Abstract
Distinction of hydatidiform moles from non-molar (NM) specimens, as well as their subclassification as complete (CHM) versus partial hydatidiform moles (PHM), is important for clinical management and accurate risk assessment for persistent gestational trophoblastic disease. Because diagnosis of hydatidiform moles based solely on morphology suffers from poor interobserver reproducibility, a variety of ancillary techniques have been developed to improve diagnosis. Immunohistochemical assessment of the paternally imprinted, maternally expressed p57 gene can identify CHMs (androgenetic diploidy) by their lack of p57 expression, but cannot distinguish PHMs (diandric monogynic triploidy) from NMs (biparental diploidy). Short tandem repeat genotyping can identify the parental source of polymorphic alleles and thus discern androgenetic diploidy, diandric triploidy, and biparental diploidy, which allows for specific diagnosis of CHMs, PHMs, and NMs, respectively. In this study, a retrospectively collected set of morphologically typical CHMs (n = 8), PHMs (n = 10), and NMs (n = 12) was subjected to an analytic validation study of both short tandem repeat genotyping and p57 immunohistochemistry. Several technical and biological problems resulted in data that were difficult to interpret. To avoid these pitfalls, we have developed an algorithm with quantitative guidelines for the interpretation of short tandem repeat genotyping data.
Distinction of hydatidiform moles (HM) from non-molar (NM) specimens and the subclassification of HMs as complete hydatidiform mole (CHM), partial hydatidiform mole (PHM), or early CHM are important for both clinical practice and investigational studies. The risk of persistent gestational trophoblastic disease and hence, clinical management of patients, differs for CHMs, PHMs, and NMs.1,2,3,4,5 However, diagnosis of HMs based solely on morphology suffers from poor interobserver reproducibility.6,7 Even among experienced pathologists, high interobserver and intraobserver variability exist.8
The unique genetic features of CHMs, PHMs, and NMs can be exploited to improve diagnosis of HMs. CHMs are most often diploid with both chromosomal complements being paternal in origin (androgenetic diploidy), whereas PHMs are triploid with one maternal chromosome complement and two paternal chromosome complements (diandric triploidy). NMs are typically diploid with one maternal and one paternal chromosome complement (biparental diploidy); some NMs can be triploid due to two maternal and one paternal chromosome complement (digynic triploidy) but these specimens do not have the morphological features of PHMs.9,10
A variety of ancillary techniques targeting these genetic differences have been used to improve diagnosis of HMs. These include formal cytogenetic analysis (karyotyping), determination of ploidy by flow cytometry,11,12,13 fluorescent in situ hybridization,14,15 PCR amplification of short tandem repeat (STR) loci,16,17,18,19 and immunohistochemistry for the paternally imprinted gene p57.18,20 Most of these techniques, including karyotyping and ploidy analyses, have limitations beyond the known technical and interpretive difficulties in that they cannot specifically discern the maternal and paternal chromosomal contributions in a specimen. Thus, while diploid and triploid specimens can be identified to improve both recognition of PHMs and distinction of PHMs from CHMs, CHMs (particularly the morphologically subtle early forms) cannot be distinguished from NMs (both yield nonspecific diploid results), and PHMs cannot be distinguished from digynic triploid NMs (both yield nonspecific triploid results). CHMs (including early forms) can be distinguished from PHMs and NMs by immunohistochemical assessment of expression of the paternally imprinted p57 gene. CHMs are characterized by lack of p57 expression in villous stromal cells and cytotrophoblast due to the lack of maternal DNA. However, p57 immunohistochemistry cannot distinguish PHMs from NMs since both retain expression of p57 due to the presence of maternal DNA.
Molecular genetic analysis of the type provided by STR genotyping offers greater diagnostic discriminatory capability than these other techniques in that CHMs, PHMs, and NMs can be specifically distinguished from one another based on identification of the parental source of polymorphic alleles and their ratios. In particular, this analysis can discern androgenetic diploidy, diandric triploidy, and biparental diploidy to rigorously diagnose and distinguish CHMs, PHMs, and NMs, respectively. Such analysis is particularly important for the diagnosis of PHMs, which continue to pose diagnostic difficulty and cannot be distinguished from NMs, especially those exhibiting abnormal villous morphology of the type associated with other (non-molar type) genetic abnormalities, due to shared p57 expression patterns.21
Several studies have demonstrated the proof of concept and utility of STR genotyping for distinction of CHMs, PHMs, and NMs.16,17,19 However, none of these reports provides a technical validation that includes specific details regarding how STR data were interpreted, criteria for accepting or rejecting data, and sources of technical and interpretive problems. In the current report we have used a set of retrospective, morphologically typical cases of HMs and NMs to develop an algorithm for the interpretation of STR genotyping data, including specific quantitative criteria for interpretation of results. To fully validate the utility of both STR genotyping and p57 immunohistochemistry and to implement these ancillary tests into routine clinical practice, this algorithm was then applied prospectively to all cases encountered on the Johns Hopkins Gynecologic Pathology Service for which a diagnosis of hydatidiform mole was considered (overwhelmingly morphologically challenging consultation cases); this prospective analysis is described in a separate report.22
Materials and Methods
Case Selection
Thirty cases were selected from the routine Gynecologic Pathology Service files of The Johns Hopkins Hospital, Baltimore, MD. These included 8 CHMs, 10 PHMs, and 12 NMs. All cases were reviewed by two pathologists (B.M.R., R.V.) at a multiheaded microscope to select only those cases for which there was complete agreement that the morphological features were fully diagnostic of the assigned diagnostic category (CHM, PHM, NM). The material from these cases was collected between the years 1995 and 2006. This study is covered under Institutional Review Board approval NA 00002948 for the use of de-identified clinical samples to evaluate new methods/technologies.
