Abstract
Protein glycosylation involves the addition of monosaccharides in a stepwise process requiring no glycan template. Therefore, identifying the numerous glycoforms, including isomers, can help elucidate the biological function(s) of particular glycans. A method to assess the diversity of the N-linked oligosaccharides released from human serum without derivatization has been developed using on-line nano-liquid chromatography (nanoLC) and high resolution time-of-flight mass spectrometry. The N-linked oligosaccharides were analyzed with matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry (MALDI FT-ICR MS) and microchip liquid chromatography mass spectrometry (HPLC-Chip/TOF MS). Two microfluidic chips were employed, the glycan chip (40 nL enrichment column, 43×0.075 mm ID analytical column) and the high capacity chip (160 nL enrichment column, 140×0.075 mm ID analytical column), both with graphitized carbon as the stationary phase. Both chips offered good sensitivity and reproducibility in separating a heterogeneous mixture of neutral and anionic oligosaccharides between injections. Increasing the length and volume of the enrichment and the analytical columns improved resolution of the peaks. Complex type N-linked oligosaccharides were the most abundant oligosaccharides in human serum accounting for ~96% of the total glycans identified, while hybrid and high mannose type oligosaccharides comprise the remaining ~4%.
Keywords: FT-ICR MS, oligosaccharides, human serum, HPLC/Chip-TOF MS, N-linked
1 INTRODUCTION
The addition of carbohydrates to proteins is an important post-translational modification given the roles of glycoproteins in many biological processes including immune defense, cell growth, and cell-cell adhesion[1, 2]. In addition to providing vital functionality to proteins, glycans also play a role in protein stability[3, 4]. When present in biological fluids, distinct glycan structures may provide information to specific pathologic states in diseases involving cancer and inflammation[5]. Glycans are often branched molecules with differing linkages between the monosaccharide residues leading to a large diversity in structures[1, 6]. The anomeric character of each linkage and the propensity to branch make oligosaccharides ideally suited for containing a broad set of biological information.
Two main types of oligosaccharides are attached to proteins, N-linked and O-linked. O-linked glycans (or O-glycans) are attached to the hydroxyl group of either serine (Ser) or threonine (Thr). N-linked glycans (or N-glycans) are attached to the amide group of asparagine (Asn) having a consensus sequence of Asn-Xxx-Ser/Thr and a less common one, Asn-Xxx-Cys (cysteine), where Xxx can be any amino acid with the exception of proline[2, 7, 8]. These oligosaccharides have a basic core comprised of three mannoses (Man) and a chitibiose or two N-acetylglucosamines (GlcNAc). Monosaccharides are subsequently attached stepwise through cellular pathways involving highly conserved enzymes[5, 6]. The N-linked glycans are mainly categorized as high mannose, complex, and hybrid type oligosaccharides, comprising approximately 90% of the glycans present in eukaryotic cells[2, 4]. High mannose types are defined by the attachment of mannose residues to the pentasaccharide core, while complex types have no mannose residues aside from those present in the core but are branched (up to five antennae) with additions of N-acetylglucosamine residues to the mannoses. Lastly, hybrid types are a combination of high mannose and complex type oligosaccharides. Addition of a bisecting GlcNAc to complex types and N-acetylneuraminic acid (sialic acid, NeuAc) and fucose (Fuc) to complex and hybrid types leads to more structural diversity within each subgroup[1].
A recent report by Packer et al. thoroughly discusses the advantages of glycomics for disease discovery[9]. Thus, glycan profiles can provide information to diagnose diseases and disorders such as congenital disorders of glycosylation[7, 10, 11], cirrhosis[12], and cancer[13]. More recently, glycan profiles from human serum using mass spectrometry (MS) were studied for breast[14], ovarian[15–17], pancreatic[18], and prostate[19] cancer diagnosis demonstrating the potential of glycomic profiling for early disease diagnosis. MS analysis of glycan mixtures released from glycoproteins by enzymatic or chemical means provides rapid and sensitive composition determination based on accurate mass measurements, although many possible structures may correspond to a given composition. Due to the structural diversity of glycans, isomers differing in linkage and connectivity must be separated and profiled to identify specific candidates for diseases. For these reasons, techniques are being developed to analyze oligosaccharide mixtures using high performance methods to separate isomers[20, 21].
Analytical size columns (2.1 to 4.6 mm diameter) traditionally used for HPLC separation of native oligosaccharides include porous graphitized carbon (PGC), amine/amide-based, and anion-exchange media as stationary phases[22–27]. Derivatization of oligosaccharides with 2-aminobenzamide (2-AB) is often employed with standard reverse-phase columns[7, 28]. Butler et al. profiled N-linked glycans labeled with 2-AB from human serum glycoproteins separating them on an amide column using normal-phase HPLC[7]. Alternatively, PGC columns have been used to determine glycan profiles from HPLC for a number of standard glycoproteins including erythropoietin[29–31], parotid gland[28], fibrin(ogens)[32], and human serum[7, 28, 33]. PGC columns have the advantages of separating a mixture of neutral and sialylated oligosaccharides in a single run, being stable over a large pH range, and having low column variability[23]. Kim et al. recently profiled N-glycans and proteins from human serum simultaneously using parallel capillary columns packed with PGC and C18 with electrospray mass spectrometry[33].
Nanoflow liquid chromatography (nanoLC) is emerging as a valuable technique offering high sensitivity, shorter analysis time, high resolution, and effective separation[21]. The ability of nanoLC to separate linkage and other structural isomers makes it a valuable technique for oligosaccharide profiling, especially when handling small amounts of sample[26, 34, 35]. Integrating mass spectrometric detection with nanoLC improves sensitivity and provides an ideal platform for structure determination[21]. NanoLC had been performed primarily with reversed-phase and normal-phase columns that made it difficult to analyze simultaneously both sialylated and neutral oligosaccharides without permethylation or desialylation[36, 37]. Recent studies from this laboratory have shown that oligosaccharides from a pooled human milk sample are readily separated using nanoLC yielding more than 200 neutral and anionic species in a single analysis while using PGC as the stationary phase[34, 35]. Employing a high performance mass analyzer yielded not only highly reproducible retention times but also high mass accuracy (1–6 ppm mass error).
