Abstract
Objective
Obese patients respond differently to weight loss interventions. No efficient diagnostic tool exists to separate obese patients into subtypes as a means to improve prediction of response to interventions. We aimed to separate obese subjects into distinct subgroups using microarray technology to identify gene expression-based subgroups to predict weight loss.
Design
A total of 72 obese men and women without family history of diabetes were enrolled in the study; 52 were treated with ephedra and caffeine (E+C) and 20 with placebo for 8 weeks. Adipose and skeletal muscle tissue biopsies were performed at baseline. RNA sample pairs were labeled and hybridized to oligonucleotide microarrays. Quantile normalization and gene shaving were performed, and a clustering algorithm was then applied to cluster subjects based on their gene expression profile. Clusters were visualized using heat maps and related to weight changes.
Results
Cluster analysis of gene expression data revealed two distinct subgroups of obesity and predicted weight loss in response to the treatment with E+C. One cluster (`red') decreased to 96.87±2.35% body weight, and the second cluster (`green') decreased to 95.59±2.75% body weight (P<0.05). `Red' cluster had less visceral adipose tissue mass (2.77±1.08 vs 3.43±1.49 kg; P<0.05) and decreased size of the very large fat cells (1.45±0.61 vs 2.16±1.74 μl; P<0.05) compared to `green' cluster. Gene expression for both skeletal muscle and adipose tissue was also different between clusters.
Conclusions
Our study provides the first evidence that the combined approach of gene expression profiling and cluster analysis can identify discrete subtypes of obesity, these subtypes have different physiological characteristics and respond differently to an adrenergic weight loss therapy. This brings us that into an era of personalized treatment in the obesity clinic.
Keywords: subtyping obesity, microarray, cluster analysis, clinical characteristics, weight loss
Introduction
The current treatment recommendations for obesity are quite simple. All patients are essentially given the same advice: less food intake, more physical activities or taking medications. In contrast, physicians and scientists realize that overweight and obese patients are unique. That is, patients respond by varying degrees to pharmacologic and other weight loss interventions. Unfortunately, our understanding of what characteristics identify patients as `responders' or `nonresponders' to a specific therapy is limited because we do not have diagnostic tools to separate obese patients into subtypes. The concept of obesity subtypes is based on the assumption that people become obese for a variety of reasons. Increased food intake (such as craving and night eating syndrome,1,2 decreased physical activity3 or defects in substrate metabolism4 may predominate in a single patient. A second assumption is that these different causes converge to produce the sine qua non of obesity, which is increased body fat, and these different physiological pathways manifest themselves at the cellular, molecular and/or tissue levels.
There is a large degree of interindividual variability in the amount of weight loss in response to obesity therapy. The concept of targeted or `tailored' therapy is common in medical disciplines other than obesity. For example, we do not treat type 1 and type 2 diabetics the same way, and an oncologist does not treat a patient with estrogen receptor-positive breast cancer the same way as a patient with estrogen receptor-negative breast cancer. In this light, obesity therapy is inadequate. The current medical and scientific community does not distinguish between the subtypes of obesity that most certainly exist.
Interindividual variation in the response to obesity therapy is in part due to differences in compliance with therapy5 but a patient's response also depends on their genetic and/or physiological makeup.6 Low baseline levels of the adipocyte hormone leptin predicted response to obesity therapies.7,8 Other factors, such as gender and degree of visceral adiposity, also predict the loss of visceral adipose tissue in relation to total body fat loss.9 However, neither leptin, gender visceral adiposity is sufficiently powerful to separate obese patients into distinct subtypes.
The sequencing of the human genome provides a fantastic or opportunity to advance our understanding of obesity subtypes. Breast cancer, lymphoma and prostate cancer can be separated into distinct phenotypic groups by simultaneously measuring the expression level for thousands of mRNAs with cDNA microarrays.10,11 The pattern of mRNAs present in an patient sample is known as `expression profiling' or `molecular fingerprinting'. The molecular fingerprint reflects the underlying pathophysiology of a tumor or a disease.10 Molecular fingerprints are then used to separate patients into distinct groups and to predict the prognosis of cancer patients and/or their response to chemotherapy.12 As an example, Alizadeh et al.13 demonstrated that patients with B-cell lymphomas that clustered into the `activated B-cell category' had a higher mortality and responded poorly to chemotherapy.
Different tissues have distinct energy needs and vary widely in gene expression patterns.14 Skeletal muscle is mainly involved in energy expenditure (EE) and glucose metabolism,15 whereas adipose tissue has been developed as a site of storage for fat and production of certain cytokines and hormones.16 A quantitative comparison of different tissues by means of cDNA microarray expression analysis highlighted functional classes of genes that are regulated in a tissue-specific manner.
Based on the two fundamental premises stated above that (1) obese patients become obese due to a variety of causes and (2) these causes manifest themselves at the levels of the tissue and gene expression pattern, we proposed that using microarray analysis of skeletal muscle and adipose tissue mRNA would separate obese patients into distinct subgroups based on their `molecular fingerprint'. By analyzing gene expression pattern in skeletal muscle and adipose tissue mRNA in obese subjects before the treatment with ephedra and caffeine (E + C), we predict that (1) clusters of genes identified will separate obese subjects into distinct subgroups, and these groups will (2) differ in their weight loss response to E + C, (3) differ in their thermogenic response to the β-adrenergic treatment (E + C), (4) differ in their clinical characteristics, including visceral fat mass and fat cell size (FCS) and (5) these findings will lead to better diagnosis and therapeutic decision making for obese patients.
