Abstract
Nucleic acids, whether designed or selected in vitro, play important roles in biosensing, medical diagnostics and therapy. Specifically, the conjugation of functional nucleic acid-based probe molecules and nanomaterials has resulted in an unprecedented improvement in the field of molecular recognition. With their unique physical and chemical properties, nanomaterials facilitate the sensing process and amplify the signal of recognition events. Thus, the coupling of nucleic acids with various nanomaterials opens up a promising future for molecular recognition. The literature offers a broad spectrum of recent advances in biosensing by employing different nano-platforms with designed nucleic acids, especially gold nanoparticles, carbon nanotubes, silica nanoparticles and quantum dots. The advantages of these novel combinations are discussed from the perspective of molecular recognition in chemistry, biology and medicine, along with the problems confronting future applications.
Keywords: Molecular Recognition, DNA, Aptamers, Molecular Beacons, DNAzyme, Gold Nanoparticles, Nanorod, Carbon Nanotubes, Silica Nanoparticles, Quantum Dots
Molecular recognition is key in the design of sensors and switches, as well as the development of clinical diagnostic tools and therapeutic modalities. In early years, various organic molecules possessing unique properties drew the attention of investigators to achieve the recognition of different targets.1,2 Particularly, since the discovery of the double helix structure of DNA,3 the Watson-Crick type of hydrogen bonds, combined with electrostatic force, π-stacking and hydrophobic forces, have made it possible to design suitable probes for signaling biomolecular interaction4 by the very nature of highly specific molecular recognition ability of nucleotide base pairs. In addition to Watson-Crick type of hydrogen bonds for base pairing in molecular recognition of nucleic acids, there are recently developed nucleic acid probes known as aptamers for the recognition of a wide array of targets ranging from small ions to proteins to cells and tissues. Aptamers are oligonucleic acids selected in vitro by a process termed systematic evolution of ligands by exponential enrichment (SELEX) for binding different targets.5, 6 Thus, with the advent of aptamers, the previous application of nucleic acids for molecular recognition took a big step forward.7 Compared with traditional chemical recognition mechanisms, such as host-guest chemistry, the interaction of nucleic acids is universal and easily modified. As a consequence, various biosensors and diagnostic methods have been developed based on the special recognition properties of nucleic acids.8 Moreover, progress in the development of nanomaterials provides nucleic acids even more flexibility as molecular recognition tools.
Unlike other biomolecules, such as proteins, nucleic acid probes are more stable and flexible when they are used with modifications. That is, the ease of handling DNA base modification and DNA strands, when combined with the different modification strategies of nanomaterials, provides a vast platform upon which to build novel molecular recognition tools. This is illustrated by the broad application of DNA and nanomaterial conjugates in the fields of spectroscopy, electrochemistry, magnetics,9 and others.10 Such conjugates offer three important improvements in molecular recognition.
First, nanomaterials can facilitate signal transduction; that is, when suitable nanomaterials work as reporters, the signal of recognition events can be amplified by several orders of magnitude.11 In this way, a number of reporters can be incorporated into a single nanoparticle, which can enhance signal transduction by thousands of times. Moreover, at the nanoscale, a single recognition event might break the balance between different nanoparticles. This event, in turn, could be accompanied by a change in the property of the whole assembly, resulting in a greatly amplified signal. This phenomenon can be demonstrated by the color change of gold nanoparticle solution caused by the unbalanced electrostatic interactions resulting from the introduction of a small interference. Second, nanomaterials may make recognition more effective. Nanomaterials can be modified according to the function of the designed DNA probes, particularly given their high ratio of surface area to volume. In addition, cooperative interaction, also known as synergism, plays a key role in molecular recognition. By definition, synergism is the combined effect of two or more like-acting components exceeding the sum of the effect of the components used alone.12 In this way, cooperative interaction can ease the challenge of recognition towards targets that have more binding sites. Furthermore, nanomaterials can participate in the molecular recognition process by interacting with the DNA probe, which may also increase the DNA binding selectivity. Third, the unusual interactions between nanomaterials and living systems make the application of functional DNA more practical for molecular recognition in medical diagnostics. For example, with the help of nanomaterials, nucleic acids can escape nuclease digestion13 and be transported across the cell membranes to recognize bioactive substances, thus allowing real-time monitoring of recognition events in vivo.14
As a consequence of these advantages, a combination of DNA molecular design and different nanomaterials will lead to enhanced, sometimes new, functions in molecular recognition. Today, there are many nanomaterials such as gold nanoparticles, carbon nanotubes, silica nanoparticles, quantum dots and magnetic nanoparticles which have been widely applied in the interdisciplinary fields of chemistry,15 biology16 and medicine.17 It is from this perspective that we focus on how recent advances in nanomaterial conjugation improve the designed nucleic acid probes for molecular recognition.
Gold Nanomaterials
Compared with either bulk metals or those of molecular compounds, metal nanomaterials display distinct physical properties18 depending on material size, shape, surface function, and interval distance. Furthermore, the special interaction between mercapto-group and Au atom facilitates the modification of nanomaterials with oligonucleotides19 and other compounds. By modifying these physical properties to meet the functional requirements of the DNA probe, metal nanomaterials-conjugated oligonucleotides become ideal platforms for achieving efficient molecular recognition.
To report the DNA hybridization event, oligonucleotides were functionalized with gold nanoparticles. The mechanism of action is based on the distance-dependent surface plasmon absorption of gold nanoparticles.10, 20 Thus upon the addition of target DNA, the probe DNA-modified dispersion of gold nanoparticles takes place, turning the solution from red to blue as a result of the hybridization of DNA functionalized gold nanoparticles by hundreds of target DNA. Due to the extremely high molar absorptivity of gold nanoparticles, 1000-times higher than that of organic dyes,21 the DNA hybridization event is signaled and amplified.
In this method, it is the dispersing to aggregating movement, or vice versa, of the oligonucleotide-functionalized gold nanoparticles that causes the obvious and sensitive color change and, at the same time, facilitates signal amplification. Using this principle as a foundation, the application the DNA conjugates could be extended to recognize different molecules. Most notably, assembled gold nanoparticles in combination with aptamers make the aptamers be efficient for colorimetric recognition of targets based on the structural change of the aptamer upon target binding.22 DNAzymes, which catalyze the hydrolysis of nucleic acids containing given sequences with cofactors, such as metal ions,23 were also demonstrated to be an effective colorimetric probe by using gold nanoparticle. With the help of Pb2+, the DNAzyme would cleave the substrate DNA, and gold nanoparticles facilitated the recognition event and signal transduction.24
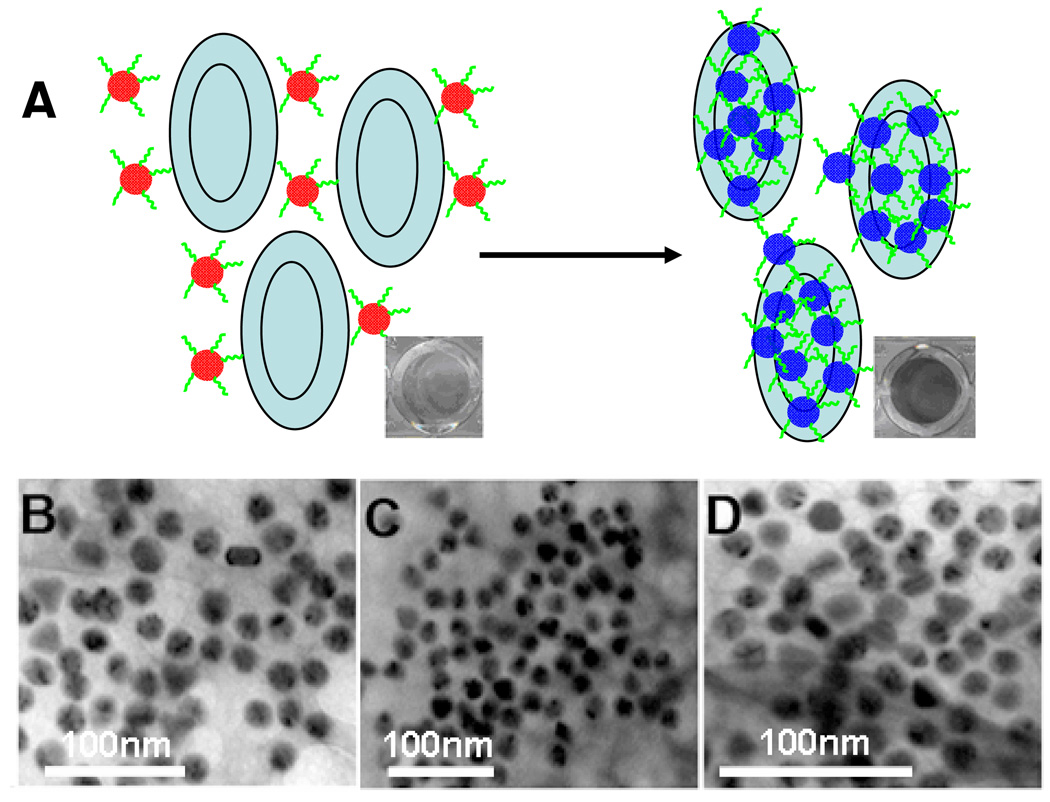
If the target is much bigger in size than the aggregate of nanoparticles or contains more binding sites, the performance of molecular recognition could be improved by the action of synergy, which, as defined previously, is the combined effect of two or more like-acting components exceeding the sum of the effect of the components used alone.12 This has been demonstrated by the gold nanoparticle-based colorimetric detection of platelet-derived growth factor (PDGF)25 and cancer cells. Specifically, there are two sites for aptamer binding on PDGF, and they act like glue to crosslink the aptamer-labeled gold nanoparticles. Since this activity results in net aggregation of the nanoparticles and target, the accompanying absorbance change of the solution is more sensitive to the target, and PDGF at the nanomolar level can therefore be detected. By using cancer cell aptamers26 and gold nanoparticles, direct colorimetric assay of cancer cells has also been achieved (Figure 1).27 Since the volume of a given cancer cell is much larger than the aptamer-functionalized nanoparticles, many aptamers immobilized on gold nanoparticles can bind with one cell very fast; thereafter, the effect of synergy greatly enhances the recognition ability of the aptamers. Thus, target binding and gold nanoparticle assembly has been achieved simultaneously. As confirmed by TEM pictures, gold particles attached to and assembled on the surface of target cells caused the color to change.