Given the potential of N-linked oligosaccharides as markers for diseases, it is necessary to fully characterize the extent and the heterogeneity of the N-linked glycome. In this report, we profile the N-linked glycome in human serum using nanoLC coupled to a high mass accuracy analyzer. The combination provides high repeatability in both retention times and mass assignments. The relative quantitation of the three major sub-classes of N-linked oligosaccharides was readily obtained. The oligosaccharides were separated on two different microfluidic chips with varying length and capacity both with graphitized carbon as the stationary phase (a glycan and high capacity chip). The HPLC-Chip was interfaced with an orthogonal time-of-flight mass spectrometer (TOF-MS). The ability to separate and simultaneously analyze neutral and anionic N-linked oligosaccharides from human serum without derivatization in a single analysis demonstrates a rapid, yet highly sensitive tool with potential for clinical applications.
2 MATERIALS AND METHODS
2.1 Human serum samples
Commercial human serum was purchased from Sigma-Aldrich (St. Louis, MO). Volunteers from the UC Davis Medical Center, Davis, California, generously provided donor samples.
2.2 Release of oligosaccharides from serum by N-glycosidase F digestion
N-linked oligosaccharides were released from human sera using N-glycosidase F, PNGase F, (EMD Biosciences, San Diego, CA). For each 50 µL of human serum; 50 µL of 0.2 M ammonium bicarbonate and 2 µL PNGase F (specific activity ≥ 10 units/mg protein) were added. The samples were then placed on a rotating carousel in a 37 °C oven for 24 hours. To the mixture, ethanol was added to a total volume of 90% ethanol and the mixture was placed into a −20 °C freezer overnight. The mixture was then centrifuged and the ethanol supernatant was removed and dried in vacuum. The sample was then reconstituted with 1 mL of nanopure water and was purified using solid-phase extraction.
2.3 Reduction of released oligosaccharides
After the N-linked oligosaccharides were released, the N-glycans were reconstituted with 100 µL of nanopure water. To the samples 100 µL of 2.0 M sodium borohydride was added and incubated at 65 °C for 2 hours. Samples were then purified by solid phase extraction.
2.4 Oligosaccharide purification using solid-phase extraction
Oligosaccharides released by PNGase F were purified using graphitized carbon cartridges, GCC (Alltech Associated Inc., Deerfield, IL). Each cartridge was conditioned with 4 mL nanopure water followed by 4 mL 0.1% trifluoroacetic acid (TFA) in 80% acetonitrile in water, (v/v) [22, 23, 27]. The oligosaccharide solution was loaded onto each cartridge and washed with 12 mL of nanopure water at a flow rate of approximately 1mL/min to remove excess salts. The oligosaccharides were eluted in the following order; 10% acetonitrile in water (v/v), 20% acetonitrile in water (v/v), and 0.05% TFA in 40% acetonitrile in water (v/v). Each eluted fraction was collected and dried in vacuum using a Centrivap Concentrator (Labconco Corp, Kansas City, MO) prior to mass spectral analysis.
2.5 High performance liquid chromatography (HPLC) - Chip/time-of-flight (TOF)-MS analysis
Oligosaccharide fractions were analyzed using an Agilent 6200 Series HPLC-Chip/TOFMS system equipped with a microwell-plate autosampler (maintained at 20.0 °C), capillary sample loading pump, nanopump, HPLC-Chip interface, and the Agilent 6210 TOF LC/MS. The HPLC-Chip (Glycan Chip) consisted of a 40 nL enrichment column and a 43×0.075 mm ID analytical column, both with graphitized carbon (5 µm) as stationary phase. For comparison, a high capacity HPLC-Chip was used and consisted of a 160 nL enrichment column and a 150×0.075 mm ID analytical column, both packed with graphitized carbon (5 µm) as the stationary phase. For sample loading, the capillary pump delivered 0.1% formic acid in 3.0% acetonitrile in water (v/v) isocratically at 4 µL/min. Injection volume was 0.2 µL for each sample. A nanoliter pump gradient was delivered at 0.3 µL/min using A) 0.1% formic acid in 3.0% acetonitrile in water (v/v) and B) 0.1% formic acid in 90% acetonitrile in water (v/v). A 45.0 minute nanoLC gradient was run from 0%–16% B, 2.5–20.0 minutes; 16%–44% B, 20.0–30.0 minutes; 44%–100% B, 30.0–35.0 minutes and held for 10.0 minutes at 100% B with an equilibration time of 20.0 minutes at 0% B. The drying gas temperature was set at 325 °C with a flow of 4.0 L/min (2.0 L nitrogen and 2.0 L dry grade compressed air). Data was acquired in the positive ionization mode within a mass range of m/z 300–3000. Mass correction was enabled using reference masses of m/z 519.139 and m/z 1221.991 as internal standards (ESI-TOF Tuning Mix G1969–85000, Agilent Technologies, Inc., Santa Clara, CA). Data analysis was performed using Analyst QS 1.1 software and the deconvoluted lists of masses were generated using Agilent Mass Hunter (Molecular Feature Extraction) software. The retention time reproducibility of the respective columns was assessed after alignment using the algorithm developed by Wong et al.[38]. Oligosaccharides were identified using a Glycan Finder software program (written in-house) in Igor Pro version 5.04B (WaveMetrics, Inc.). The algorithm was designed to examine a list of experimentally measured masses and searched for all possible monosaccharide combinations matching the experimental mass within a specified tolerance level (mass error). In addition to providing information regarding the possible monosaccharide composition, the program output sorts each measured mass based upon its retention time and relative intensity.
2.6 Fourier transform ion cyclotron resonance (FTICR) MS analysis
Oligosaccharide fractions were initially analyzed on a ProMALDI-FT-ICR-MS (IonSpec, Lake Forest, CA) system equipped with hexapole accumulation cell, 355 nm pulsed Nd:YAG laser, and 7.0 Tesla magnet. The sample was prepared by mixing analyte, 1 µM NaCl, and 0.3 M 2,5-dihydroxybenzoic acid in 50% acetonitrile in water (v/v) (Sigma Aldrich, St. Louis, MO) in a 1:1:1 ratio on a 100 well MALDI sample plate. The mixture was then dried similar to the fast evaporation method. Tandem mass spectrometry was performed using sustained off resonance irradiation (SORI) collision induced dissociation (CID). The precursor ion was isolated and excited 1000 Hz of their cyclotron frequency at SORI amplitude of 2.55 V. Nitrogen gas was pulsed in to maintain a pressure of 10−6 Torr.