Research design and methods
Overview and study population
After signing the informed written consent approved by the Pennington Biomedical Research Center ethical review board, 72 healthy obese men and women without type 2 diabetes or a family history of type 2 diabetes, aged 44.0±10.6 (range 20–64) years with a body mass index (BMI) of 30.3±2.8 kg/m2, were enrolled in the study. Subjects underwent routine fasting blood work including a complete blood count, glucose, insulin, and lipids as well as a physical exam and medical history. Baseline body composition was measured by dual energy X-ray absorptiometry (DEXA) and visceral adipose tissue (VAT) mass by multislice CT as previously described.17 Body weight was measured in a gown after voiding and waist circumference measured using a standardized protocol. Abdominal subcutaneous adipose tissue biopsy was obtained during fasting state, using a Bergstrom needle. Volunteers with chronic illnesses such as heart disease, hypothyroidism, renal, lung and liver diseases were excluded. The use of β-blockers and other drugs known to affect body weight or adrenergic tone were also exclusionary.
Resting energy expenditure and the thermic effect of ephedra plus caffeine
After abstinence from alcohol, exercise and caffeine for 48 h, EE and respiratory quotient (RQ) were measured for 30 min with a ventilated hood system (Deltatrac II; DATEX-Ohmeda, Helsinki, Finland).18 EE was adjusted for lean body mass (LBM), body fat mass (by DEXA) and gender.
Subjects were randomized in a blinded fashion and then consumed their first dose of ephedra (20 mg) plus caffeine (200 mg; MetaboLife, San Diego, CA, USA) or placebo depending on their treatment assignment. This was followed by measurements of EE hourly for 30 min per hour over the next 4h. The last 20min of each measurement period were used to calculate EE for that hour. The thermic response to caffeine and ephedra was calculated as the 4-h area under the curve over baseline EE. Oxygen consumption (ml min−1) over the last 20min was converted to total EE (kcal) and multiplied up to 24 h.19 All metabolic measures were made at baseline only.
Skeletal muscle and adipose tissue biopsy and RNA extraction
After an overnight fast and local anesthesia with lidocaine/bupivicaine, volunteers were biopsied by using Bergstrom technique with suction (Micrins Inc., Lake Forest, IL, USA) from the vastus lateralis skeletal muscle (approximately 100 mg) and abdominal subcutaneous adipose tissue (approximately 500 mg) for microarray expression analysis before beginning the E + C treatment phases as previously described.20,21 Total RNA was extracted by Trizol (Invitrogen, Carlsbad, CA, USA) followed by a clean up on a RNA binding column (RNAEasy; Qiagen, Valencia, CA, USA). The quantity and integrity of the RNA was analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Oligonucleotide cDNA microarray
Gene expressions were measured using the 18 861-gene (Compugen, Bethesda, MD, USA) oligonucleotide microarray chips as previously described.21 Briefly, RNA sample pairs (adipose tissue and skeletal muscle 2 mg for each) from each subject were labeled with dCTP-Cy5 and dCTP-Cy3, respectively, using MICROMAX TSA Labeling kit (Perkin-Elmer, Wellesley, MA, USA). Equal amounts of labeled cDNA probes were hybridized to oligonucleotide slides in hybridization chambers (GenomicSolutions, Ann Arbor, MI, USA) for up to 72 h at 42 °C. Detection and washing were performed at room temperature according to the manufacturer's protocol using MICROMAX TSA Detection kit (Perkin-Elmer). Microarray slides were scanned using a GSI Lumonics ScanArray 5000 scanner (Perkin-Elmer) at medium laser intensities (~80% for Cy3, ~70% for Cy5) applying ScanArray Express software and quantified using QuantArray (GenomicSolutions). The correct assignment of adipose tissue to Cy5 and skeletal muscle to Cy3 was confirmed by tissue-specific gene expression unique to that dye channel. For example, expressions of adipose tissue genes are highly dyed on the Cy5 channel but not highly dyed on the Cy3 channel, and vice versa (data not shown).
Fat cell size
FCS was determined as previously described.22 Briefly, adipose tissue (50 mg) was fixed in osmium tetrachloride/collidine-HCl followed by disassociation by urea digestion. Cells were counted on a Multisizer-3 (Beckman Coulter, Fullerton, CA, USA) using a 400-μm aperture (dynamic linear range, 12–320 μm) and reported as the mean of all adipocytes >22.5 μm. Subdistributions of FCS (small, medium, large and very large) were determined using a previously published statistical method for each subjects FCS dataset (n≈2000 cells).23
Herbal ephedra/caffeine treatment
Subjects were randomized to receive a pill containing ephedra (20 mg standardized extract)/caffeine (200 mg; E+C) or a matching placebo in a 3:1 ratio. Subjects were dosed once daily for 1 week, twice daily for the next week and increased to three times daily thereafter. Treatment was planned for a total of 6 months; however, all herbal E+C products were removed from the market by the FDA in the middle of the study, therefore leaving complete 8-week data for only 72 subjects.