Figure 1.
(A) Schematic of the colorimetric assay for cancer cells recognition based on the aptamer functionalized gold nanoparticle. (B–D) TEM images show the binding and assembling of aptamer-functionalized gold nanoparticles on different regions of the target cancer cell surface. Adapted from ref 27.
The interactions between DNA strands and bare gold nanoparticles provide a convenient way for gold nanoparticles to not only signal and amplify the recognition event, but also participate in the recognition process. For example, gold nanoparticles show more strong affinity to single-stranded (ssDNA) than that of double-stranded DNA (dsDNA). The negatively charged backbones of adsorbed ssDNA provide more electrostatic repulsion to stabilize gold nanoparticles, while dsDNA has less ability to stabilize gold nanoparticles in high salt solution.28 This different propensity of ssDNA and dsDNA to adsorb onto gold nanoparticles could enable the design of a label-free colorimetric approach for DNA hybridization assay.29 Specifically, since the electrostatic balance is easily broken by the small disturbance caused by the hybridization of DNA, recognition events can be amplified by the aggregation of the whole nanoparticles.30 Metal ions, 31–33 protein, 34 and other molecules35, 36 can also be detected by the noncovalent assembly of gold nanoparticles and functional oligonucleotides. .
Gold nanoparticles also play an important role in overcoming difficulties encountered in using nucleic acids-based fluorescent probes. One challenge in designing DNA fluorescent probes, such as molecular beacon (MB) and fluorescence signaling aptamer, is the several variables that can compromise the increment of signal change upon interacting with the targets. These primarily include (1) selection of dye-quencher properties, (2) means of attachment of dye-quencher groups, (3) unidentifiable target binding sites, and (4) unforeseen conformational changes. As a consequence of specific electronic properties, gold nanoparticles are good quencher of a fluorophore.37 By applying gold nanoparticles as a substitute for organic quenchers, using either covalent or noncovalent modification with the DNA, the quenching efficiency could be improved greatly, providing a more efficient method for fluorescent detection of DNA,38–40 protein41 or metal ion.42
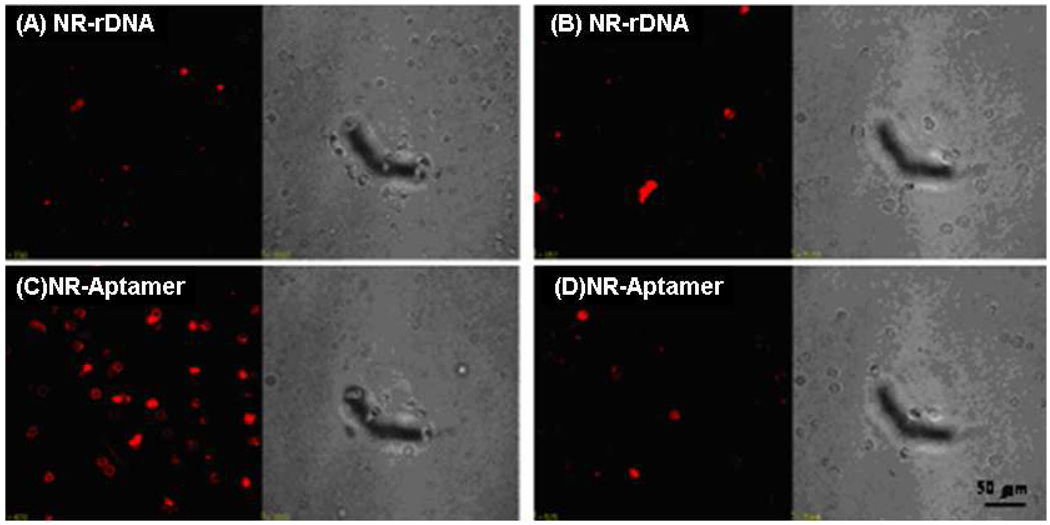
As anisotropic nanoparticles with different aspect ratios, gold nanorods can be easily synthesized and immobilized with huge numbers of functional oligonucleotides. The advantage of coupling gold nanorods with a DNA molecular probe design is the large absorption cross section at the near-infrared (NIR) range, which provides for the development of a novel photothermal transformer for therapy.43 Moreover, compared to individual oligonucleotide probes, functionalizing one nanorod with several oligonucleotides significantly improves the ability to signal the binding event. This improved performance was confirmed by conjugating an aptamer, with only weak binding affinity to cancer cells, and a gold nanorod.44 Flow cytometry analysis showed 300-fold fluorescence intensity enhancement was achieved by using the nanorod-conjugated DNA probe in comparison with single aptamer molecule. The evolved recognition ability of the aptamer-conjugated nanorod was further demonstrated by its photothermal effect (Figure 2).45 With excellent absorption in the NIR range, which overlaps the spectrum of minimum extinction of animal tissues, the aptamer-functionalized Au-Ag nanorod conjugate selectively bound to the target cell with enhanced affinity. After exposure to NIR light irradiation, the nanorod-bound cancer cells were killed by the localized heat produced by photothermal conversion, while the control cells remained live.
Figure 2.
Binding assay of nanorod-aptamer conjugates (NR-Aptamer) towards target cells (C) and control cells (D). The confocal images of target cells (A) and control cells (B) stained by random DNA conjugated nanorod (NR-rDNA) show less fluorescence. The scale bars are 50µm. Fluorescence images (left) and optical images (right). Adapted from ref 45.