P57 Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections (5-micron) were stained using a Ventana BenchMark XT automated immunostaining system (Tucson, AZ) with mouse monoclonal antibodies against p57 protein (Neomarkers, Fremont, CA) from a ready to use preparation without dilution. The presence or absence of nuclear positivity was assessed in villous stromal cells, cytotrophoblast, intermediate trophoblast, and maternal decidua. Based on the staining patterns reported in the literature indicating that virtually all CHMs entirely lack p57 expression in villous stromal cells and cytotrophoblast whereas PHMs and NMs diffusely express p57 in these cell types, the quantity of these cells expressing p57 was estimated by routine light microscopic assessment of all of the villi present in the stained sections, without resorting to actual cell counting. Staining was semiquantitatively assessed as follows: negative (no staining in these cell types), limited (staining in <10% of these cell types), focally positive (staining in >10% but <50% of these cells types), and diffusely positive (staining in >50% of these cell types). The p57 immunostain result was then interpreted as “positive” when the extent of staining was diffusely positive in these cell types. This pattern of expression is consistent with all forms of NMs as well as PHMs and cannot distinguish among these entities. The p57 immunostain was interpreted as “negative” and satisfactory when maternal decidua and/or intermediate trophoblastic cells exhibited nuclear expression of p57 (serving as internal positive control) but villous stromal cells and cytotrophoblast were either entirely negative or demonstrated only limited expression (<10%). This negative result was then interpreted as consistent with a diagnosis of CHM. Nuclear expression in villous stromal cells and cytotrophoblast in the focally positive range (>10% but <50%) was considered an equivocal result, encompassing the 30% value used in one study as a cut-off for positive and negative results but allowing for a wider range to enable use of quick visual estimation rather than cell counting to determine a result.6 Of note, we did not encounter any equivocal results in this validation study but have encountered a few examples in a prospective study applying this immunohistochemical assay to diagnostic cases in our laboratory.22 Thus, in our overall experience, the vast majority of cases exhibited either diffuse positivity for p57 or complete lack of expression of p57 in the cytotrophoblast and villous stromal cells, so equivocally staining results were a minimal problem.
Molecular Genotyping
The AmpFlSTR Profiler kit (Applied Biosystems, Foster City, CA) was used for this analysis. An H&E-stained 5-micron section of formalin-fixed paraffin-embedded tissue was used to identify well-separated areas of maternal decidua and villous tissue. A serial, unstained 10-micron slide was superimposed on top of the marked H&E slide and Pinpoint solution (Zymo Research, Orange CA) was applied to the areas of interest. The tissue was removed from the slide and digested per the manufacturer's protocol. DNA was further purified using QIAamp DNA spin columns (Qiagen, Valencia, CA). PCR amplification of nine microsatellite loci and the amelogenin locus was performed in duplicate. Thermal cycling conditions and capillary electrophoresis performed according to the manufacturer's instructions. In brief, the PCR conditions were 95°C for 11 minutes followed by 28 cycles of 94°C for 1 minute, 59°C for 1 minute, and 72°C for 1 minute, followed by a final extension at 60°C for 45 minutes. After amplification, 1 microliter of multiplex PCR product was mixed with 9 μL of deionized formamide/GeneScan 500 [ROX] size standard (Applied Biosystems). Samples were denatured at 95°C for 2 minutes and placed on ice for at least 1 minute before analysis on the ABI3100 Genetic Analyzer (Applied Biosystems). Capillary electrophoresis data from both the maternal and villous tissues were analyzed to identify alleles at each locus. The paternal genetic pattern was not known. For each locus with two alleles identified, the allelic ratio was calculated by dividing the peak height of the longer allele by the peak height of the shorter allele. Allelic ratios between 0.61 and 1.17 were considered to be consistent with diploidy. Allelic ratios between 0.33 and 0.60 or 1.5 and 2.0 were considered to be consistent with triploidy. Loci with three alleles identified were also considered consistent with triploidy. The origin of triploidy was determined when possible based on combined evaluation of allele ratios and source for those alleles with sufficient polymorphism.