Statistical analysis
All microarray and clinical data were analyzed in SAS v8.2. (SAS, Cary, NC, USA; the flow chart of analysis in Supplementary Figure 1). Genes with saturated signals and those less than three times the standard deviation (s.d.) of the local background were filtered out. The data were then log transformed as previously described and normalized using the quantile normalization method as discussed by Bolstad et al.24 The quantile normalizations were applied to slides between subjects among same tissue samples. `Gene shaving' was used as previously described21,25 to remove genes that do not contribute to interpatient variance. A clustering algorithm (Wards method) was then applied to the remaining genes to cluster subjects based on their gene expression profile. Power curves for the goodness-of-fit of the clustering were constructed by the statistics of the R-squared. Clusters (subjects) were visualized in JMP 5.0.1 (SAS) as heat maps (Figure 1) where the variations in the expression of genes across patients are shown in red (downregulated) or green (upregulated) for each gene and each subject. Principal component analysis was used to choose the genes playing a substantial role in subgrouping the subjects. Using PANTHER,26 we identified pathways in which the 448 transcripts of interest were involved (Supplementary Table 2). The ratios were determined as the ratio of skeletal muscle gene expression to adipose tissue gene expression (Supplementary Table 3). Weight changes, BMI changes, adjusted EE, RQ and other clinical data in the whole population and different clusters were analyzed by ANOVA. Results are expressed as means±s.d. Differences were assumed as significant if P<0.05.
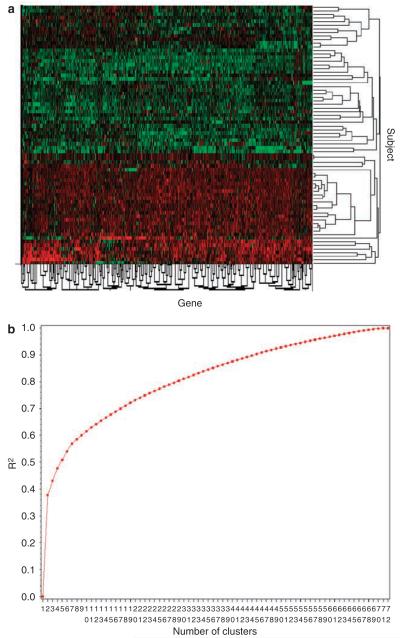
Figure 1.
Cluster analysis of gene expression data reveals two distinct subgroups of obesity. Microarrays were used to measure gene expression in both adipose tissue and skeletal muscle in a population of 72 healthy obese patients. On the basis of the ratio of adipose to skeletal muscle tissue gene expression, the subjects were clustered into two distinct subgroups (see text for details), which are represented as a heat map (Figure 1a). Heat maps depict the variations in the expression of genes across patients and are shown in red (downregulated) or green (upregulated) for each gene and each subject. The decision to limit the clusters to two categories was based on the power curve (Figure 1b), which shows a trivial increase in power as the number of clusters increases. Power was determined as the goodness of fit, R2, for each iteration of the clustering; that is, the amount of variance explained with the addition of subsequent principal components.
Statement of ethics
We certify that all applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research. The study was approved by the Pennington Biomedical Ethics Committee.
Results
Overall clinical characteristics at baseline
The baseline clinical characteristics of the study population are shown in Table 1. A total of 8 men and 64 women participated in the study. The average BMI was 30.3± 2.8 kg/m2 and body fat percentage averaged 37.4±5.6%. FCS averaged 0.77±0.21 μl and fasting glucose averaged 96.4±8.1 mg per 100 ml. EE, adjusted by LBM averaged 1492 kcal kg1 LBM per day. E+C increased oxygen consumption an average of 8.1±8.5%.
Table 1.
Clinical characteristics at baseline
| Variable | Means ± s.d. | Min | Max |
|---|---|---|---|
| N (male/female) | 72 (8/64) | N/A | N/A |
| Weight (kg) | 81.91 ± 10.24 | 61.20 | 114.60 |
| BMI (kg/m2) | 30.25 ± 2.75 | 25.06 | 35.36 |
| SBP (mmHg) | 118.21 ± 10.07 | 100.00 | 147.00 |
| DBP (mmHg) | 77.80 ± 6.67 | 62.00 | 90.00 |
| HR (bpm) | 69.29 ± 6.79 | 48.00 | 90.00 |
| VAT mass (kg) | 3.15 ± 1.37 | 0.96 | 8.07 |
| Fat cell size (μl) | 0.77 ± 0.21 | 0.27 | 1.39 |
| Body fat (%) | 37.40 ± 5.58 | 18.62 | 45.25 |
| GLU (mg per 100 ml) | 96.35 ± 8.13 | 80.00 | 125.00 |
| CHO (mg per 100 ml) | 204.38 ± 32.38 | 137.00 | 288.00 |
| TG (mg per 100 ml) | 118.35 ± 70.81 | 19.00 | 338.00 |
| HDL (mg per 100 ml) | 56.61 ± 15.16 | 30.50 | 114.30 |
| LDL (mg per 100 ml) | 124.03 ± 31.99 | 42.80 | 208.60 |
| Delta EEa | 8.12 ± 8.46 | −10.21 | 26.92 |
| Adj EE (kcal per 24 h) | 2.37 ± 157.61 | −384.10 | 357.73 |
| RQ (au) | 0.86 ± 0.05 | 0.77 | 0.98 |
Abbreviations: Adj EE, lean body mass adjusted energy expenditure presented as the residuals from predicted; BMI, body mass index; CHO, cholesterol; DBP, diastolic blood pressure; Delta EE, energy expenditure change; GLU, glucose; HDL, high-density lipoprotein cholesterol; HR, heart rate; LDL, low-density lipoprotein cholesterol; RQ, respiratory quotient; SBP, systolic blood pressure; TG, triglycerides; VAT, visceral adipose tissue. N = 72 for the whole population.