2.7 N-linked Oligosaccharide Data Analysis
The output obtained from IGOR was further refined through a “biological filter” adapted from Cooper et al.[39]. The filter sorts compositions based upon the quantities of each monosaccharide present for a composition and classifies the oligosaccharide as neutral complex, anionic complex, and hybrid/high-mannose type. For instance, to categorize hybrid and high mannose oligosaccharides the presence of deoxyhexose (fucose) has been omitted because fucosylation of high mannose and hybrid types are less evident in humans[40]. Hybrid and high mannose differentiation was based upon knowledge that high mannose glycans are comprised of Man5–9GlcNAc2 and the remaining oligosaccharides would be classified as hybrid. The criteria used for each oligosaccharide category is outlined in Table 1.
Table 1.
Criteria for N-linked oligosaccharide composition filter
| Hex | HexNAc | dHex | NeuAc | |
|---|---|---|---|---|
| Hybrid/High Mannose | ≥ 3 | ≥ 2 | = 0 | = 0 |
| Complex (Neutral) | ≥ 3 | ≥ 2 | ≤ HexNAc | = 0 |
| Complex (Anionic) | ≥ HexNAc | ≥ 3 | ≤ HexNAc | ≤ 0 |
3 RESULTS AND DISCUSSION
3.1 Mass profile of N-linked glycans from human serum
Oligosaccharides were obtained from serum through the procedure described above. Before the liquid chromatography (LC) analyses, mass profiles of the samples were obtained using MALDI FT-ICR MS. The results provided confirmation of the presence of oligosaccharides with an independent method that is complementary to the HPLC/Chip TOF. Tandem mass spectrometry was also performed using MALDI FT-ICR MS to confirm the compositions of several of the more abundant species. For glycomic analysis, mass-profile procedures are often employed, however suppression effects complicate the spectra. More abundant glycans suppress the signals of less abundant glycans among neutral oligosaccharides, moreover neutral oligosaccharides often suppress the signals of anionic oligosaccharides, particularly those containing sialic acids, in the positive ion mode [41]. However the loss of sialic acid(s) and fucose can also occur, as previously demonstrated by our laboratory [42].
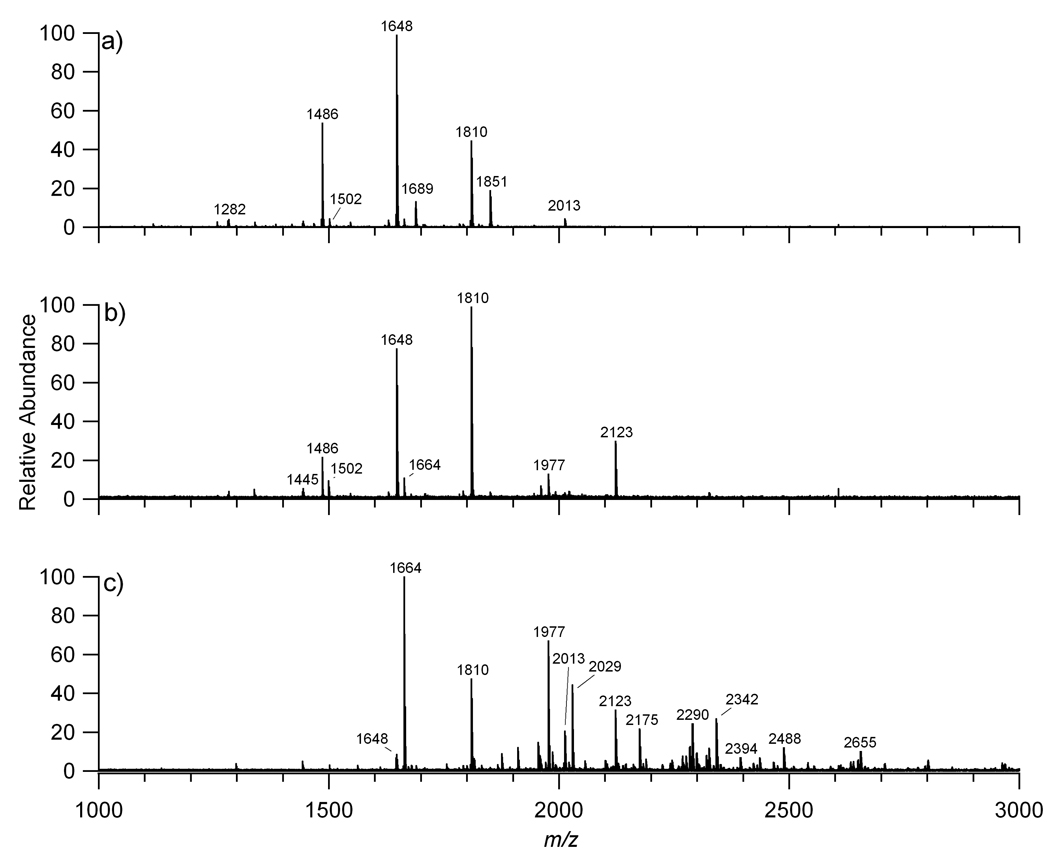
Prefractionation is necessary to mitigate some of the suppression effects particularly those between neutral and anionic oligosaccharides. Three fractions (10%, 20%, and 40% of acetonitrile in water) were used to elute the oligosaccharides from the graphitized carbon cartridges by solid phase extraction (SPE). Figure 1 shows the MALDI FT-ICR MS analysis of the unreduced N-glycans released by PNGase F from commercial serum eluted with the different concentrations of acetonitrile solutions. Figure 1a shows the MALDI FT-ICR MS of the 10% fraction, which contains neutral oligosaccharides. Figure 1b illustrates the MALDI FT-ICR MS mass spectrum of the 20% fraction containing neutral and acidic oligosaccharides. The acidic oligosaccharides had lower abundances than expected due to suppression by abundant neutral oligosaccharide, primarily m/z 1647.595 (1 Deoxyhexose (dHex), 4 Hexoses (Hex), and 4 N-acetylhexosamines (HexNAc)) and m/z 1809.655 (1 dHex, 5 Hex, and 4 HexNAc). Figure 1c shows the MALDI FT-ICR MS mass spectrum of the 40% fraction, which consists mainly of anionic oligosaccharides, however some neutral oligosaccharides are still present such as the abundant ion m/z 1663.599 with composition 5 Hex and 4 HexNAc. The effect of signal suppression will be discussed in further detail below.
