banks RE. Circulating markers of biliary malignancy: opportunities in proteomics? Lancet Oncol. 2008;9:149–158. doi: 10.1016/S1470-2045(08)70027-5. [DOI] [PubMed] [Google Scholar]
- 25.Albert MB, Steinberg WM, Henry JP. Elevated serum levels of tumor marker CA19–9 in acute cholangitis. Dig Dis Sci. 1988;33:1223–1225. doi: 10.1007/BF01536670. [DOI] [PubMed] [Google Scholar]
- 26.Kristiansen TZ, Harsha HC, Gronborg M, Maitra A, Pandey A. Differential membrane proteomics using 18O-labeling to identify biomarkers for cholangiocarcinoma. J Proteome Res. 2008;7:4670–4677. doi: 10.1021/pr800215n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kristiansen TZ, Bunkenborg J, Gronborg M, Molina H, Thuluvath PJ, Argani P, Goggins MG, Maitra A, Pandey A. A proteomic analysis of human bile. Mol Cell Proteomics. 2004;3:715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Koopmann J, Thuluvath PJ, Zahurak ML, Kristiansen TZ, Pandey A, Schulick R, Argani P, Hidalgo M, Iacobelli S, Goggins M, Maitra A. Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer. 2004;101:1609–1615. doi: 10.1002/cncr.20469. [DOI] [PubMed] [Google Scholar]
- 29.Caldas C. Biliopancreatic malignancy: screening the at risk patient with molecular markers. Ann Oncol. 1999;10:153–156. [PubMed] [Google Scholar]
- 30.Saurin JC, Joly-Pharaboz MO, Pernas P, Henry L, Ponchon T, Madjar JJ. Detection of Ki-ras gene point mutations in bile specimens for the differential diagnosis of malignant and benign biliary strictures. Gut. 2000;47:357–361. doi: 10.1136/gut.47.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubicka S, Kuhnel F, Flemming P, Hain B, Kezmic N, Rudolph KL, Manns M, Meier PN. K-ras mutations in the bile of patients with primary sclerosing cholangitis. Gut. 2001;48:403–408. doi: 10.1136/gut.48.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]