Only subjects treated with ephedra and caffeine (E+C).
Cluster analysis of gene expression data reveals two distinct subgroups of obesity
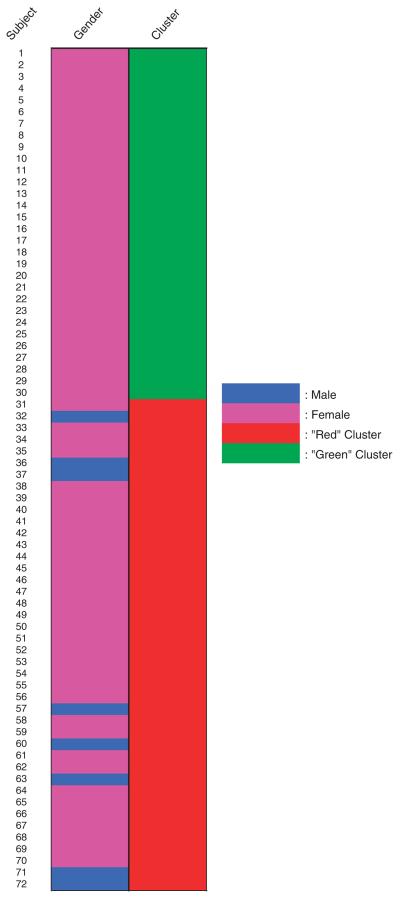
In an effort to subgroup the obese subjects based on their molecular fingerprint, we measured gene expression at baseline within skeletal muscle and adipose tissue by using microarrays. Based on the ratio of skeletal muscle (muscle) to adipose (fat) tissue gene expression and its power curve, the subjects were clustered into two distinct subgroups, which are represented as a heat map in Figure 1a (also listed in Supplementary Table 3). The decision to limit the clusters to two categories was based on the power curve analysis (Figure 1b), which showed a trivial increase in power according to the R-squared value (the amount of variance explained with the addition of subsequent principal components) as the number of clusters increased. Subjects with similar gene expression profiles grouped together and are shown in the cluster tree (on the right (y) axis) and are then separated by the genes (along the left (x) axis). To clearly define the two subgroups, one was termed `green' and graphed in green and the other termed `red' and graphed in red (Figure 1a). This point is further illustrated graphically in Figure 2, where the subgroup identity is mapped for all subjects. Strikingly, the heat map reveals a segregation of gender between the two clusters in which no men are present in the `red' cluster.
Figure 2.
Subgroups of subjects.
Clinical baseline characteristics in different groups
To explore the possible clinical factors that differed in the distinct subgroups of obesity, we further analyzed the clinical baseline data across clusters (Table 2). VAT mass is higher in the `green' cluster than in the `red' cluster (P<0.05). Systolic blood pressure and diastolic blood pressure are both higher in the `green' cluster compared to those in the `red' cluster (P<0.05). There is no significant difference for average FCS, adjusted EE, oxygen consumption, serum lipids or cholesterol between clusters; however, there was a significant difference in very large (VL) FCS between clusters (P<0.05; Table 2).
Table 2.
Clinical data in different clusters
| Variable | Subject cluster red |
Subject cluster green |
P value | ||||
|---|---|---|---|---|---|---|---|
| Mean ± s.d. | Min | Max | Mean ± s.d. | Min | Max | ||
| N | 30 | N/A | N/A | 42 | N/A | N/A | N/A |
| Body weight (kg) | 80.40 ± 8.90 | 65.40 | 102.80 | 83.04 ± 11.10 | 61.20 | 114.60 | 0.28 |
| BMI (kg/m2) | 30.23 ± 3.00 | 25.67 | 35.30 | 30.26 ± 2.58 | 25.06 | 35.36 | 0.97 |
| SBP (mmHg) | 115.03 ± 8.34 | 100.00 | 130.00 | 120.39 ± 10.63 | 100.00 | 147.00 | 0.02 |
| DBP (mmHg) | 75.80 ± 5.88 | 62.00 | 86.00 | 79.22 ± 6.83 | 62.00 | 90.00 | 0.03 |
| HR (bpm) | 69.00 ± 8.48 | 48.00 | 90.00 | 69.41 ± 5.24 | 60.00 | 84.00 | 0.81 |
| VAT mass (kg) | 2.77 ± 1.08 | 0.96 | 4.94 | 3.43 ± 1.49 | 1.22 | 8.07 | 0.03 |