Figure 1.
MALDI FT MS of a) the neutral oligosaccharides (10% acetonitrile fraction), b) the neutral and anionic oligosaccharides (20% acetonitrile fraction), and c) the anionic oligosaccharides (40% acetonitrile fraction) released from human serum.
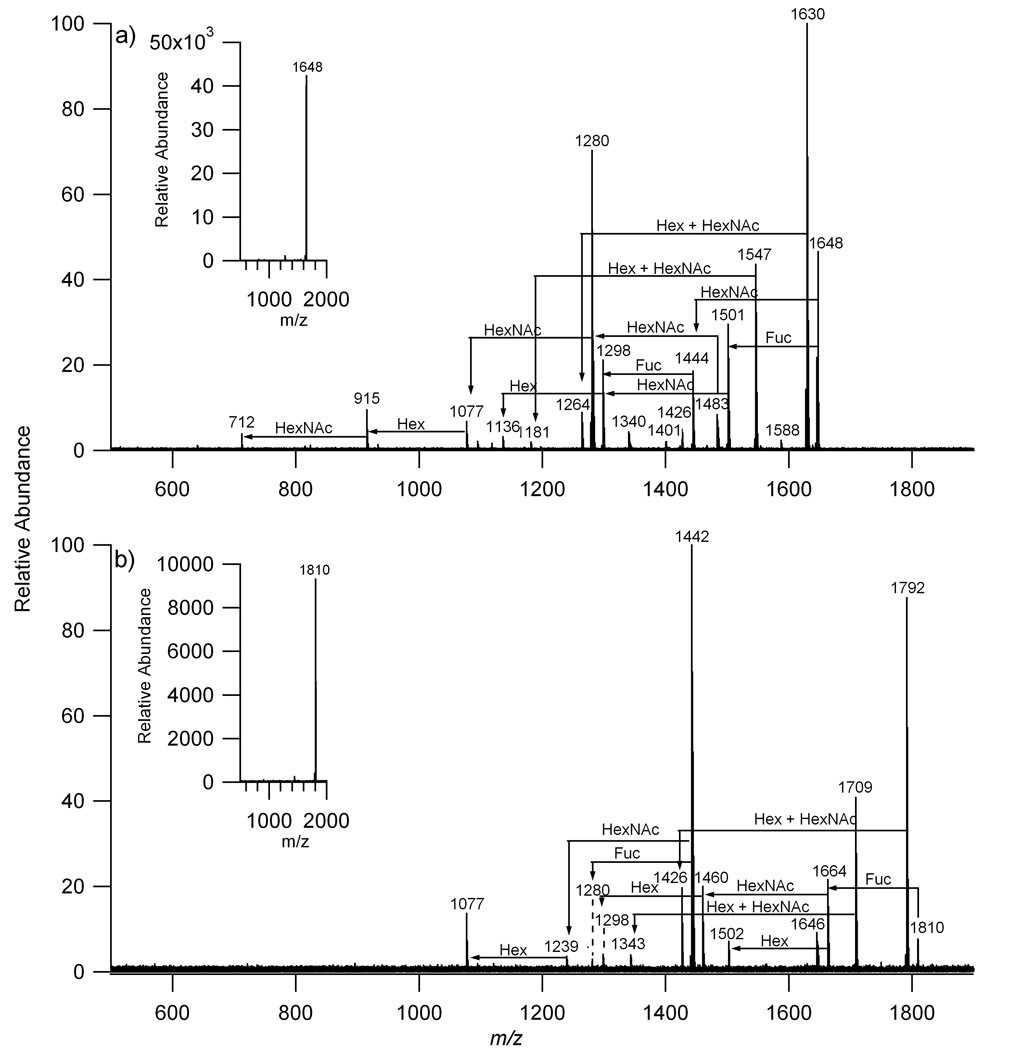
While accurate masses can rapidly provide glycan composition such as the number of hexoses, N-acetylhexosamines, sialic acids (NeuAc), and deoxyhexoses (fucose), it is necessary to validate these assignments using a separate method. Tandem mass spectrometry was used to probe several of the more abundant species using collision-induced dissociation (CID). Figure 2a is the fragmentation pattern of a neutral oligosaccharide with m/z 1647.595. The CID spectrum shows fragment ions consistent with a composition corresponding to 4Hex, 4HexNAc, 1 dHex. The N-glycan with m/z 1809.655 is shown in Figure 2b confirming the composition, 5Hex, 4HexNAc, 1 dHex.
Figure 2.
Collision induced dissociation of a) m/z 1648 with composition 1 deoxyhexose (dHex), 4 hexoses (Hex), and 4 N-acetylhexosamines (HexNAc) and b) m/z 1810 with composition 1 deoxyhexose (dHex), 5 hexoses (Hex), and 4 N-acetylhexosamines (HexNAc).
Using a mass profile one can obtain compositions accurately even in complicated mixtures. Fractionating the oligosaccharide mixture into three fractions minimizes but does not fully eliminate suppression. Based upon the mass spectra of the three fractions in Figure 1, 20 distinct compositions were identified from the m/z values in the three fractions and are listed in Table 2. This number corresponds to masses of N-linked glycans, however no information on the number of isomers associated with each N-glycan was obtained.
Table 2.