Results
STR Analysis and Expected Patterns
Thirty cases with typical morphological features of CHM, PHM, or NM were selected from our archives for analysis. DNA isolated from villous and decidual tissue was subjected to amplification of nine STR loci and the amelogenin locus in the Profiler assay. When possible, alleles in the villous tissue were identified as paternal (non-maternal) or likely maternal (also possibly paternal due to shared alleles). Figure 1 demonstrates the typical, straightforward patterns for biparental diploidy (NM) (Figure 1A), diandric triploidy (PHM) (Figure 1B), and androgenetic diploidy (CHM) (Figure 1C). For each locus with two alleles identified, the allelic ratio was calculated by dividing the peak height of the longer allele by the peak height of the shorter allele. For diploid cases, an allele ratio of 1.0 is expected, while for triploid cases with two alleles, ratios of either 0.5 or 2.0 are expected. Because of the tendency for the longer allele (allele to the right) to be shorter in height than the shorter allele (allele to the left), ratios are generally slightly lower, particularly when the alleles differ significantly in size. Each locus was scored as being consistent with diploidy or triploidy or not informative. The interpretation of all loci had to be concordant for final interpretation. On first analysis, several cases demonstrated patterns that were not consistent with the patterns expected for NM, PHM, or CHM. We identified several problems that result in patterns that are difficult to interpret. These problems were caused by both technical as well as biological reasons. The problems we encountered and the solutions we developed are outlined below.
Figure 1.
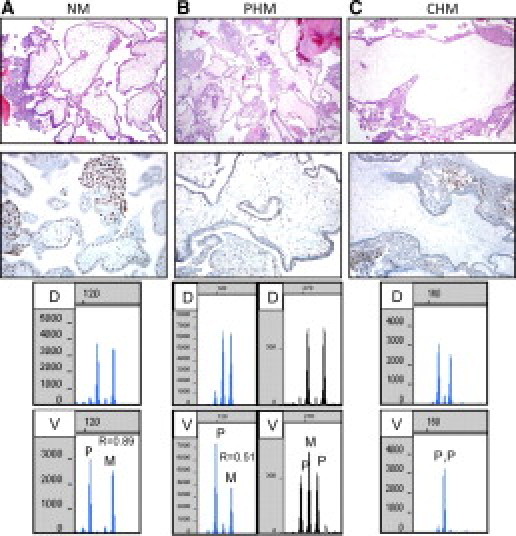
Examples of H&E, p57 immunohistochemistry, and molecular genotyping for typical cases of NM, PHM, and CHM. A: NM. Immature villi are small and have a polarized trophoblast. P57 is expressed in the nuclei of villous stromal cells and the cytotrophoblast. Molecular genotyping demonstrates bi-parental diploidy. B: PHM. Variably sized villi have highly irregularly shapes (“scalloping”) and trophoblastic inclusions but minimal trophoblastic hyperplasia. P57 is expressed in the nuclei of villous stromal cells and the cytotrophoblast. Molecular genotyping demonstrates diandric triploidy. C: CHM. Enlarged hydropic villus exhibits circumferential trophoblastic hyperplasia. Villous stromal cells and cytotrophoblast lack nuclear expression of p57 (intermediate trophoblastic cells are positive and serve as an internal positive control). Molecular genotyping demonstrates androgenetic diploidy. D = decidua, V = villous. Paternal (P) and likely maternal (M) alleles are identified in the villous tissue. The allele ratio (R) in the villous tissue is shown when two alleles are present.
Problem: Poor Amplification
Poor PCR amplification occurred occasionally, most likely because the quantity and/or quality of the DNA used for analysis was suboptimal. This was not particularly surprising since DNA isolated from formalin-fixed paraffin-embedded tissues can be significantly damaged/degraded, resulting in poor PCR amplification, especially with older specimens, and particularly of longer amplicons.23 Of note, some of the specimens we analyzed were over 10 years old. In our sample cohort, six samples yielded minimal amplification, generating PCR products from just one or two loci with the shortest amplicons. These cases were considered PCR failures and could not be analyzed. In approximately half of the cases that yielded interpretable data, one or two loci with the longest amplicons amplified poorly; these loci were excluded from the analysis. Poor PCR amplification can result in inaccurate/biased allele ratios, which can confound interpretation. Figure 2 demonstrates results from two different PCR reactions of a diploid sample, which yielded either poor amplification (Figure 2A) or robust amplification (Figure 2B). The allelic ratio was skewed when the amplification was poor and could be misinterpreted as consistent with triploidy. For this reason, we instituted a requirement of peak heights of at least 300 relative fluorescent units at a 30 second injection in order for the technologist to interpret the locus. We recognize that this requirement is somewhat arbitrary, and that peak heights can vary for reasons other than amplification efficiency (eg, age of capillary, laser, buffer). However, we feel that this criterion will allow the technologist to avoid calculating ratios from loci that may yield erroneous results and to identify specimens that require additional amplification (if all/most of the loci yield low amplification). Inclusion or exclusion of loci from the final interpretation of the case is decided by the individual responsible for reviewing and interpreting the data, and signing the report.