| Body fat (%) | 37.74 ± 5.27 | 24.30 | 43.62 | 36.97 ± 5.73 | 18.62 | 45.25 | 0.56 |
| Fat cell size (μl) | 0.76 ± 0.22 | 0.32 | 1.39 | 0.76 ± 0.20 | 0.27 | 1.19 | 0.92 |
| Very large fat cell size (μl) | 1.45 ± 0.61 | 0.51 | 3.42 | 2.16 ± 1.74 | 0.43 | 6.26 | 0.04 |
| GLU (mg per 100 ml) | 94.56 ± 6.76 | 80.00 | 113.00 | 97.65 ± 8.85 | 83.00 | 125.00 | 0.11 |
| CHO (mg per 100 ml) | 198.03 ± 28.74 | 137.00 | 245.00 | 209.02 ± 34.41 | 153.00 | 288.00 | 0.15 |
| TG (mg per 100 ml) | 103.87 ± 61.33 | 19.00 | 229.00 | 128.95 ± 75.99 | 36.00 | 338.00 | 0.14 |
| HDL (mg per 100ml) | 54.83 ± 16.02 | 35.30 | 114.30 | 57.01 ± 14.41 | 30.50 | 93.80 | 0.40 |
| LDL (mg per 100 ml) | 122.27 ± 26.84 | 68.20 | 190.80 | 125.32 ± 35.56 | 42.80 | 208.60 | 0.68 |
| Delta EEa (%) | 10.31 ± 9.01 | −2.42 | 26.92 | 6.45 ± 6.89 | −9.96 | 17.96 | 0.09 |
| Adj EE (kcal per 24 h) | −3.47 ± 175.51 | −374.30 | 357.73 | 14.25 ± 143.90 | −384.10 | 288.56 | 0.47 |
| RQ (au) | 0.86 ± 0.04 | 0.77 | 0.95 | 0.85 ± 0.04 | 0.78 | 0.98 | 0.61 |
Abbreviations: Adj EE, lean body mass adjusted energy expenditure; BMI, body mass index; CHO, cholesterol; DBP, diastolic blood pressure; Delta EE, energy expenditure change with ephedra and caffeine treatment; GLU, glucose; HDL, high-density lipoprotein cholesterol; HR, heart rate; LDL, low-density lipoprotein cholesterol; RQ, respiratory quotient; SBP, systolic blood pressure; TG, triglycerides; VAT mass, weight of visceral adipose tissue mass. n = 42 for green cluster and 30 for red cluster.
Only subjects treated with ephedra and caffeine (E+C).
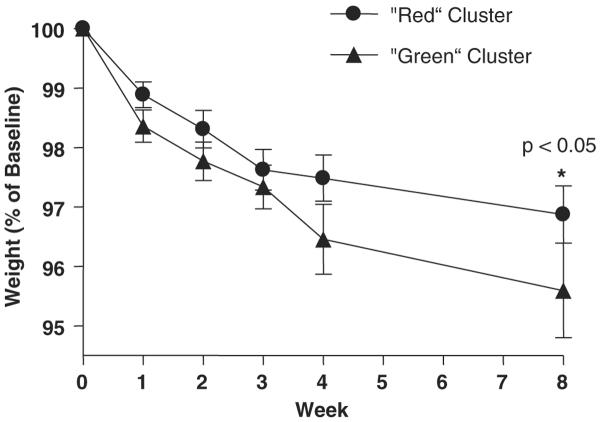
Cluster identity predicts weight loss after treatment with ephedra and caffeine
To test the hypothesis that subgrouping obese subjects with microarrays would have clinical significance, we treated 52 subjects with ephedra and caffeine and measured the weight loss over 8 weeks (Table 3). Beginning at week 4, subjects in the `green' cluster lost more weight than subjects in the `red' cluster and this difference continued to the time point of week 8 (Table 3; Figure 3; P<0.05 at week 8). In the `cluster analysis', 2497 genes were used.
Table 3.
Weight loss for ephedra + caffeine treated subjects
| Variable | `Red' cluster (N = 26) | `Green' cluster (N = 26) | P value |
|---|---|---|---|
| Gender (female/male) | 26/0 | 20/6 | N/A |
| %Weight at baseline | 100 | 100 | N/A |
| %Weight at 1 week | 98.88 ± 1.10 | 98.35 ± 1.32 | 0.22 |
| %Weight at 2 week | 98.30 ± 1.57 | 97.76 ± 1.55 | 0.22 |
| %Weight at 3 week | 97.62 ± 1.58 | 97.33 ± 1.59 | 0.27 |
| %Weight at 4 week | 97.48 ± 1.93 | 96.45 ± 2.19 | 0.06 |
| %Weight at 8 week | 96.87 ± 2.35 | 95.59 ± 2.75 | 0.03 |
A total of 52 subjects were treated with ephedra and caffeine (E+C) for 8 weeks. The weight losses are reported as % weight from baseline. %Weight is body weight/baseline. All the P values were adjusted by using Tukey-Kramer method.
Figure 3.
Weight losses by clusters.