In summary, by their unique structural and optical features, DNA- functionalized gold nanomaterials make a highly useful platform for molecular recognition with promising applications in optical detection,46 plasmonic imaging,47 and surface-enhanced resonant Raman analysis.48, 49
Carbon Nanotubes
Carbon nanotubes (CNTs),50 which can be divided into single-walled nanotubes (SWNTs) and multi-walled nanotubes (MWNTs), are another important type of nanomaterials with which to improve DNA molecular recognition, in this case, by their perfect cylindrical structure and unique mechanical, electrical, and optical characteristics.51–53
Various complexes, including DNA strands, can be adsorbed noncovalently onto the sidewalls of CNTs by means of π−π stacking interaction between nucleotide bases and the sidewalls of SWNTs,54–56 which facilitates the application of CNTs-conjugated oligonucleotides for molecular recognition. Because the native fluorescence of the nanotube57 is influenced by adsorbed DNA, SWNTs was employed to signal the DNA hybridization in aqueous solution,58 even though the DNA hybridizing process was slow. Moreover, with the unique optical property of SWNTs, ordinary environmental interference against selective recognition was weakened, making it possible to apply this technique for DNA conformational polymorphism detection, even in whole blood, tissue, and inside living cells.59
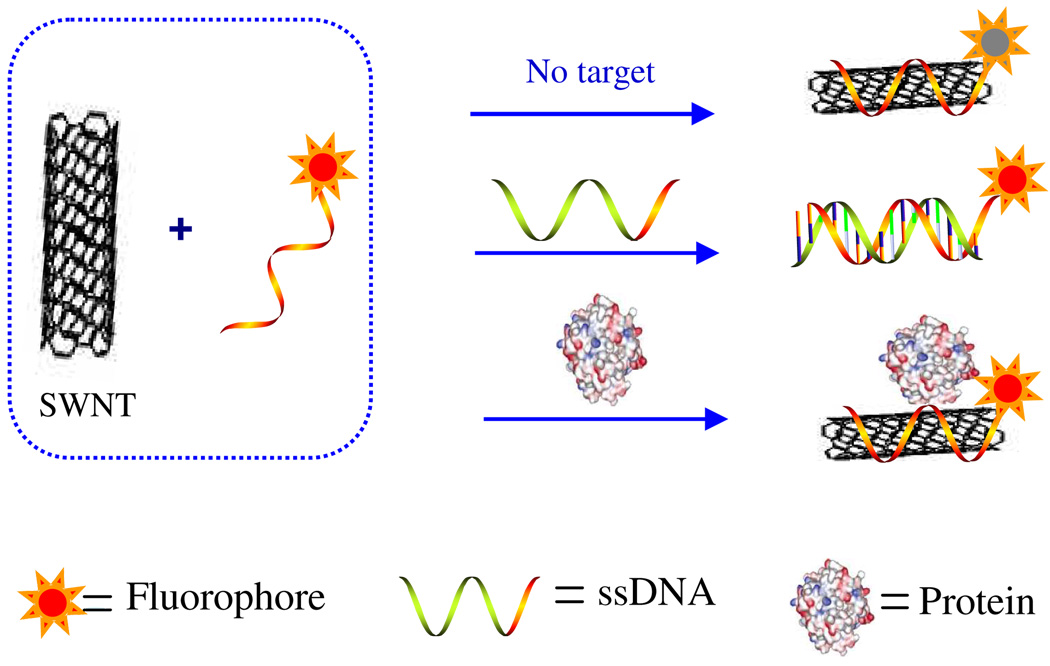
CNTs are also good candidates to improve the recognition performance of fluorescent DNA probes. Photophysical studies have demonstrated that SWNTs can act collectively as quenchers for fluorophores or fluorophore- labeled ssDNA14, 60, 61 by SWNTs, through energy-transfer and electron-transfer processes.51 With their rigid structure and hybridized bases, dsDNA, however, shows less adsorption to SWNTs than does ssDNA.55 When combined with the quenching effect of CNTs, this difference could be used to improve the molecular recognition performance for DNA and protein. The key features of this design are as follows (Figure 3).62, 63 First, as noted above, ssDNA molecules wrap around individual SWNTs by means of π-stacking interactions between the nucleotide bases and the SWNT sidewalls. Next, because the SWNTs act as both a “nanoscaffold” for the ssDNA and a “nanoquencher” of the fluorophore, only one end of the ssDNA must be labeled with a fluorophore. Under these conditions, the ssDNA molecules self-organize on the surface of the carbon nanotubes, completely quenching the fluorophore. Finally, in the presence of a target, competitive binding of the target and the carbon nanotubes with the ssDNA suppresses the fluorescence quenching, allowing fluorescence-signal enhancement that is large relative to that without a target. This combination of properties results in fluorescence enhancement that is sensitive and specific to the perfectly complementary ssDNA. Furthermore, this design, which is based on a simple, cost-effective synthesis, was shown to have a large signal-to-background ratio, high thermostability, and exceptional DNA-binding selectivity. Therefore, from the standpoints of design and engineering, production, and overall function, self-assembled ssDNA-SWNT complexes can easily replace conventional MBs, which provided new opportunities in the design of nanodevices for molecular recognition. For instance, the recognition event can also be reported by light scattering signals,64 moreover the performance of this method could be improved by employing a non-labeled DNA fluorescent dye, such as ethidium bromide (EB).65 As a planar molecule, EB can absorb on the sidewall of SWNTs, reducing the background fluorescence as much as the quenching effect of SWNTs. The adsorbed EB preferred to intercalate the hybridized bases, and the fluorescence recovered after hybridization, thus greatly enhancing S/B.
Figure 3.
Scheme for signaling biomolecular interactions by the assembly of SWNTs and fluorophore-labeled ssDNA. Adapted from ref 63
The novel interaction between CNTs and DNA increases the application of the conjugates for molecular recognition in other areas. For instance, the aptamer/SWNTs conjugate has been used to regulate the generation of singlet oxygen.66 In this case, the excited state of a photosensitizer can be quenched by SWNTs, and such quenching effect then inhibits the generation of singlet oxygen. However, upon binding with target, the photosensitizer-labeled aptamers are released from the sidewall of SWNTs, generating a considerable amount of singlet oxygen. The target protein-directed singlet oxygen generation is thereby accomplished, which demonstrates how DNA-functionalized SWNTs, with their excellent photothermal properties, have great potential for diagnostics and therapy.14
The possibility of using the CNTs-conjugated nucleic acids for diagnostics in cells was also illustrated by the satisfactory performance of a DNA probe-conjugated CNT for the recognition of specific cellular RNA.13 Since DNA is easily digested by cellular enzymes,67 a fluorescent DNA probe for the detection of manganese superoxide dismutase (MnSOD) mRNA was used to complex with SWNTs. The result of PAGE indicated that SWNTs protected ssDNA from cleaving, even after incubating 60 minutes with DNase I,13 which can unselectively cleave ssDNA or dsDNA. The capability of the complex probe was further demonstrated in a cellular environment compared with free DNA probe.