Figure 2.
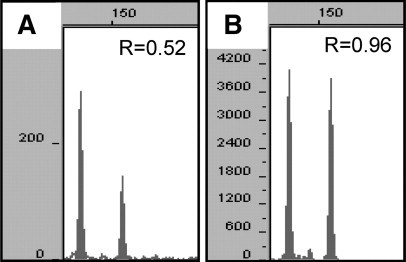
Capillary electrophoresis data from one STR locus demonstrating the effect of poor amplification (A) on allele ratios (R) compared with robust amplification (B).
Problem: Contamination of Villous Tissue with Maternal Tissue
After reviewing the initial data, we determined that the largest, most significant problem was due to the specimens not being pure populations (mixtures of decidual and villous tissue). These cases often gave conflicting interpretations from different STR loci. Contamination with decidual tissue is particularly problematic for loci at which the decidua is homozygous. For example, if the true allelic ratio of a heterozygous locus in the villous is 1:1, the presence of just 20% decidual cells that are homozygous for an allele shared with the villous would yield a ratio of (0.4 + 0.1 + 0.1): 0.4, or 1.5:1. This could be erroneously interpreted as trisomy/triploidy. Figure 3 demonstrates examples of problems we have encountered due to decidual contamination of the villous tissue. Case 1 represents a NM specimen for which, on first analysis, the allele ratio at one locus was consistent with triploidy (2.0). After more careful microdissection, the ratio of the alleles in the villous tissue was 1.1, which is consistent with diploidy. Case 2 represents a PHM specimen for which, on first analysis, the ratio of the alleles in the villous tissue appeared to be consistent with diploidy (0.73). After more careful microdissection, the ratio of the alleles in the villous tissue was 0.55, which is consistent with triploidy. Also shown is a hypothetical case demonstrating how contamination with maternal tissue can result in allele ratios that appear to be consistent with triploidy, when in fact the case is androgenetic diploid. Thus, it is very important to identify when contamination is present to avoid misinterpretation. Villous specimens with decidual contamination can be identified by the presence of loci that yield discordant interpretations and/or the presence of loci that yield allelic patterns and ratios that are not consistent with diploidy or triploidy (see below).
Figure 3.
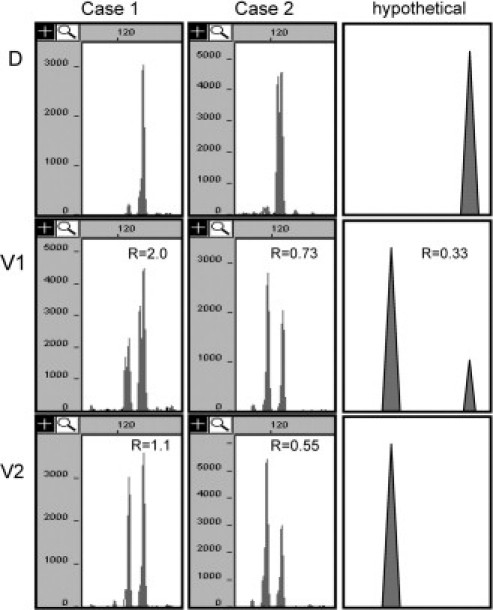
Capillary electrophoresis data (Case 1 and 2) and hypothetical data demonstrating problems due to decidual tissue (D) admixed into the villous specimen. In all cases the decidua is homozygous. On first analysis of Case 1, the ratio of the alleles in the villous tissue (V1) appeared to be consistent with triploidy (2.0). After more careful microdissection (V2), the ratio of the alleles in the villous tissue was 1.2, which is consistent with diploidy. On first analysis of Case 2, the ratio of the alleles in the villous tissue (V1) appeared to be consistent with diploidy (0.73). After more careful microdissection (V2), the ratio of the alleles in the villous tissue was 0.55, which is consistent with triploidy. The hypothetical data demonstrate that androgenetically diploid villous tissue with decidual contamination (V1) can have allele ratios that appear to be consistent with triploidy (0.33).