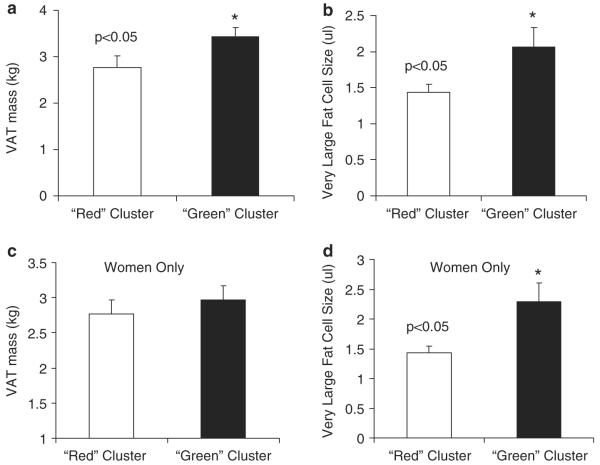
Cluster dimorphism in visceral adipose tissue and very large fat cell size
To further explore the differences in VAT mass and VL FCS between the `green' and the `red' clusters, we examined relationships without the influence of the men in the `green' cluster. There was no difference in VAT mass between clusters once the men were removed; however, the women in the `green' cluster had increased VL FCS compared to the women in the `red' cluster (P<0.05; Figure 4). We also examined other clinical characteristics such as waist and hip circumference and found no significant difference between the women in the two different clusters (Supplementary Table 1).
Figure 4.
Cluster dimorphism in visceral adipose tissue and very large fat cell size. Gene expression was measured in both adipose and skeletal muscle tissue in a population of 72 healthy obese patients using microarrays. On the basis of their gene expression profile, subjects were clustered into two distinct subgroups of either `green' or `red'. All eight men were grouped into the `green' cluster based on their gene expression profile. Visceral adipose tissue (VAT) mass (a) and very large fat cell size (b) were significantly different between the `green' and `red' clusters. These relationships were reanalyzed without the men and VAT mass (c) was no longer different, but very large fat cell size (d) remained significantly different in the `green' and `red' clusters.
To further investigate the influence of clinical characteristics on FCS in these two clusters, we performed a stepwise linear regression analysis for `green' and `red' clusters separately, using the clinical characteristics from the database (body weight, BMI, fasting blood glucose and lipids, total adipose tissue (TAT) mass, superficial subcutaneous adipose tissue mass, deep subcutaneous adipose tissue (DSAT) mass and visceral adipose tissue mass). The three major clinical correlates of FCS in women of the `green' cluster were serum low-density lipoprotein (LDL), TAT mass and body weight explaining 44% of the variance. In the women of the `red' cluster, body weight and DSAT explain 28% of the variance (data not shown).
Thermogenic response to a single dose of ephedra and caffeine
The `red' cluster had a somewhat increased EE after the first dose of ephedra and caffeine, and it approached traditional statistical significance (P = 0.09; Table 2; Figure 1a).
Genes separating subjects into subgroups
To identify the genes that play a substantial role in subgrouping the subjects, we performed a principal component analysis. Based on the eigenvalues of the first principal component, six skeletal muscle genes and six adipose tissue genes were selected as major contributions to the overall variance. KCNMA1, MARCH-VI, TYR, HIST1H4E, CDC7 and PDE8B are skeletal muscle genes that are involved in cation transport (related to muscle contraction), mRNA polyadenylation and other protein metabolism, amino-acid metabolism, chromatin packaging and remodeling, cell-cycle control and signal transduction, respectively. NT5C2, SLC26A3, TNFAIP6, IL1F5, GPRASP1 and LMAN2 are adipose tissue genes that are related to purine metabolism, anion transport, cell adhesion, signaling pathway, cell sorting and protein targeting, respectively (Supplementary Table 1).
Discussion
People become obese from a variety of physiological, environmental, psychological and genetic causes. In an effort to improve our diagnostic and therapeutic acumen in obesity, we subdivided obese subjects into discrete diagnostic categories based on microarray analysis of gene expression in the adipose tissue and skeletal muscle tissue from 72 healthy obese men and women. Using the micro-array gene expression data, we clustered subjects in a process similar to that employed by oncology research scientists to subtype cancerous tumors. Our analyses revealed that adipose tissue and skeletal muscle mRNA expression separated obese subjects into two distinct subgroups. The analysis of the tissues identified a subgroup of obese patients who lost significantly more weight when treated with the adrenergic herbal pill containing ephedra and caffeine (E+C) as compared to the remaining patients. This study provides the first evidence that gene expression profiling can identify subtypes of obesity, that these subtypes have different physiological characteristics and that these sub-types respond differently to an adrenergic obesity therapy.