Silica Nanoparticles
With well-defined morphology and porosity, silica colloid was first prepared and characterized by Stöber et al.68 As the native feature of silica, various hybrid silica nanoparticles, such as dye-doped fluorescent silica nanoparticles69 and magnetic silica nanoparticles,70 with distinct properties are prepared, making silica nanoparticles good candidates for constructing hybrid materials which can load and transport different agents for applications in different fields.71
The combination of hybrid silica nanoparticles with functional oligonucleotides offers great improvements in molecular recognition, particularly for sensitive reporting. Because large numbers of luminescent dyes can be encapsulated inside, the as-prepared dye-doped silica nanomaterials have promising advantages for amplifying the recognition signal over their counterparts with high intensity and excellent optical stability.72 More important, unlike quantum dots and metal nanoparticles, the luminescent nanoparticles are more hydrophilic, biocompatible, and relatively more stable under different conditions, all of which make them excellent candidates for applications with functional nucleic acids in vivo.73
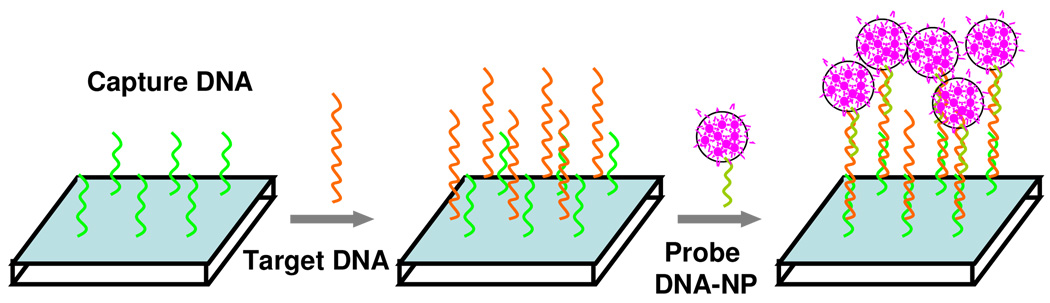
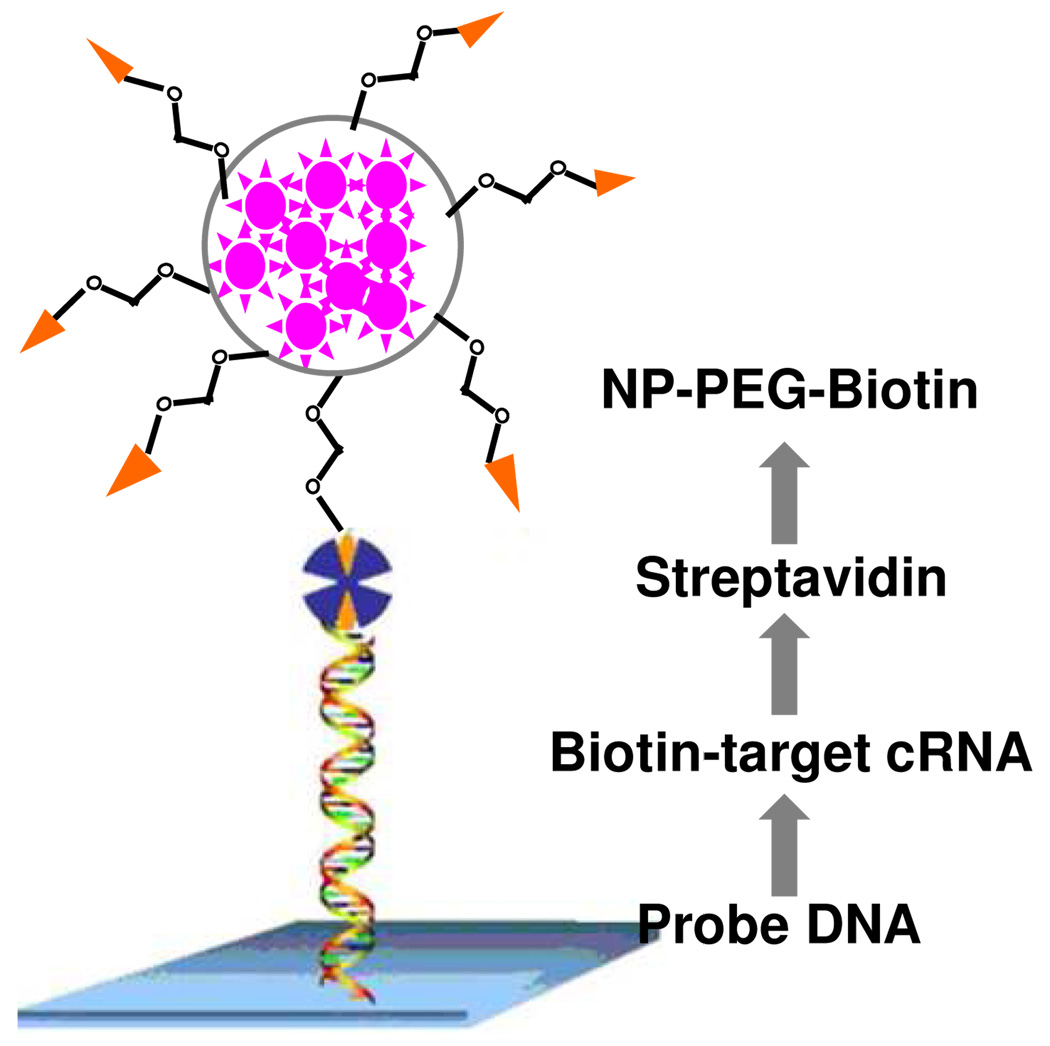
As a consequence of their signal amplification, dye-doped silica nanoparticles modified with oligonucleotides can be applied for ultrasensitive DNA detection (Figure 4).74 Traditionally, one DNA probe could be labeled with only one or a few fluorophores, resulting in limited fluorescent signal. By contrast, since one silica nanoparticle can trap hundreds of fluorophores, an intense fluorescent signal, which is approximately 104 times higher compared with that of the single fluorophore-labeled DNA probe, from the trapped fluorophores can be obtained upon target recognition.75 In addition, the silica shells can protect doped dyes from photodamage by minimizing oxygen through the outer environment. Based on the designed sandwich assay, target DNA in the subfemtomolar range can be detected. The signal amplification effect of the hybrid nanoparticle was also proved in genechips (Figure 5).76 The DNA immobilized doped silica nanoparticle was used as a staining probe, which greatly enhanced detection sensitivity and photostability when compared to the traditional fluorescent protein streptavidin-phycoerythrin.
Figure 4.
Schematic of a sandwich DNA assay based on dye-doped silica nanoparticles. Adapted from ref 74
Figure 5.
Strategy of dye-doped silica nanoparticle based labeling for genechip technology. Adapted from ref 76
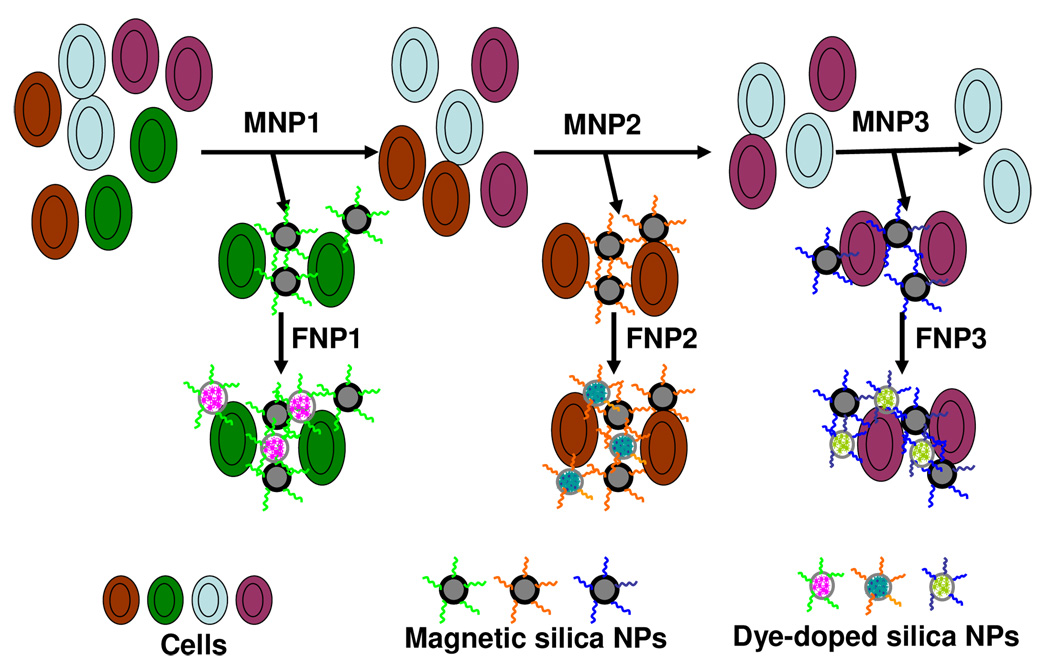
Moreover, fluorophores with different emission properties can be trapped in the matrix of silica nanoparticles for multiplex signaling.77 By varying the ratio of doped fluorophores, nanoparticles exhibit multiple colors with one single wavelength excitation based on fluorescence resonance energy transfer,78 which could be applied for multiple cancer cell recognition. For instance, aptamers for different cancer cells were immobilized onto the surface of silica-coated magnetic nanoparticles. Meanwhile, different dye-doped silica nanoparticles were also labeled with aptamers to report the binding of particular types of cancer cells. After magnetic washes, the collected samples were imaged, and the amounts of three different types of cancer cells were determined simultaneously (Figure 6).79
Figure 6.
Schematic representation of the multiple extraction procedure with the magnetic silica nanoparticles being added and extracted stepwise and the corresponding dye-doped silica nanoparticles being added post-magnetic extraction of cell samples. Adapted from ref 79.