Education of both pathologists and technologists is the key to avoiding the problem of decidual contamination of the villous specimen. In our initial analysis, when larger areas of villous and decidual tissue were marked, eight cases yielded mixed genetic patterns. Each of these cases was re-dotted by a pathologist, with marking of smaller areas of tissue and careful attention to avoid inclusion of even minute fragments of decidual tissue within areas designated as villous tissue. DNA extracted from these more rigorously selected areas yielded interpretable data. Pathologists must be educated to very carefully identify areas of decidual and villous tissue that are essentially pure, since the presence of just 20% maternal material can confound interpretation. In particular, pathologists should be aware that the purity of the tissue chosen is much more important than the amount of tissue analyzed. In addition, the technologists must be educated to perform careful, accurate microdissection, and to be aware of the possibility of contamination when interpreting the data. Training and education has significantly reduced this problem in our laboratory. Mixed genetic patterns were encountered in none (0/28) of the cases analyzed as part of our prospective validation,22 and are only rarely a problem in clinical cases. An alternative solution would be the use of laser capture microdissection to obtain a pure population of cells for analysis. For clinical use, this was not a practical option for us.
Problem: Number of Informative Loci
Although the Profiler assay evaluates nine STR loci, often many of the loci are not informative. This occurs for several reasons. First, as noted above, poor DNA quality may result in a lack of amplification, or inferior amplification, particularly of longer amplicons, which may not be sufficient for interpretation. Second, shared alleles and an unknown paternal pattern can make interpretation difficult. By definition, PHMs and NMs share at least one allele at each locus with maternal DNA. If the villous tissue is homozygous for an allele shared with maternal tissue, the locus is completely noninformative. If the villous tissue shares both alleles with the maternal tissue with approximately equal height, one can determine diploidy but cannot be certain of the source (maternal versus paternal) of the alleles. Several scenarios result in patterns that are consistent with triploidy but for which the source (maternal versus paternal) cannot be determined. This occurs when the allele(s) in double dosage in the villous tissue is shared with maternal allele(s).
In our experience, the average number of informative and interpretable STR loci for CHM specimens was 5 (range, 2 to 8). Amelogenin was not informative for any of the CHMs because all arose from monospermy and were XX. For PHMs, the average number of loci that demonstrated triploidy was 6 (range, 6 to 7), while the average number of loci that demonstrated that the triploidy was diandric, monogynic was 3 (range 2 to 5). For NMs, the average number of fully informative loci was 4 (range, 2 to 6). Amelogenin was informative (XY) in approximately half of the NM cases (6/13) as would be expected.
Analysis of multiple loci is essential for an accurate interpretation of the data. Individual trisomies and other genetic alterations could lead to misinterpretation of the data. For example, if only one locus was informative and showed a pattern consistent with three alleles, one could not determine whether the specimen had an isolated trisomy or was triploid. Thus, a NM specimen with an isolated trisomy could be misinterpreted as PHM (triploid). This is especially important since specimens with trisomies can have some morphological features that overlap with PHMs, making them diagnostically challenging and in need of ancillary molecular analysis. In addition, rare examples of CHMs that demonstrate androgenetic diploidy at most loci but retain one or a few maternal chromosomes (trisomy for these chromosomes) have been reported.24,25 Analysis of only these loci would lead to misinterpretation as a triploid rather than a diploid specimen. This would result in misclassification as PHM rather than CHM, with the potential for insufficient clinical follow-up to monitor for persistent gestational trophoblastic disease. For these reasons, we have instituted the criterion that at least two loci on separate chromosomes must be used to make a final interpretation. The pattern for all loci must be concordant, even if not fully informative (eg, demonstrate triploidy even if the origin is not determinable). If the patterns for all loci are not concordant, further investigation is required.
Development of an Interpretation Algorithm for STR Data
We have developed an algorithm for interpretation of STR data (Figure 4). At each locus, the observed pattern falls into 1 of 4 categories: one allele, two alleles, three alleles, or greater than three alleles. A single allele in the villous tissue is interpreted as evidence of androgenicity if not shared with the decidua and is not informative if it is shared with the decidua. When two alleles are present, the allelic ratio must be calculated to correctly interpret the pattern. Our analysis of samples in duplicate showed good reproducibility of allelic ratios between different amplifications and between multiple capillary injections of the same amplification. Specifically, the ratios for replicate amplifications always fell within the same interpretation category, as long as the amplification was sufficient. Only when different areas of tissue were analyzed were different ratios obtained, which was due to the relative mixture of decidual and villous tissue (see Problem: Contamination of villous tissue with maternal tissue). For clinical practice, we amplify each sample just once, not in replicates.
Figure 4.
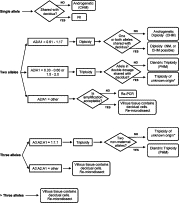
STR interpretation algorithm. NI = not informative; * = diandric, not digynic, triploidy is consistent with PHM. If the source of the triploidy cannot be determined from any of the loci, the data should be interpreted carefully in the context of the morphology.