Interestingly, all of the men in this study clustered into the `green' cluster. A priori, we predicted that the presence of mRNAs from the male Y chromosome would place men into a discrete cluster; however, this was not the case. We speculate that women in the `green' cluster have a more `masculine' gene profile or share a pattern of gene expressions that is similar to that of men in the `green' cluster. This interpretation must be investigated further using a candidate gene approach. Clinically, men rarely display a glutealfemoral or `pear shape' pattern of fat, whereas women exhibit a wide variation in fat patterning. Adipose tissue distribution is regulated by both genetic and environmental factors,27 and the differences among these factors between men and women may explain the segregation by gender we observed in our data. Gender differences include a larger subcutaneous adipose tissue in women than men, explainable at least partly by a depot in the gluteal-femoral region in women, which is essentially absent in nonobese men.17 Men, on the other hand, seem to have a larger proportion of their adipose tissue organ localized intra-abdominally.28 In addition, the gluteal-femoral fat cells are specifically enlarged in women and have a higher lipoprotein lipase activity. In premenopausal women, subcutaneous abdominal adipose tissue has a higher lipid turnover than femoral adipose tissue. Results of studies in vitro indicate that this difference is diminished at the menopause, and restored by estrogen substitution, suggesting that the functional effects of estrogens in women are similar to those of testosterone in men.29
VAT mass has been found to have a strong relationship with insulin resistance and type 2 diabetes.30 Our study showed that subjects with greater VAT mass in the `green' cluster lost more weight than those with less VAT mass in the `red' cluster when treated with ephedra and caffeine (E+C). VAT mass is responsive to weight reduction because the VAT adipocyte is more metabolically active and sensitive to β-adrenergic stimulated lipolysis.9 Two major differences between the `green' and the `red' clusters were VAT mass and VL FCS. Our results indicate that the `green' cluster had more VAT mass and greater VL fat cells compared to the `red' cluster. However, after excluding all men from the `green' cluster, women in that cluster had no difference in VAT mass, but did have larger fat cells, compared to the women in the `red' cluster. Further analysis showed that in addition to a gender dimorphism in FCS, there is also a dimorphism among women in different clusters for the determinants of FCS and suggested that women having greater serum LDL level, TAT and body weight, also have larger fat cells. Historically, it has been shown that the long-term prognosis for weight reduction is worse for those patients with higher fat cell number compared to those patients with larger FCS.31
Our study showed that of the six skeletal muscle genes that contributed to the overall variance in both clusters, three have been connected to the cAMP/PKA pathway (TYR, CDC7 and PDE8B), and others are involved in muscle contraction (KCNMA1) and protein metabolism (MARCH-VI), respectively. Some of the adipose tissue genes are adipocytokines (TNFAIP6, IL1F5), others are involved in extracellular transport (solute carrier family 26, member 3), purine metabolism (NT5C2) and anion transport (SLC26A3), respectively.
This investigation does not imply that there are only two subcategories of obesity. More categories may be uncovered as transcriptome analysis/microarray technology improves (splice-chips, increasing number of genes, and so on), or as additional technologies are brought online that characterize the genome, proteome or lipidome within tissues and cells. Furthermore, the changes that follow the different patterns of gene expression (vis-à-vis changes in protein content and function) will manifest themselves at the structural level of the cells and tissues, which will provide additional opportunities. A combined approach of examining tissues from multiple perspectives simultaneously, for example, may reveal additional subtypes of obesity or strengthen the diagnostic utility of the current subtype definition.
Taken together, the oncology microarray literature demonstrates that knowing the exact genes that discriminate these subtypes is not necessary for the purposes of diagnosis, prognosis of disease risk or treatment planning.13 However, there are several reasons it is important to discover the identity of the genes that discriminate between subtypes. First, the list may provide clues as to why obese patients are different and why some patients have abdominal obesity and others have a gluteal-femoral pattern. Does one subtype have a defect in food intake control and the other in peripheral fat metabolism? Further exploration of these questions should include consideration of the results presented herein. Second, there may be single genes that discriminate between these two predominant `flavors' of obesity. Mini-aspirates of adipose tissue, like routine phlebotomy, are minimally invasive and provide enough material to measure a handful of genes. If the proper genes are chosen, this candidate gene approach could provide a rapid classification of a patient and assist in the choice of an optimal obesity therapy. Clearly more work is needed to answer these questions; however, this investigation provides support for the notion that this approach might yield insights into the diagnosis and/or pharmacogenomics of obesity. Other tissues (for example, liver and hypothalamus) probably contribute in terms of different subtypes of obesity and different skeletal muscle and adipose tissue depots might also respond or contribute differently. The orthogonal question `does the adipose tissue gene expression at baseline influence the response to treatment?' is interesting and merits investigation; however, this investigation was limited to the question of whether the baseline characteristics of the adipose tissue and muscle influence the response to a weight loss treatment. It is logical to consider that the identified `subtypes' might determine the tissue response to a treatment. This concept is being explored in subsequent clinical studies.
In summary, this study provides the first evidence that the combined approach of gene expression profiling plus cluster analysis can identify discrete subtypes of obesity, these subtypes have different physiological characteristics and these subtypes respond differently to an adrenergic therapy in vivo. This opens the door to the development of new clinical diagnostic tools that may lead us to discard the current onesize-fits-all advice given to obese patients in the clinic and enter an era of personalized treatment in the obesity clinic.
Supplementary Material
Acknowledgements
This work was supported by Community Foundation of Southeast Michigan and the PBRC CNRU NIH P30-DK072476. We thank David Hymel for the RNA extractions. We are grateful to the volunteers who participated in this study and the staff of the clinic at Pennington Biomedical Research Center for their assistance.