List of the distinct N-linked oligosaccharides observed in human serum using MALDI FT-ICR MS
| Mass |
Oligosaccharide Composition |
|||||||
|---|---|---|---|---|---|---|---|---|
| Expt m/z | Calc m/z | Delta | Hex | dHex | HexNAc | NeuAc | Na+ | |
| 1 | 1136.404 | 1136.397 | 0.007 | 3 | 3 | 1 | ||
| 2 | 1257.433 | 1257.423 | 0.010 | 5 | 2 | 1 | ||
| 3 | 1282.465 | 1282.455 | 0.010 | 3 | 1 | 3 | 1 | |
| 4 | 1298.462 | 1298.450 | 0.012 | 4 | 3 | 1 | ||
| 5 | 1339.488 | 1339.476 | 0.011 | 3 | 4 | 1 | ||
| 6 | 1419.490 | 1419.476 | 0.014 | 6 | 2 | 1 | ||
| 7 | 1444.521 | 1444.508 | 0.014 | 4 | 1 | 3 | 1 | |
| 8 | 1485.541 | 1485.534 | 0.007 | 3 | 1 | 4 | 1 | |
| 9 | 1501.546 | 1501.529 | 0.017 | 4 | 4 | 1 | ||
| 10 | 1542.572 | 1542.556 | 0.016 | 3 | 5 | 1 | ||
| 11 | 1647.595 | 1647.587 | 0.008 | 4 | 1 | 4 | 1 | |
| 12 | 1663.599 | 1663.582 | 0.017 | 5 | 4 | 1 | ||
| 13 | 1688.630 | 1688.614 | 0.016 | 3 | 1 | 5 | 1 | |
| 14 | 1704.629 | 1704.609 | 0.020 | 4 | 5 | 1 | ||
| 15 | 1743.605 | 1743.582 | 0.024 | 8 | 2 | 1 | ||
| 16 | 1809.655 | 1809.640 | 0.015 | 5 | 1 | 4 | 1 | |
| 17 | 1850.686 | 1850.666 | 0.020 | 4 | 1 | 5 | 1 | |
| 18 | 1866.674 | 1866.661 | 0.013 | 5 | 5 | 1 | ||
| 19* | 1976.728 | 1976.660 | −0.068 | 5 | 4 | 1 | 2 | |
| 20 | 2012.748 | 2012.719 | 0.029 | 5 | 1 | 5 | 1 | |
[M+2Na-H]+
3.2 NanoLC of N-linked glycans in human serum
NanoLC was performed on the reduced and unreduced N-glycans released from human serum. Each individual N-glycan SPE fraction was initially analyzed prior to combining the fractions to obtain an N-glycan profile for a particular serum sample. Separating the glycans by chromatography into single components alleviates the suppression problem often encountered when electrospraying solutions by direct infusion. A microchip device, employed in this study, was developed for separating glycans using nanoLC and graphitized carbon. Two chips with varying column lengths were utilized, a commercial glycan chip corresponding to a chip with a 40 nL enrichment column and a 43×0.075 mm ID analytical column, and a high capacity chip with a 160 nL enrichment column and a 150×0.075 mm ID analytical column produced specifically for this project. Both chips were packed with graphitized carbon as described previously[43, 44]. The microchips were interfaced with an Agilent TOF mass analyzer that routinely provided a mass measurement accuracy of less than 3 ppm.
NanoLC-MS of each SPE fraction and the combined fractions were performed. Analysis of each individual fraction was necessary because the SPE fractionation was used to improve partitioning of oligosaccharides in highly complicated mixtures. Once each SPE fraction was profiled and characterized, an aliquot of each SPE fraction was combined and the mixture was also analyzed by nanoLC MS. All mass spectra were obtained in the positive mode. Figure 3 shows the base peak chromatograms of the three fractions 10% (Figure 3a), 20% (Figure 3b), and 40% fractions (Figure 3c). The 10% fraction contains neutral oligosaccharides, which elute between 15.0 and 25.0 minutes. The 20% fraction contains both neutral and anionic oligosaccharides eluting between 15.0 and 30.0 minutes, while the acidic oligosaccharides in the 40% fraction elute much later between 25.0 and 35.0 minutes. There is considerable overlap in the signals between the 10% and 20% fractions, however primarily core fucosylated oligosaccharides were present in the 20% fractions based on the intensities of each N-glycan. (data not shown) The total ion chromatograms of the three fractions can be found in under Supporting Information.
Figure 3.
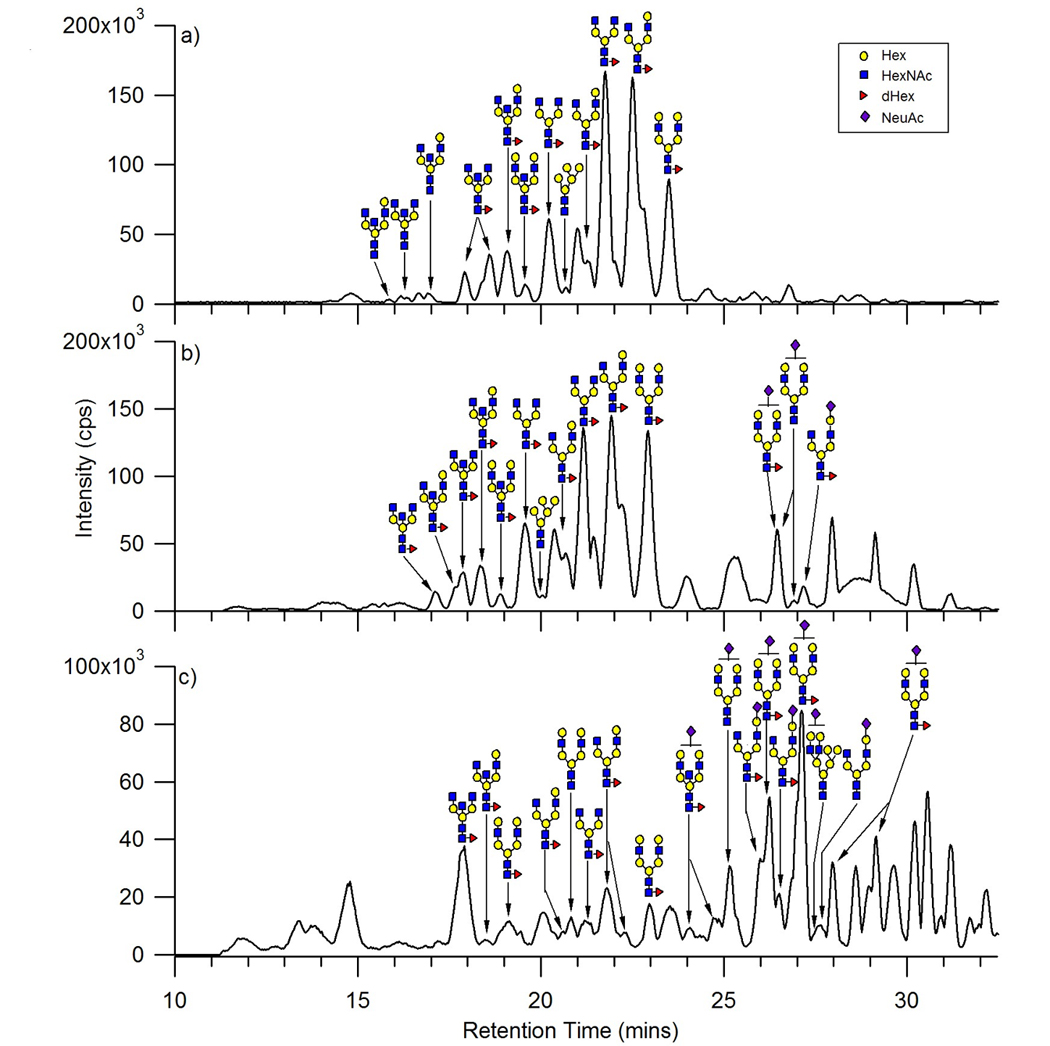
Base peak chromatograms of a) the neutral oligosaccharides, b) the neutral and anionic oligosaccharides, and c) the anionic oligosaccharides released from human serum. Each fraction has been annotated with the proposed structure corresponding to the most abundant oligosaccharide at that retention time.