Review of our data from the NM cases identified 42 loci that were informative and fulfilled our amplification criteria. From these 42 loci, we determine an average allelic ratio to be 0.89 with a SD of 0.14. Thus when the allelic ratio is determined to be between 0.61 and 1.17 (2 SD range), this is interpreted as consistent with diploidy. Diploidy can be biparental (consistent with NM) or androgenetic (consistent with CHM). Review of our data from the PHM cases identified 26 loci that were informative and fulfilled our amplification criteria. From these loci, we determined allelic ratios consistent with triploidy to be 1.50 to 2.0 and 0.33 to 0.6. Loci that are consistent with triploidy must be further reviewed to determine whether they can be categorized as diandric. Loci with three alleles that are approximately equivalent are also interpreted as consistent with triploidy and the source should be determined if possible. If the source of the triploidy cannot be defined as diandric from any of the loci analyzed, this should be reflected in the report, and the data should be interpreted carefully in the context of the morphology. Loci with two or three alleles that have allele ratios outside of the expected ranges for diploidy or triploidy require further investigation. When more than three alleles are present, this usually indicates that there is decidual tissue admixed with the villous tissue. This pattern can arise when decidual tissue is admixed in a villous specimen that is a PHM or a dispermic CHM.
Validation
Using the interpretation criteria and algorithm we developed, 24 cases yielded satisfactory and informative results for both p57 immunostaining and molecular genotyping. Six cases failed to yield adequate amplification for molecular genotyping interpretation, the majority of which were greater than 5 years old. Although we had a significant number of PCR failures in our validation study, in our experience using this assay for clinical testing, less than 5% of cases have failed to sufficiently amplify, suggesting that the amplification problems may have been due to the older age of the validation specimens.
Seven specimens diagnosed as CHM demonstrated a negative p57 result and androgenetic diploidy. All of these (7/7) were homozygous at each locus indicating that the CHM arose from a single sperm that had duplicated its genome. Although this homozygous pattern does not definitively demonstrate diploidy, it does demonstrate complete androgenicity, which is probably the most important information needed. A minority of CHMs are tetraploid, and possibly aneupolid; however these entities appear to have the same risk of gestational trophoblastic disease as diploid CHMs.26,27 In addition to the seven CHMs analyzed here, we have analyzed 10 more CHMs as part of our prospective study,22 and an additional five CHMs to date in our subsequent ongoing clinical application of this technique. Of these 22 CHMs analyzed thus far, we have found only one case of androgenetic diploidy arising from dispermy. Thus, at our institution, this appears to be less common than expected from the literature, which suggest that 10% to 20% of CHMs arise from dispermy.5,28
Eight specimens diagnosed as PHMs demonstrated a positive p57 result and diandric monogynic triploidy. For all of these cases (8/8) there was at least one locus that demonstrated two unique paternal alleles indicating that the PHM arose from dispermy. Nine specimens diagnosed as NMs demonstrated a positive p57 result and biparental diploidy.
Discussion
The value of STR analysis, in contrast to the other methods of ploidy determination, is that the exact maternal and paternal chromosomal contributions can be determined specifically to distinguish androgenetic diploid CHMs from biparental diploid NMs and to distinguish diandric triploid PHMs from digynic triploid NMs. Here we have established quantitative criteria for interpreting allelic ratios and have demonstrated that robust amplification, purity of the tissues analyzed, and use of data from two or more loci on separate chromosomes are essential for obtaining accurate results. The criteria developed in this validation study have been applied in a separate prospective analysis of challenging cases, derived largely from a gynecologic pathology consultation practice. Those results and our algorithm for ancillary testing of HMs are reported elsewhere.22
In addition to the interpretive pitfalls described above, one potential limitation of this analysis that should be noted concerns the rare cases of biparental CHM. This disorder, which appears to result from inheritance of an autosomal recessive pattern mapped to 19q13.4, presents clinically, morphologically, and immunohistochemically (negative p57 result) as multiple CHMs, which appear to have a risk of persistent gestational trophoblastic disease similar to that of conventional CHM (uniparental androgenetic diploidy).29,30,31 Molecular genotypic analysis in such a case would result in biparental diploidy that could be misinterpreted as a non-molar gestation in the absence of correlation with morphological features and p57 results. Furthermore, infrequent yet well-documented cases of mosaicism and chimerism found in HMs might lead to results that are difficult to interpret.20
Our experience in this validation study indicates that Profiler STR analysis is an accurate and practical ancillary molecular technique applicable to routine practice for classifying molar specimens. This assay is one of several commercially available STR assays that analyze multiple (approximately 4 to 15) STR loci in single multiplex PCR reactions. Many of these assays have been created for forensic and paternity applications and thus are highly robust and informative. In our experience, known STR PCR artifacts including stutter and null alleles created no interpretation difficulties. We chose to use the Profiler assay because it is used in our laboratory for bone marrow engraftment analysis and therefore was easily implemented. When determining which reagents to choose, considerations include the number and informativity of the loci, and the length of the amplicons. Although it seems logical that more loci would be preferred, in general, increasing the number of loci requires that larger amplicons (>300 bases) are produced for some loci, which may not yield sufficient amplification for interpretation. Regardless of which STR loci are used, rigorous validation of the assay is required if the results are to be reported for clinical use. Communication between the pathologist choosing tissue for analysis and the laboratory performing the assay is also essential for quality results.