Another important hybrid material conjugated with DNA used for molecular recognition is magnetic silica nanoparticles. Magnetic nanomaterials are excellent tools as contrast agents for magnetic resonance imaging (MRI) and for use in separation and drug delivery.80 Silica-coated magnetic nanoparticles have been synthesized in water/oil microemulsion.70 Disulfide bond and biotin-avidin linkage81 were successfully used for labeling nucleic acids with silica magnetic nanoparticles to concentrate, separate and diagnose different targets. MBs could be combined with magnetic silica nanoparticles to extend their application in separation. MBs-labeled magnetic nanoparticles are efficient tools for separation and collection of DNA trace amounts from a complex mixture. At the same time, the collection process can be monitored in real time from the fluorescence enhancement of the MBs.
The stable and facile synthesis of aptamers makes this versatile DNA-functionalized silica nanoparticle-based method ideal for biological applications and diagnostics. Particularly, magnetic separation, coupled with the highly selective properties of aptamers, is efficient for target collection and enrichment in complex clinical samples. In addition to efficient separation, dye-doped silica nanoparticles can be used to monitor the recognition event and provide signal enhancement. Recently, this novel magnetic separation method was applied to rapid cancer cell collection and detection.82 To achieve both separation and monitoring, carboxyl-functionalized dye-doped nanoparticles were covalently linked with an aptamer for the cancer cells, while silica-coated magnetic nanoparticles were immobilized with aptamers via biotin-avidin interaction. After three magnetic extractions, 40% of spiked target cancer cells were counted, consistent with extraction efficiency values of the immunomagnetic method.
Quantum Dots
Quantum dots (QDs) or inorganic semiconductor nanocrystals are another kind of important nanomaterials, which have involved deeply in biological analysis83 and imaging owing to their distinct photophysical properties, such as size-depended stable luminescence properties, high quantum yields, broad absorbance bands but narrow emission spectrums.84 In 1998, two pioneer works about water-soluble QDs inspired the succedent applications of quantum dots for molecular recognitions.85, 86 QDs integrated with functional nucleic results in obvious evolution of molecular recognition, especially in the fields of multiplexed targets detection and single molecule/particle analysis.
Hydroxylated QDs were first immobilized with probe DNA to fluorescent monitor the in situ hybridization event with Y chromosome in human sperm cells.87 The organic fluorophores of MBs can be replaced by QDs, to achieve better photostability, which could be used for longer time imaging.88 Preliminary siRNA screening was also demonstrated by using the DNA conjugated QDs.89 The specific but stable optical features make QDs based DNA probes superior in molecular recognition. Incorporate with aptamers, the functional QDs were successfully applied to fluorescent detect ATP,90 thrombin,91, 92 PDGF, 93 and cancer cells.94–96
Compared with the stable but complicated covalent procedures for immobilizing of functional nucleic acids with of QDs, electrostatic self-assembling of DNA probes and QDs provides more versatile scaffold to establish molecular recognition. The prepared negatively charged QDs could form compact complex with probe DNA in the presence of cationic polymer, exhibiting the strong FRET between QDs and the dyes on the DNA probes. After the hybridization, as the rigid dsDNA, the distance of energy transfer changed which reduced the efficiency of FRET. The hybridization event was detected by the changes of FRET between QDs and dye labeled DNA.97 Modifying the surface of QDs with positively charged groups simplified the process of assemble, and the cationic polymer linker was avoided.98
Due to the broad absorption bands, QDs with different emissions can be excited simultaneously, which provides chances to recognize and monitor different targets at the same time after excitation by one source.99 ZnS-capped CdSe QDs of different sizes were successfully incorporated into polymer beads with different ratios. Conjugated with different DNA probes, multiplexed DNA recognitions were demonstrated by using triple-color encoded QDs-tagged beads. 100 When QDs with different emissions were combined with aptamers, different target molecules could be recognized at the same time. Simultaneously cocaine and adenosine detection has realized by using the aptamers conjugated QDs assembles.101 The electrochemical multiplex targets analysis was also achieved by the DNA functionalized QDs.102, 103
With high quantum yields and anti-photobleaching properties, conjugations of QDs and functional nucleic acids are wonderful candidates to investigate the molecular recognition events in single molecule/particle level, which supply more real time information and improved sensitivities.104 In a designed sandwiched assay, QDs not only acted as donors of FRET pairs, but also provided a nanoplatform to confine numbers of captured targets and amplify the signals, 100-fold greater than the conventional assays detected by confocal fluorescence spectroscopy. Similar aptamer coupled single-particle based assays were used to detect cocaine,105 and to study the interaction between RNA and protein to screen inhibitors.106
Conclusion and Perspective
The combination of nanomaterials and functional nucleic acids is universal and shows excellent performance for molecular recognition. The powerful functional nucleic acids act as the recognition part,107 and different nanomaterials supply a powerful nano-platform to assist oligonucleotides to improve their ability in recognizing target molecules from complex samples with high sensitivity and selectivity.
Most of the presented advances are in vitro studies, succeeding in solving some problems of molecular recognition, including, for example, ultrasensitive detection, signal amplification, and enhanced recognition. However, existing shortcomings remain to be addressed. For instance, it has been found that aggregation of nanomaterials in a complex environment, especially living systems, greatly depressed the effectiveness and the nonspecific adsorption of some molecules, such as proteins, consequently producing a disturbance that leads to false results. Moreover, the potential toxicity of some nanomaterials to the human body is still not very clear.108
To overcome these challenges, surface modifications and improved hybrid nanomaterials are considered. After immobilization with different functional and biocompatible compounds, nanomaterials would produce fewer agglomerations and less injury towards cells, while still showing more effectiveness for research in vivo.109, 110 Meanwhile, the ordinary procedure for assembling nanomaterials and oligonucleotides should help to facilitate the recognition of target molecules. DNA-templated or as-prepared hybrid nanomaterials might also be a solution.111, 112
Some novel detection techniques also emerge and show advantages when fabricated with the functional hybrid DNA nanomaterials in the field of molecular recognition. For example, the semiconductor CNTs113 and silica nanowires114 based field-effect transistors (FETs) are label-free, reusable and high sensitive, which get good performances in the detection of DNA,115, 116 proteins117, 118 and Escherichia coli119 by modification with different DNA or aptamers. This kind of newly developed methods greatly supports the wider application of the functional nucleic acid conjugated nanomaterials.
With further improvements, these hybrid materials will have more significant impact in bioanalysis and exhibit attractive potential for further applications, such as diagnostics, drug screening, molecular therapy, and efficient drug delivery.
Vocabulary
Molecular beacons
artificial single stranded oligonucleotides designed with stem-loop structures, which comprise a fluorophore and a quencher moiety at two opposite ends. Without target molecules, the base pairs of stem portion hybridize to hold the fluorophore and quencher close and the fluorescence is quenched. In the presence of target, the loop DNA region can bind to it and cause the stem-loop structure open, which would spatially separate the fluorophore from the quencher and the fluorescence increase.
Aptamer
in vitro selected short single stranded DNA or RNA with high binding affinity and specificity to various target molecules by folding into defined tertiary structures. Aptamers for different targets can be produced from random-sequence DNA or RNA libraries by a process called SELEX after a few rounds of affinity selection and amplification.
DNAzyme
catalytic DNA molecule, also called DNA enzyme or deoxyribozymes, which is selected in vitro from random sequence DNA pools. With the help of particular cofactors, such as metal ions and hemin, this DNA-based biocatalyst facilitates the chemical reaction of the substrates.
ACKNOWLEDGEMENTS
We acknowledge financial support through U.S. NIH, NSF, and China NSFC (20525518, 20775005) grants.
Contributor Information
Ronghua Yang, Email: yangrh@hnu.cn.
Weihong Tan, Email: tan@chem.ufl.edu.