Footnotes
Supplementary Information accompanies the paper on International Journal of Obesity website (http://www.nature.com/ijo)
References
- 1.White MA, Whisenhunt BL, Williamson DA, Greenway FL, Netemeyer RG. Development and validation of the food-craving inventory. Obes Res. 2002;10:107–114. doi: 10.1038/oby.2002.17. [DOI] [PubMed] [Google Scholar]
- 2.Stunkard A. Two eating disorders: binge eating disorder and the night eating syndrome. Appetite. 2000;34:333–334. doi: 10.1006/appe.1999.0337. [DOI] [PubMed] [Google Scholar]
- 3.Levine JA, Eberhardt NL, Jensen MD. Role of nonexercise activity thermogenesis in resistance to fat gain in humans (see comments) Science. 1999;283:212–214. doi: 10.1126/science.283.5399.212. [DOI] [PubMed] [Google Scholar]
- 4.Ukropcova B, McNeil M, Sereda O, de Jonge L, Xie H, Bray GA, et al. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J Clin Invest. 2005;115:1934–1941. doi: 10.1172/JCI24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guy-Grand B, Apfelbaum M, Crepaldi G, Gries A, Lefebvre P, Turner P. International trial of long-term dexfenfluramine in obesity. Lancet. 1989;2:1142–1145. doi: 10.1016/s0140-6736(89)91499-2. [DOI] [PubMed] [Google Scholar]
- 6.Lefevre M, Greenway FL, Smith SR, Ryan DH, Bray GA, Morales S, et al. Genetic Mutations That Predict Weight Loss In An Obesity Treatment Program. Int J Obes. 1998;22(Suppl 3):P61. [Google Scholar]
- 7.Torgerson JS, Carlsson B, Stenlof K, Carlsson LM, Bringman E, Sjostrom L. A low serum leptin level at baseline and a large early decline in leptin predict a large 1-year weight reduction in energy-restricted obese humans. J Clin Endocrinol Metab. 1999;84:4197–4203. doi: 10.1210/jcem.84.11.6089. [DOI] [PubMed] [Google Scholar]
- 8.Verdich C, Toubro S, Buemann B, Holst JJ, Bulow J, Simonsen L, et al. Leptin levels are associated with fat oxidation and dietary-induced weight loss in obesity. Obes Res. 2001;9:452–461. doi: 10.1038/oby.2001.59. [DOI] [PubMed] [Google Scholar]
- 9.Smith SR, Zachwieja JJ. Visceral adipose tissue: a critical review of intervention strategies. Int J Obes Relat Metab Disord. 1999;23:329–335. doi: 10.1038/sj.ijo.0800834. [DOI] [PubMed] [Google Scholar]
- 10.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Alizadeh AA, Ross DT, Perou CM, van de Rijn M. Towards a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195:41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 14.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 16.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- 17.Smith SR, Lovejoy JC, Greenway F, Ryan D, Dejonge L, De La Bretonne J, et al. Contributions of total body fat, abdominal subcutaneous adipose tissue compartments, and visceral adipose tissue to the metabolic complications of obesity. Metabolism. 2001;50:425–435. doi: 10.1053/meta.2001.21693. [DOI] [PubMed] [Google Scholar]
- 18.de Jonge L, Agoues I, Garrel DR. Decreased thermogenic response to food with intragastric vs. oral feeding. Am J Physiol. 1991;260:E238–E242. doi: 10.1152/ajpendo.1991.260.2.E238. [DOI] [PubMed] [Google Scholar]
- 19.Elia M, Livesey G. Theory and validity of indirect calorimetry during net lipid synthesis. Am J Clin Nutr. 1988;47:591–607. doi: 10.1093/ajcn/47.4.591. [DOI] [PubMed] [Google Scholar]
- 20.Smith SR, Gawronska-Kozak B, Janderova L, Nguyen T, Murrell A, Stephens JM, et al. Agouti expression in human adipose tissue: functional consequences and increased expression in type 2 diabetes. Diabetes. 2003;52:2914–2922. doi: 10.2337/diabetes.52.12.2914. [DOI] [PubMed] [Google Scholar]
- 21.Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 22.Harris RB, Ramsay TG, Smith SR, Bruch RC. Early and late stimulation of ob mRNA expression in meal-fed and overfed rats. J Clin Invest. 1996;97:2020–2026. doi: 10.1172/JCI118637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith SR, Xie H, Baghian S, Needham A, McNeil M, Bogacka I, et al. Pioglitazone chnages the distribution of adipocyte size in type 2 diabetics. Adipocytes. 2006;2:11–22. [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Hastie T, Tibshirani R, Eisen MB, Alizadeh A, Levy R, Staudt L, et al. `Gene shaving' as a method for identifying distinct sets of genes with similar expression patterns. Genome Biol. 2000;1(2) doi: 10.1186/gb-2000-1-2-research0003. RESEARCH0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjorntorp P. Adipose tissue distribution and function. Int J Obes. 1991;15(Suppl 2):67–81. [PubMed] [Google Scholar]
- 28.Vague J. Sexual differentiation. A determinant factor of the forms of obesity. Obes Res. 1996;4:201–203. doi: 10.1002/j.1550-8528.1996.tb00535.x. 1947 (classical article) (see comments) [DOI] [PubMed] [Google Scholar]
- 29.Rebuffe-Scrive M, Lonnroth P, Marin P, Wesslau C, Bjorntorp P, Smith U. Regional adipose tissue metabolism in men and postmenopausal women. Int J Obes. 1987;11:347–355. [PubMed] [Google Scholar]
- 30.Gastaldelli A, Sironi AM, Ciociaro D, Positano V, Buzzigoli E, Giannessi D, et al. Visceral fat and beta cell function in non-diabetic humans. Diabetologia. 2005;48:2090–2096. doi: 10.1007/s00125-005-1891-3. [DOI] [PubMed] [Google Scholar]
- 31.Krotkiewski M, Sjostrom L, Bjorntorp P, Carlgren G, Garellick G, Smith U. Adipose tissue cellularity in relation to prognosis for weight reduction. Int J Obes. 1977;1:395–416. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.