References
- 1.Berkowitz RS, Goldstein DP. Diagnosis and management of the primary hydatidiform mole. Obstet Gynecol Clin North Am. 1988;15:491–503. [PubMed] [Google Scholar]
- 2.Berkowitz RS, Tuncer ZS, Bernstein MR, Goldstein DP. Management of gestational trophoblastic diseases: subsequent pregnancy experience. Semin Oncol. 2000;27:678–685. [PubMed] [Google Scholar]
- 3.Sebire NJ, Fisher RA, Foskett M, Rees H, Seckl MJ, Newlands ES. Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. Bjog. 2003;110:22–26. [PubMed] [Google Scholar]
- 4.Sebire NJ, Foskett M, Fisher RA, Lindsay I, Seckl MJ. Persistent gestational trophoblastic disease is rarely, if ever, derived from non-molar first-trimester miscarriage. Med Hypotheses. 2005;64:689–693. doi: 10.1016/j.mehy.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 5.Garner EI, Goldstein DP, Feltmate CM, Berkowitz RS. Gestational trophoblastic disease. Clin Obstet Gynecol. 2007;50:112–122. doi: 10.1097/GRF.0b013e31802f17fc. [DOI] [PubMed] [Google Scholar]
- 6.Javey H, Borazjani G, Behmard S, Langley FA. Discrepancies in the histological diagnosis of hydatidiform mole. Br J Obstet Gynaecol. 1979;86:480–483. doi: 10.1111/j.1471-0528.1979.tb10793.x. [DOI] [PubMed] [Google Scholar]
- 7.Howat AJ, Beck S, Fox H, Harris SC, Hill AS, Nicholson CM, Williams RA. Can histopathologists reliably diagnose molar pregnancy? J Clin Pathol. 1993;46:599–602. doi: 10.1136/jcp.46.7.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukunaga M, Katabuchi H, Nagasaka T, Mikami Y, Minamiguchi S, Lage JM. Interobserver and intraobserver variability in the diagnosis of hydatidiform mole. Am J Surg Pathol. 2005;29:942–947. doi: 10.1097/01.pas.0000157996.23059.c1. [DOI] [PubMed] [Google Scholar]
- 9.Redline RW, Hassold T, Zaragoza MV. Prevalence of the partial molar phenotype in triploidy of maternal and paternal origin. Hum Pathol. 1998;29:505–511. doi: 10.1016/s0046-8177(98)90067-3. [DOI] [PubMed] [Google Scholar]
- 10.Zaragoza MV, Surti U, Redline RW, Millie E, Chakravarti A, Hassold TJ. Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. Am J Hum Genet. 2000;66:1807–1820. doi: 10.1086/302951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lage JM, Mark SD, Roberts DJ, Goldstein DP, Bernstein MR, Berkowitz RS. A flow cytometric study of 137 fresh hydropic placentas: correlation between types of hydatidiform moles and nuclear DNA ploidy. Obstet Gynecol. 1992;79:403–410. doi: 10.1097/00006250-199203000-00016. [DOI] [PubMed] [Google Scholar]
- 12.Lage JM, Popek EJ. The role of DNA flow cytometry in evaluation of partial and complete hydatidiform moles and hydropic abortions. Semin Diagn Pathol. 1993;10:267–274. [PubMed] [Google Scholar]
- 13.Lage JM, Bagg A, Berchem GJ. Gestational trophoblastic diseases. Curr Opin Obstet Gynecol. 1996;8:79–82. [PubMed] [Google Scholar]
- 14.Yver M, Carles D, Bloch B, Bioulac-Sage P, Martin Negrier ML. Determination of DNA ploidy by fluorescence in situ hybridization (FISH) in hydatidiform moles: evaluation of FISH on isolated nuclei. Hum Pathol. 2004;35:752–758. doi: 10.1016/j.humpath.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Lescoat D, Jouan H, Loeuillet-Olivo L, Le Calve M. Fluorescent in situ hybridization (FISH) on paraffin-embedded placental tissues as an adjunct for understanding the etiology of early spontaneous abortion. Prenat Diagn. 2005;25:314–317. doi: 10.1002/pd.1132. [DOI] [PubMed] [Google Scholar]
- 16.Bell KA, Van Deerlin V, Addya K, Clevenger CV, Van Deerlin PG, Leonard DG. Molecular genetic testing from paraffin-embedded tissue distinguishes nonmolar hydropic abortion from hydatidiform mole. Mol Diagn. 1999;4:11–19. doi: 10.1016/s1084-8592(99)80045-9. [DOI] [PubMed] [Google Scholar]