The base peak chromatograms of the three fractions (Figure 3) were labeled with the putative structure for the most abundant glycan for each chromatographic peak. The structures were based solely on composition and profiles of human serum N-glycans from literature[33, 45]. The analysis of each component in the three fractions showed that the 10% contained a mixture of the neutral complex, hybrid, and high mannose, while the 20% fraction contained a mixture of the neutral complex, hybrid, high mannose, and some anionic complex oligosaccharides. The 40% fraction was predominantly anionic complex oligosaccharides, however some neutral complex, hybrid, and high mannose oligosaccharides were observed. Moreover in all three fractions a biantennary N-glycan with a possible bisecting HexNAc was observed, however based upon composition this N-glycan can also be a triantennary N-glycan. Further studies will be needed to confirm the structure of this N-glycan and has been labeled as biantennary with a bisecting HexNAc due to the rigid isomer having a lower retention time on the PGC in comparison to a more planar triantennary N-glycan.
Based upon the chromatograms and deconvoluted data, the retention order of the oligosaccharides were determined. The hybrid type oligosaccharides are the first to elute followed by the high mannose type, both found in the 10% fraction. The high mannose oligosaccharides observed were low in abundance in comparison to the complex types. However based on the deconvoluted data, the oligosaccharide with the larger molecular mass among the high mannose oligosaccharides eluted first, while the lower molecular mass oligosaccharides eluted at later times. These findings were in agreement with those observed by Itoh et al. in their study of model oligosaccharides released from standard glycoproteins[29]. The neutral fucosylated complex type oligosaccharides were found in all three fractions. The acidic oligosaccharides that eluted at later times were seen only in the 20% and 40% fractions with the predominance in the latter fraction. Among the acidic complex N-glycans, the monosialylated biantennary with and without core fucosylation were the most abundant. These results are consistent with previous observations on PGC, that anionic oligosaccharides have a higher affinity to the graphitized carbon compared to neutral oligosaccharides[22, 23]. It should be noted that the precise nature of the interaction between the oligosaccharides and the graphite sheets of the PGC remains poorly understood and therefore, retention patterns are not easy to predict[22, 23, 25, 27].
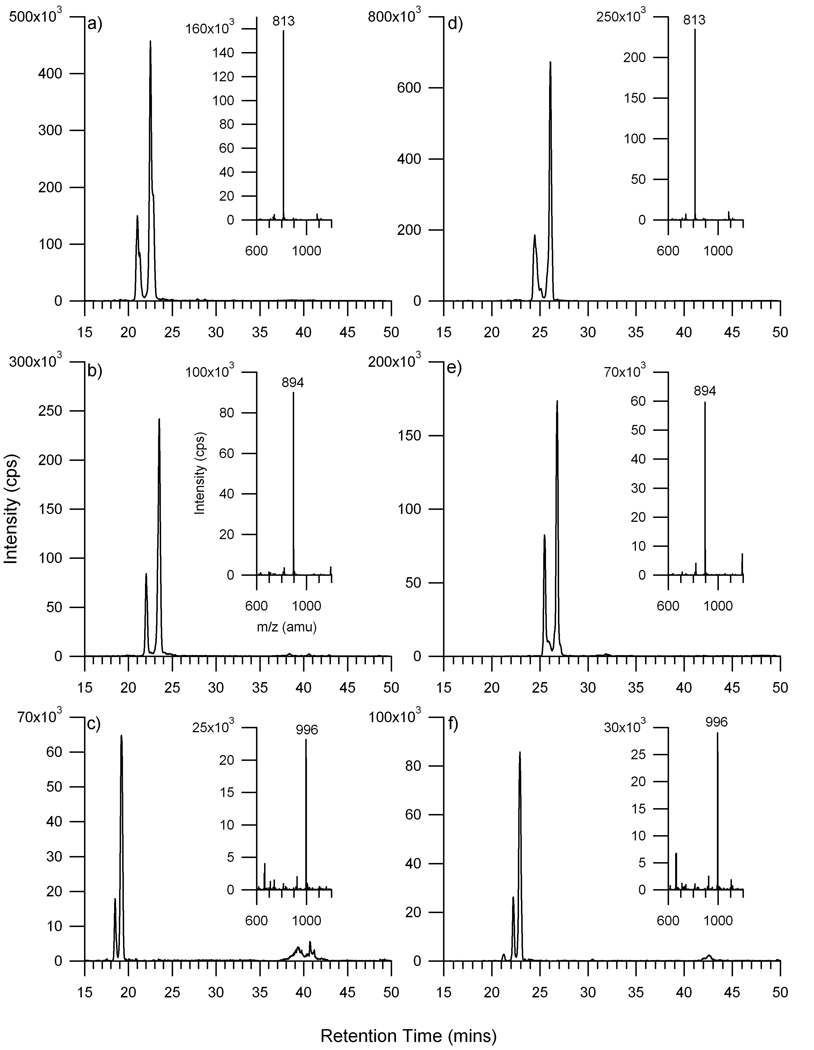
Representative extracted ion chromatograms (XIC) of the neutral and acidic oligosaccharides are shown in Figure 4 to illustrate the separation and the number of isomers that may comprise a specific composition. Five oligosaccharides (three neutral and two sialylated) were selected and compared between the glycan and the high capacity chip. The prototype, high capacity chip, has an analytical column that is 3.5 times longer than the standard glycan chip. These experiments were performed to determine whether better separation of isomers could be obtained with the high capacity chip. The three neutral oligosaccharides chosen for this purpose were composed of fucose, hexoses, and N-acetylhexosamines but with varying quantities of the latter two monosaccharides. The extracted ion chromatograms from both chips of neutral N-glycans in human serum are illustrated in Figure 4. The three neutral oligosaccharides showed two isomers for each composition. The results from the glycan chip were in agreement with the results from the high capacity chip. Mass spectra corresponding to the maximum of the dominant peak for each XIC are shown to the right of each XIC. For all three glycans, the doubly protonated species were the dominant peaks with the singly protonated species being minor signals. There appears to be only minor differences in the chromatograms between the glycan chip and the high capacity chip.