References and Notes
- 1.Gellman SH. Introduction: Molecular Recognition. Chem. Rev. 1997;97:1231–1232. doi: 10.1021/cr970328j. [DOI] [PubMed] [Google Scholar]
- 2.de Silva AP, Gunaratne HQN, Gunnlaugsson T, Huxley AJM, McCoy CP, Rademacher JT, Rice TE. Signaling Recognition Events with Fluorescent Sensors and Switches. Chem. Rev. 1997;97:1515–1566. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]
- 3.Watson JD, Crick FHC. A Structure for Deoxyribose Nucleic Acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 4.Fang XH, Li JJ, Perlette J, Tan WH, Wang KM. Peer Reviewed: Molecular Beacons: Novel Fluorescent Probes. Anal. Chem. 2000;72:747 A–753 A. doi: 10.1021/ac003001i. [DOI] [PubMed] [Google Scholar]
- 5.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 6.Ellington AD, Szostak JW. In Vitro Selection of RNA Molecules that Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Famulok M, Hartig JS, Mayer G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Cao Z, Lu Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Josephson L, Perez JM, Weissleder R. Magnetic Nanosensors for the Detection of Oligonucleotide Sequences. Angew. Chem. Int. Ed. 2001;40:3204–3206. doi: 10.1002/1521-3773(20010903)40:17<3204::AID-ANIE3204>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 10.Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 11.Wang J. Nanomaterial-Based Amplified Transduction of Biomolecular Interactions. Small. 2005;1:1036–1043. doi: 10.1002/smll.200500214. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, Cao ZH, Tan WH. Molecular Assembly for High-Performance Bivalent Nucleic Acid Inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu YR, Phillips JA, Liu HP, Yang RH, Tan WH. Carbon Nanotubes Protect DNA Strands during Cellular Delivery. ACS Nano. 2008;2:2023–2028. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kam NWS, O'Connell M, Wisdom JA, Dai H. Carbon Nanotubes as Multifunctional Biological Transporters and Near-Infrared Agents for Selective Cancer Cell Destruction. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Lu Y. Functional DNA Directed Assembly of Nanomaterials for Biosensing. J. Mater. Chem. 2009;19:1788–1798. doi: 10.1039/B813939C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De M, Ghosh PS, Rotello VM. Applications of Nanoparticles in Biology. Adv. Mater. 2008;20:4225–4241. [Google Scholar]
- 17.Liu Z, Winters M, Holodniy M, Dai H. siRNA Delivery into Human T cells and Primary Cells with Carbon-Nanotube Transporters. Angew. Chem. Int. Ed. 2007;46:2023–2027. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh SK, Pal T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory to Applications. Chem. Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- 19.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 20.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 21.Zhao W, Brook MA, Li YF. Design of Gold Nanoparticle-Based Colorimetric Biosensing Assays. ChemBiochem. 2008;9:2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Lu Y. Smart Nanomaterials Responsive to Multiple Chemical Stimuli with Controllable Cooperativity. Adv. Mater. 2006;18:1667–1671. [Google Scholar]
- 23.Silverman SK. In Vitro Selection, Characterization, and Application of Deoxyribozymes that Cleave RNA. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Lu Y. A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 25.Huang CC, Huang YF, Cao ZH, Tan WH, Chang HT. Aptamer-Modified Gold Nanoparticles for Colorimetric Determination of Platelet-Derived Growth Factors and Their Receptors. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 26.Shangguan DH, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Aptamers Evolved From Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medley CD, Smith JE, Tang ZW, Wu YR, Bamrungsap S, Tan WH. Gold Nanoparticle-Based Colorimetric Assay for the Direct Detection of Cancerous Cells. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 28.Sato K, Hosokawa K, Maeda M. Rapid Aggregation of Gold Nanoparticles Induced by Non-Cross-Linking DNA Hybridization. J. Am. Chem. Soc. 2003;125:8102–8103. doi: 10.1021/ja034876s. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Rothberg L. Colorimetric Detection of DNA Sequences Based on Electrostatic Interactions with Unmodified Gold Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Rothberg LJ. Label-Free Colorimetric Detection of Specific Sequences in Genomic DNA Amplified by the Polymerase Chain Reaction. J. Am. Chem. Soc. 2004;126:10958–10961. doi: 10.1021/ja048749n. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Liu X, Hu X, Song S, Fan C. Unmodified Gold Nanoparticles as A Colorimetric Probe for Potassium DNA Aptamers. Chem. Commun. 2006:3780–3782. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- 32.Liu CW, Hsieh YT, Huang CC, Lina ZH, Chang HT. Detection of Mercury(II) Based on Hg2+-DNA Complexes Inducing the Aggregation of Gold Nanoparticles. Chem. Commun. 2008:2242–2244. doi: 10.1039/b719856f. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Wang Z, Liu J, Lu Y. Highly Sensitive and Selective Colorimetric Sensors for Uranyl (UO22+): Development and Comparison of Labeled and Label-Free DNAzyme-Gold Nanoparticle Systems. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei H, Li B, Li J, Wang E, Dong S. Simple and Sensitive Aptamer-based Colorimetric Sensing of Protein Using Unmodified Gold Nanoparticle Probes. Chem. Commun. 2007:3735–3737. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Wang L, Pan D, Song S, Boey FYC, Zhang H, Fan C. Visual Cocaine Detection with Gold Nanoparticles and Rationally Engineered Aptamer Structures. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 36.Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. DNA Aptamer Folding on Gold Nanoparticles: From Colloid Chemistry to Biosensors. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 37.Fan C, Wang S, Hong JW, Bazan GC, Plaxco KW, Heeger AJ. Beyond Superquenching: Hyper-Efficient Energy Transfer from Conjugated Polymers to Gold Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6297–6301. doi: 10.1073/pnas.1132025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubertret B, Calame M, Libchaber AJ. Single-Mismatch Detection Using Gold-Quenched Fluorescent Oligonucleotides. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell DJ, Taylor JR, Nie S. Self-Assembled Nanoparticle Probes for Recognition and Detection of Biomolecules. J. Am. Chem. Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Rothberg LJ. DNA Sequence Detection Using Selective Fluorescence Quenching of Tagged Oligonucleotide Probes by Gold Nanoparticles. Anal. Chem. 2004;76:5414–5417. doi: 10.1021/ac049173n. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Chen C, Qian M, Zhao XS. Aptamer Biosensor for Protein Detection Using Gold Nanoparticles. Anal. Biochem. 2008;373:213–219. doi: 10.1016/j.ab.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Wang YX, Jin JY, Yang RH. Gold Nanoparticle-Based Colorimetric and "Turn-On" Fluorescent Probe for Mercury(II) Ions in Aqueous Solution. Anal. Chem. 2008;80:9021–9028. doi: 10.1021/ac801382k. [DOI] [PubMed] [Google Scholar]
- 43.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Noble Metals on the Nanoscale: Optical and Photothermal Properties and Some Applications in Imaging, Sensing, Biology, and Medicine. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 44.Huang YF, Chang HT, Tan WH. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 45.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan WH. Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 46.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous Detection of Unamplified Genomic DNA Sequences Based on Colorimetric Scatter of Gold Nanoparticle Probes. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart ME, Anderton CR, Thompson LB, Maria J, Gray SK, Rogers JA, Nuzzo RG. Nanostructured Plasmonic Sensors. Chem. Rev. 2008;108:494–521. doi: 10.1021/cr068126n. [DOI] [PubMed] [Google Scholar]