- 17.Lai CY, Chan KY, Khoo US, Ngan HY, Xue WC, Chiu PM, Tsao SW, Cheung AN. Analysis of gestational trophoblastic disease by genotyping and chromosome in situ hybridization. Mod Pathol. 2004;17:40–48. doi: 10.1038/modpathol.3800010. [DOI] [PubMed] [Google Scholar]
- 18.Popiolek DA, Yee H, Mittal K, Chiriboga L, Prinz MK, Caragine TA, Budimlija ZM. Multiplex short tandem repeat DNA analysis confirms the accuracy of p57(KIP2) immunostaining in the diagnosis of complete hydatidiform mole. Hum Pathol. 2006;37:1426–1434. doi: 10.1016/j.humpath.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 19.Bifulco C, Johnson C, Hao L, Kermalli H, Bell S, Hui P. Genotypic analysis of hydatidiform mole: an accurate and practical method of diagnosis. Am J Surg Pathol. 2008;32:445–451. doi: 10.1097/PAS.0b013e3181520034. [DOI] [PubMed] [Google Scholar]
- 20.Hoffner L, Dunn J, Esposito N, Macpherson T, Surti U. P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum Pathol. 2008;39:63–72. doi: 10.1016/j.humpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Chew SH, Perlman EJ, Williams R, Kurman RJ, Ronnett BM. Morphology and DNA content analysis in the evaluation of first trimester placentas for partial hydatidiform mole (PHM) Hum Pathol. 2000;31:914–924. doi: 10.1053/hupa.2000.9085. [DOI] [PubMed] [Google Scholar]
- 22.McConnell TG, Murphy KM, Hafez M, Vang R, Ronnett BM. Diagnosis and subclassification of hydatidiform moles using p57 immunohistochemistry and molecular genotyping: validation and prospective analysis in routine and consultation practice settings with development of an algorithmic approach. Am J Surg Pathol. 2009;33:805–817. doi: 10.1097/PAS.0b013e318191f309. [DOI] [PubMed] [Google Scholar]
- 23.Siwoski A, Ishkanian A, Garnis C, Zhang L, Rosin M, Lam WL. An efficient method for the assessment of DNA quality of archival microdissected specimens. Mod Pathol. 2002;15:889–892. doi: 10.1097/01.MP.0000024288.63070.4F. [DOI] [PubMed] [Google Scholar]
- 24.Fisher RA, Nucci MR, Thaker HM, Weremowicz S, Genest DR, Castrillon DH. Complete hydatidiform mole retaining a chromosome 11 of maternal origin: molecular genetic analysis of a case. Mod Pathol. 2004;17:1155–1160. doi: 10.1038/modpathol.3800175. [DOI] [PubMed] [Google Scholar]
- 25.McConnell TG, Norris-Kirby A, Hagenkord JM, Ronnett BM, Murphy KM. Complete hydatidiform mole with retained maternal chromosomes 6 and 11. Am J Surg Pathol. 2009;33:1409–1415. doi: 10.1097/PAS.0b013e3181a90e01. [DOI] [PubMed] [Google Scholar]
- 26.Bewtra C, Frankforter S, Marcus JN. Clinicopathologic differences between diploid and tetraploid complete hydatidiform moles. Int J Gynecol Pathol. 1997;16:239–244. doi: 10.1097/00004347-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 27.Fukunaga M. Flow cytometric and clinicopathologic study of complete hydatidiform moles with special reference to the significance of cytometric aneuploidy. Gynecol Oncol. 2001;81:67–70. doi: 10.1006/gyno.2000.6100. [DOI] [PubMed] [Google Scholar]
- 28.Van den Veyver IB, Al-Hussaini TK. Biparental hydatidiform moles: a maternal effect mutation affecting imprinting in the offspring. Hum Reprod Update. 2006;12:233–242. doi: 10.1093/humupd/dmk005. [DOI] [PubMed] [Google Scholar]
- 29.Ambani LM, Vaidya RA, Rao CS, Daftary SD, Motashaw ND. Familial occurrence of trophoblastic disease-report of recurrent molar pregnancies in sisters in three families. Clin Genet. 1980;18:27–29. doi: 10.1111/j.1399-0004.1980.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 30.Fisher RA, Hodges MD, Newlands ES. Familial recurrent hydatidiform mole: a review. J Reprod Med. 2004;49:595–601. [PubMed] [Google Scholar]
- 31.Feltmate CM, Growdon WB, Wolfberg AJ, Goldstein DP, Genest DR, Chinchilla ME, Lieberman ES, Berkowitz RS. Clinical characteristics of persistent gestational trophoblastic neoplasia after partial hydatidiform molar pregnancy. J Reprod Med. 2006;51:902–906. [PubMed] [Google Scholar]