Figure 4.
Extracted ion chromatograms of neutral N-linked oligosaccharides released from human serum a & d) m/z 813, [M+2H]2+ with a composition of 1 dHex, 4 Hex, 4 HexNAc, b & e) m/z 894, [M+2H]2+ with a composition of 1 dHex, 5 Hex, 4 HexNAc, and c & f) m/z 996, [M+2H]2+ with a composition of 1 dHex, 5 Hex, 5 HexNAc. The chromatograms on the left column were obtained using the glycan chip (a–c) and those on the right column were obtained using the high capacity chip (d–f).
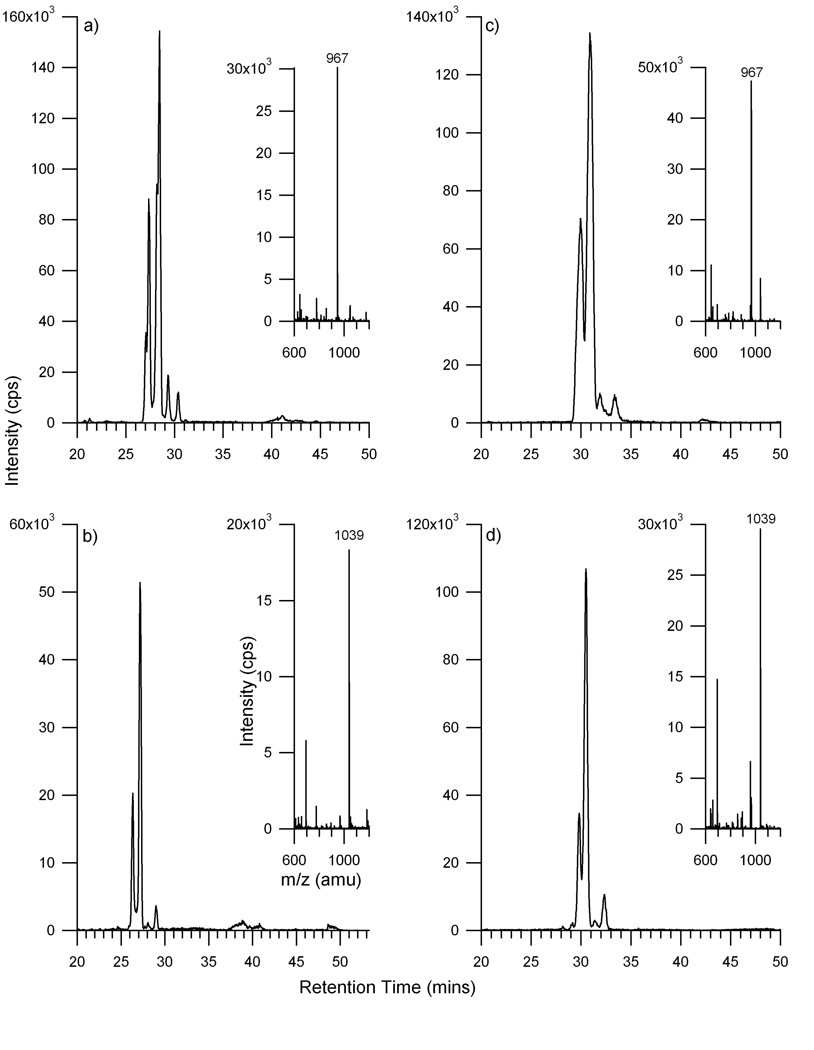
For comparison, the anionic oligosaccharides were examined. The XICs of two anionic oligosaccharides are illustrated in Figure 5. Results from the standard glycan chip are shown with the high capacity chip, both yielding similar results. The two anionic oligosaccharides have the same compositions of 1 N-acetyl neuraminic acid (sialic acid), 5 hexoses, and 4 N-acetylhexosamines (Figure 5a), with the larger differing only by the presence of a fucose (Figure 5b). Several peaks with different retention times were observed for each of the two ions, indicative of possible isomers for those specific compositions. As with the neutral glycans, the mass spectra show the doubly protonated species as the most intense peaks with the corresponding singly protonated species with significantly lower intensity peak. Interestingly, it appears that the sialylated oligosaccharides may have more possible isomers than the neutral oligosaccharides. Three to four potential isomeric species were observed for the sialylated glycans compared to the two observed for the neutral glycans. It is not confirmed whether the isomers of the sialylated oligosaccharides are from α2,3- and α2,6-linked sialic acids or from the possibility of partial fragmentation of the N-glycans due to the loss of a sialic acid or fucose and will be the subject of future publications.
Figure 5.
Extracted ion chromatograms of anionic N-linked oligosaccharides released from human serum a & c) m/z 967, [M+2H]2+ with a composition of 1 NeuAc, 5 Hex, 4 HexNAc and b & d) m/z 1039, [M+2H]2+ with a composition of 1 NeuAc, 1 dHex, 5 Hex, 4 HexNAc. The chromatograms on the left column were obtained using the glycan chip (a & b) and those on the right column were obtained using the high capacity chip (c & d).