- 48.Qian X, Zhou X, Nie S. Surface-Enhanced Raman Nanoparticle Beacons Based on Bioconjugated Gold Nanocrystals and Long Range Plasmonic Coupling. J. Am. Chem. Soc. 2008;130:14934–14935. doi: 10.1021/ja8062502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho H, Baker BR, Wachsmann-Hogiu S, Pagba CV, Laurence TA, Lane SM, Lee LP, Tok JBH. Aptamer-Based SERRS Sensor for Thrombin Detection. Nano Lett. 2008;8:4386–4390. doi: 10.1021/nl802245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iijima S. Helical Microtubules of Graphitic Carbon. Nature. 1991;354:56–58. [Google Scholar]
- 51.Tasis D, Tagmatarchis N, Bianco A, Prato M. Chemistry of Carbon Nanotubes. Chem. Rev. 2006;106:1105–1136. doi: 10.1021/cr050569o. [DOI] [PubMed] [Google Scholar]
- 52.Sinha N, Ma J, Yeow JTW. Carbon Nanotube-Based Sensors. J. Nanosci. Nanotechnol. 2006;6:573–590. doi: 10.1166/jnn.2006.121. [DOI] [PubMed] [Google Scholar]
- 53.Prato M, Kostarelos K, Bianco A. Functionalized Carbon Nanotubes in Drug Design and Discovery. Acc. Chem. Res. 2008;41:60–68. doi: 10.1021/ar700089b. [DOI] [PubMed] [Google Scholar]
- 54.Guldi DM, Rahman GMA, Zerbetto F, Prato M. Carbon Nanotubes in Electron Donor-Acceptor Nanocomposites. Acc. Chem. Res. 2005;38:871–878. doi: 10.1021/ar040238i. [DOI] [PubMed] [Google Scholar]
- 55.Zheng M, Jagota A, Semke ED, Diner BA, McLean RS, Lustig SR, Richardson RE, Tassi NG. DNA-Assisted Dispersion and Separation of Carbon Nanotubes. Nat. Mater. 2003;2:338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 56.Zheng M, Jagota A, Strano MS, Santos AP, Barone P, Chou SG, Diner BA, Dresselhaus MS, McLean RS, Onoa GB, Samsonidze GG, Semke ED, Usrey M, Walls DJ. Structure-Based Carbon Nanotube Sorting by Sequence-Dependent DNA Assembly. Science. 2003;302:1545–1548. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- 57.O'Connell MJ, Bachilo SM, Huffman CB, Moore VC, Strano MS, Haroz EH, Rialon KL, Boul PJ, Noon WH, Kittrell C, Ma J, Hauge RH, Weisman RB, Smalley RE. Band Gap Fluorescence from Individual Single-Walled Carbon Nanotubes. Science. 2002;297:593–596. doi: 10.1126/science.1072631. [DOI] [PubMed] [Google Scholar]
- 58.Jeng ES, Moll AE, Roy AC, Gastala JB, Strano MS. Detection of DNA Hybridization Using the Near-Infrared Band-Gap Fluorescence of Single-Walled Carbon Nanotubes. Nano Lett. 2006;6:371–375. doi: 10.1021/nl051829k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heller DA, Jeng ES, Yeung TK, Martinez BM, Moll AE, Gastala JB, Strano MS. Optical Detection of DNA Conformational Polymorphism on Single-Walled Carbon Nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 60.Nakayama-Ratchford N, Bangsaruntip S, Sun X, Welsher K, Dai H. Noncovalent Functionalization of Carbon Nanotubes by Fluorescein-Polyethylene Glycol: Supramolecular Conjugates with pH-Dependent Absorbance and Fluorescence. J. Am. Chem. Soc. 2007;129:2448–2449. doi: 10.1021/ja068684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin SJ, Keskar G, Wu YN, Wang X, Mount AS, Klaine SJ, Moore JM, Rao AM, Ke PC. Detection of Phospholipid-Carbon Nanotube Translocation Using Fluorescence Energy Transfer. Appl. Phys. Lett. 2006;89:143118. [Google Scholar]
- 62.Yang RH, Jin JY, Chen Y, Shao N, Kang HZ, Xiao ZY, Tang ZW, Wu YR, Zhu Z, Tan WH. Carbon Nanotube-Quenched Fluorescent Oligonucleotides: Probes that Fluoresce upon Hybridization. J. Am. Chem. Soc. 2008;130:8351–8358. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 63.Yang RH, Tang ZW, Yan JL, Kang HZ, Kim Y, Zhu Z, Tan WH. Noncovalent Assembly of Carbon Nanotubes and Single-Stranded DNA: An Effective Sensing Platform for Probing Biomolecular Interactions. Anal. Chem. 2008;80:7408–7413. doi: 10.1021/ac801118p. [DOI] [PubMed] [Google Scholar]
- 64.Zhang L, Huang CZ, Li YF, Xiao SJ, Xie JP. Label-Free Detection of Sequence-Specific DNA with Multiwalled Carbon Nanotubes and Their Light Scattering Signals. J. Phys. Chem. B. 2008;112:7120–7122. doi: 10.1021/jp800092r. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Wang YX, Jin JY, Wang H, Yang RH, Tan WH. Fluorescent Assay of DNA Hybridization with Label-Free Molecular Switch: Reducing Background-Signal and Improving Specificity by Using Carbon Nanotubes. Chem. Commun. 2009:665–667. doi: 10.1039/b819526a. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Z, Tang ZW, Phillips JA, Yang RH, Wang H, Tan WH. Regulation of Singlet Oxygen Generation Using Single-Walled Carbon Nanotubes. J. Am. Chem. Soc. 2008;130:10856–10857. doi: 10.1021/ja802913f. [DOI] [PubMed] [Google Scholar]
- 67.Cao ZH, Huang CC, Tan WH. Nuclease Resistance of Telomere-like Oligonucleotides Monitored in Live Cells by Fluorescence Anisotropy Imaging. Anal. Chem. 2006;78:1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 68.Stöber W, Fink A, Bohn E. Controlled Growth of Monodisperse Silica Spheres in the Micron Size Range. J. Colloid Interface Sci. 1968;26:62–69. [Google Scholar]
- 69.Wang L, Wang KM, Santra S, Zhao XJ, Hilliard LR, Smith JE, Wu YR, Tan WH. Watching Silica Nanoparticles Glow in the Biological World. Anal. Chem. 2006;78:646–654. [Google Scholar]
- 70.Santra S, Tapec R, Theodoropoulou N, Dobson J, Hebard A, Tan WH. Synthesis and Characterization of Silica-Coated Iron Oxide Nanoparticles in Microemulsion: The Effect of Nonionic Surfactants. Langmuir. 2001;17:2900–2906. [Google Scholar]
- 71.Trewyn BG, Slowing II, Giri S, Chen HT, Lin VSY. Synthesis and Functionalization of a Mesoporous Silica Nanoparticle Based on the Sol-Gel Process and Applications in Controlled Release. Acc. Chem. Res. 2007;40:846–853. doi: 10.1021/ar600032u. [DOI] [PubMed] [Google Scholar]
- 72.Yao G, Wang L, Wu YR, Smith J, Xu JS, Zhao WJ, Lee EJ, Tan WH. FloDots: Luminescent Nanoparticles. Anal. Bioanal. Chem. 2006;385:518–524. doi: 10.1007/s00216-006-0452-z. [DOI] [PubMed] [Google Scholar]
- 73.He XX, Nie HL, Wang KM, Tan WH, Wu X, Zhang PF. In Vivo Study of Biodistribution and Urinary Excretion of Surface-Modified Silica Nanoparticles. Anal. Chem. 2008;80:9597–9603. doi: 10.1021/ac801882g. [DOI] [PubMed] [Google Scholar]
- 74.Zhao XJ, Tapec-Dytioco R, Tan WH. Ultrasensitive DNA Detection Using Highly Fluorescent Bioconjugated Nanoparticles. J. Am. Chem. Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 75.Zhao XJ, Hilliard LR, Mechery SJ, Wang YP, Bagwe RP, Jin SG, Tan WH. A Rapid Bioassay for Single Bacterial Cell Quantitation Using Bioconjugated Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15027–15032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, Lofton C, Popp M, Tan WH. Using Luminescent Nanoparticles as Staining Probes for Affymetrix GeneChips. Bioconjugate Chem. 2007;18:610–613. doi: 10.1021/bc060365u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Yang CY, Tan WH. Dual-Luminophore-Doped Silica Nanoparticles for Multiplexed Signaling. Nano Lett. 2005;5:37–43. doi: 10.1021/nl048417g. [DOI] [PubMed] [Google Scholar]
- 78.Wang L, Tan WH. Multicolor FRET Silica Nanoparticles by Single Wavelength Excitation. Nano Lett. 2006;6:84–88. doi: 10.1021/nl052105b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith JE, Medley CD, Tang ZW, Shangguan DH, Lofton C, Tan WH. Aptamer-Conjugated Nanoparticles for the Collection and Detection of Multiple Cancer Cells. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 80.Jun YW, Seo JW, Cheon J. Nanoscaling Laws of Magnetic Nanoparticles and Their Applicabilities in Biomedical Sciences. Acc. Chem. Res. 2008;41:179–189. doi: 10.1021/ar700121f. [DOI] [PubMed] [Google Scholar]
- 81.Zhao XJ, Tapec-Dytioco R, Wang KM, Tan WH. Collection of Trace Amounts of DNA/mRNA Molecules Using Genomagnetic Nanocapturers. Anal. Chem. 2003;75:3476–3483. doi: 10.1021/ac034330o. [DOI] [PubMed] [Google Scholar]
- 82.Herr JK, Smith JE, Medley CD, Shangguan DH, Tan WH. Aptamer-Conjugated Nanoparticles for Selective Collection and Detection of Cancer Cells. Anal. Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 83.Gill R, Zayats M, Willner I. Semiconductor Quantum Dots for Bioanalysis. Angew. Chem. Int. Ed. 2008;47:7602–7625. doi: 10.1002/anie.200800169. [DOI] [PubMed] [Google Scholar]
- 84.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 86.Chan WCW, Nie SM. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 87.Pathak S, Choi SK, Arnheim N, Thompson ME. Hydroxylated Quantum Dots as Luminescent Probes for in Situ Hybridization. J. Am. Chem. Soc. 2001;123:4103–4104. doi: 10.1021/ja0058334. [DOI] [PubMed] [Google Scholar]
- 88.Kim JH, Morikis D, Ozkan M. Adaptation of Inorganic Quantum Dots for Stable Molecular Beacons. Sens. Actuators B. 2004;102:315–319. [Google Scholar]
- 89.Bakalova R, Zhelev Z, Ohba H, Baba Y. Quantum Dot-Conjugated Hybridization Probes for Preliminary Screening of siRNA Sequences. J. Am. Chem. Soc. 2005;127:11328–11335. doi: 10.1021/ja051089h. [DOI] [PubMed] [Google Scholar]
- 90.Chen Z, Li G, Zhang L, Jiang JF, Li Z, Peng ZH, Deng L. A New Method for the Detection of ATP Using A Quantum-Dot-Tagged Aptamer. Anal. Bioanal. Chem. 2008;392:1185–1188. doi: 10.1007/s00216-008-2342-z. [DOI] [PubMed] [Google Scholar]
- 91.Levy M, Cater SF, Ellington AD. Quantum-Dot Aptamer Beacons for the Detection of Proteins. ChemBioChem. 2005;6:2163–2166. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 92.Choi JH, Chen KH, Strano MS. Aptamer-Capped Nanocrystal Quantum Dots: A New Method for Label-Free Protein Detection. J. Am. Chem. Soc. 2006;128:15584–15585. doi: 10.1021/ja066506k. [DOI] [PubMed] [Google Scholar]
- 93.Kim GI, Kim KW, Oh MK, Sung YM. The Detection of Platelet Derived Growth Factor Using Decoupling of Quencher-Oligonucleotide from Aptamer/Quantum Dot Bioconjugates. Nanotechnology. 2009;20:175503. doi: 10.1088/0957-4484/20/17/175503. [DOI] [PubMed] [Google Scholar]
- 94.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Labeling Tumor Cells with Fluorescent Nanocrystal-Aptamer Bioconjugates. Biosens. Bioelectron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 95.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum Dot-Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 96.Chen XC, Deng YL, Lin Y, Pang DW, Qing H, Qu F, Xie HY. Quantum Dot-Labeled Aptamer Nanoprobes Specifically Targeting Glioma Cells. Nanotechnology. 2008;19:235105. doi: 10.1088/0957-4484/19/23/235105. [DOI] [PubMed] [Google Scholar]
- 97.Peng H, Zhang L, Kjallman THM, Soeller C. DNA Hybridization Detection with Blue Luminescent Quantum Dots and Dye-Labeled Single-Stranded DNA. J. Am. Chem. Soc. 2007;129:3048–3049. doi: 10.1021/ja0685452. [DOI] [PubMed] [Google Scholar]
- 98.Lee J, Choi Y, Kim J, Park E, Song R. Positively Charged Compact Quantum Dot-DNA Complexes for Detection of Nucleic Acids. ChemPhysChem. 2009;10:806–811. doi: 10.1002/cphc.200800504. [DOI] [PubMed] [Google Scholar]
- 99.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Luminescent Quantum Dots for Multiplexed Biological Detection and Imaging. Curr. Opin. Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 100.Han MY, Gao XH, Su JZ, Nie S. Quantum-Dot-Tagged Microbeads for Multiplexed Optical Coding of Biomolecules. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 101.Liu J, Lee JH, Lu Y. Quantum Dot Encoding of Aptamer-Linked Nanostructures for One-Pot Simultaneous Detection of Multiple Analytes. Anal. Chem. 2007;79:4120–4125. doi: 10.1021/ac070055k. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, Liu G, Merkoci A. Electrochemical Coding Technology for Simultaneous Detection of Multiple DNA Targets. J. Am. Chem. Soc. 2003;125:3214–3215. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 103.Hansen JA, Wang J, Kawde A-N, Xiang Y, Gothelf KV, Collins G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006;128:2228–2229. doi: 10.1021/ja060005h. [DOI] [PubMed] [Google Scholar]
- 104.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-Quantum-Dot-Based DNA Nanosensor. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 105.Zhang C, Johnson LW. Single Quantum-Dot-Based Aptameric Nanosensor for Cocaine. Anal. Chem. 2009;81:3051–3055. doi: 10.1021/ac802737b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang CY, Johnson LW. Quantifying RNA-Peptide Interaction by Single-Quantum Dot-Based Nanosensor: An Approach for Drug Screening. Anal. Chem. 2007;79:7775–7781. doi: 10.1021/ac071225w. [DOI] [PubMed] [Google Scholar]
- 107.Breaker RR. Natural and Engineered Nucleic Acids as Tools to Explore Biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- 108.AshaRani PV, Mun LKG, Hande MP, Valiyaveettil S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 109.Prencipe G, Tabakman SM, Welsher K, Liu Z, Goodwin AP, Zhang L, Henry J, Dai H. PEG Branched Polymer for Functionalization of Nanomaterials with Ultralong Blood Circulation. J. Am. Chem. Soc. 2009;131:4783–4787. doi: 10.1021/ja809086q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. Compact Biocompatible Quantum Dots Functionalized for Cellular Imaging. J. Am. Chem. Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berti L, Burley GA. Nucleic Acid and Nucleotide-Mediated Synthesis of Inorganic Nanoparticles. Nat. Nanotechnol. 2008;3:81–87. doi: 10.1038/nnano.2007.460. [DOI] [PubMed] [Google Scholar]
- 112.Ma N, Sargent EH, Kelley SO. One-Step DNA-Programmed Growth of Luminescent and Biofunctionalized Nanocrystals. Nat. Nanotechnol. 2009;4:121–125. doi: 10.1038/nnano.2008.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Allen BL, Kichambare PD, Star A. Carbon Nanotube Field-Effect-Transistor-Based Biosensors. Adv. Mater. 2007;19:1439–1451. [Google Scholar]
- 114.Gao ZQ, Agarwal A, Trigg AD, Singh N, Fang C, Tung CH, Fan Y, Buddharaju KD, Kong JM. Silicon Nanowire Arrays for Label-Free Detection of DNA. Anal. Chem. 2007;79:3291–3297. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 115.Star A, Tu E, Niemann J, Gabriel JCP, Joiner CS, Valcke C. Label-Free Detection of DNA Hybridization Using Carbon Nanotube Network Field-Effect Transistors. Proc. Natl. Acad. Sci. U.S.A. 2006;103:921–926. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gui EL, Li LJ, Zhang KK, Xu YP, Dong XC, Ho XN, Lee PS, Kasim J, Shen ZX, Rogers JA, Mhaisalkar SG. DNA Sensing by Field-Effect Transistors Based on Networks of Carbon Nanotubes. J. Am. Chem. Soc. 2007;129:14427–14432. doi: 10.1021/ja075176g. [DOI] [PubMed] [Google Scholar]
- 117.Maehashi K, Katsura T, Kerman K, Takamura Y, Matsumoto K, Tamiya E. Label-Free Protein Biosensor Based on Aptamer-Modified Carbon Nanotube Field-Effect Transistors. Anal. Chem. 2007;79:782–787. doi: 10.1021/ac060830g. [DOI] [PubMed] [Google Scholar]
- 118.Lee HS, Kim KS, Kim CJ, Hahn SK, Jo MH. Electrical Detection of VEGFs for Cancer Diagnoses Using Anti-Vascular Endotherial Growth Factor Aptamer-Modified Si Nanowire FETs. Biosens. Bioelectron. 2009;24:1801–1805. doi: 10.1016/j.bios.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 119.So HM, Park DW, Jeon EK, Kim YH, Kim BS, Lee CK, Choi SY, Kim SC, Chang H, Lee JO. Detection and Titer Estimation of Escherichia Coli Using Aptamer-Functionalized Single-Walled Carbon-Nanotube Field-Effect Transistors. Small. 2008;4:197–201. doi: 10.1002/smll.200700664. [DOI] [PubMed] [Google Scholar]