There were concerns that α, β anomers would separate under nanoLC conditions. Davies et al. reported the separation of reduced oligosaccharides (alditol form) on PGC to reduce complication of anomers[22]. Therefore the released N-glycans were reduced to their alditol forms and fractionated by SPE. The reduced fractions were analyzed with the HPLC-Chip/TOF MS. Reducing the oligosaccharides increased the resolution of the chromatograms. Extracted ion chromatograms and deconvoluted lists of masses of the reduced oligosaccharides were obtained and compared with its native form to determine the degree of isomerization. In comparison to the native form, a decrease in the number of isomers was not observed for the larger N-linked oligosaccharides, however for small biantennary N-glycans a decrease in the number of isomers was observed and will be the subject of future work.
An attempt was made to determine the total number of oligosaccharides that can be identified using this method. After deisotoping the LC-MS data for each component, the possible N-linked glycan composition(s) for each m/z ion was determined within 10 ppm mass error using an in-house program (Glycan Finder) written in IGOR Pro. The program was written to determine potential combinations of user specified monosaccharides for a particular m/z within a defined mass error tolerance. Two sets of criteria were used to verify the composition of each glycan. First, the mass tolerance was selected at 10 ppm and the composition must have the pentasaccharide core (minimum of 3 Hex and 2 HexNAc). Compositions outside that range were rejected. Second, the composition was examined to determine whether it corresponded to one of the three types of N-linked oligosaccharides; high mannose, hybrid, and complex. This “biological filter” followed the conditions outlined in Table 1. For example, hybrid and high mannose oligosaccharides rarely contain sialic acids. A list of the N-glycans and the compositions observed in human serum are provided under Supporting Information. The N-glycans observed were primarily in the doubly charged, [M+2H]2+ state and have been deconvoluted to their neutral masses.
3.3 Reproducibility of the method and instrumentation
Triplicate experiments were performed to assess the method and to determine the general performance of the HPLC-Chip/TOF for N-linked glycans analysis. In these experiments three aliquots of commercial serum were processed separately and the resulting glycan mixture were analyzed with the glycan chip. Figure 6a shows the base peak chromatogram for the neutral glycans. The base peak chromatogram is sensitive to discrepancies in mixtures, which can affect the profile. Figure 6a shows the variations in the glycan release and the total abundances of the monitored glycans. The retention times of these glycans were in agreement with under 1% relative standard deviation. These variations are likely a result of the release and purification method of the oligosaccharides prior to MS analysis.
Figure 6.
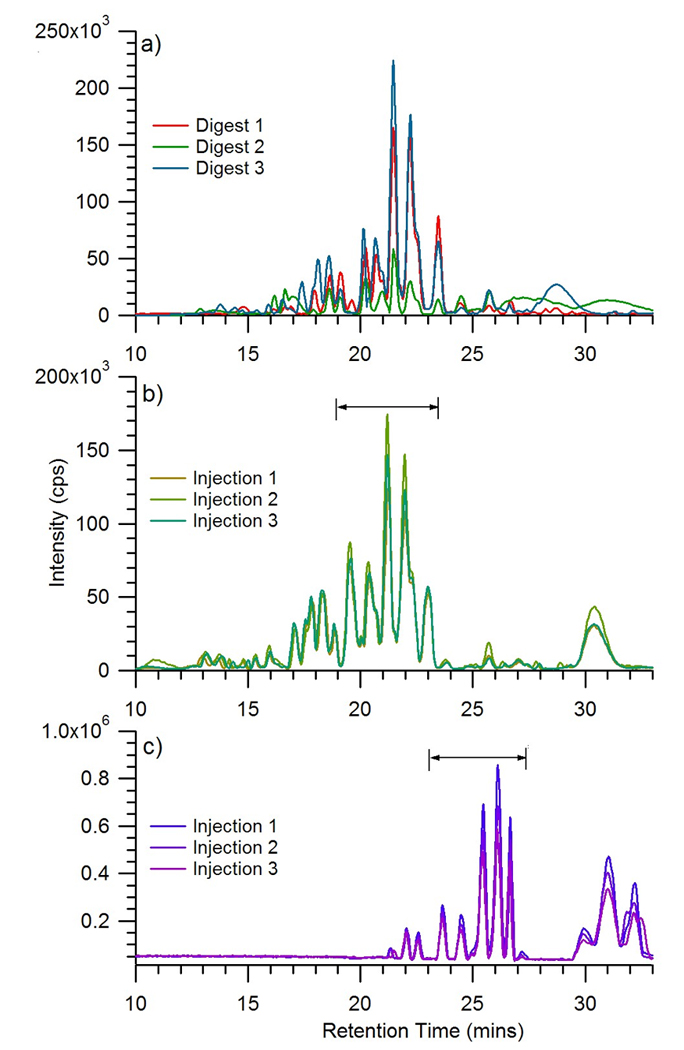
Base peak chromatograms of a) the neutral oligosaccharides from triplicate digestion experiments, b) the neutral oligosaccharides from a single experiment after repeated injections of three using the glycan chip, and c) the neutral and anionic oligosaccharides after repeated injections of three using the high capacity chip.
Multiple injections of the same volume from a single fraction were also analyzed using the glycan chip (Figure 6b) and the high capacity chip (Figure 6c). The retention times, peak heights, and peak areas of the double charged ions m/z 732, m/z 813, and m/z 894 were monitored. Both chips performed well in retention time reproducibility having less than 1% relative standard deviation, however the high capacity chip showed less retention time shifts compared to the glycan chip. The glycan chip did perform better in peak height and peak area reproducibility than the high capacity chip, however both performed generally well.
3.4 Variations in serum glycans of different individuals
N-glycans released from two individual donors were purified by SPE and the three N-linked oligosaccharide fractions were combined and analyzed by nanoLC MS. The individual sera samples analyzed corresponded to a female (Donor 1) and a male (Donor 2). Figure 7 is representative of the total ion chromatograms of the released oligosaccharides in the sera samples using the high capacity chip. A similar glycan profile was observed using the glycan chip (data not shown). The three sera samples, commercial human serum, Donor 1, and Donor 2, had similar profiles with overlapping of the higher abundance peaks. Many of the same oligosaccharides were found in the three sera samples. Differences between the three sera samples were expected and include differences in intensity and minor shifts of the same peak. Factors most likely affecting variability in sera samples include age[46] and possibly gender.
Figure 7.
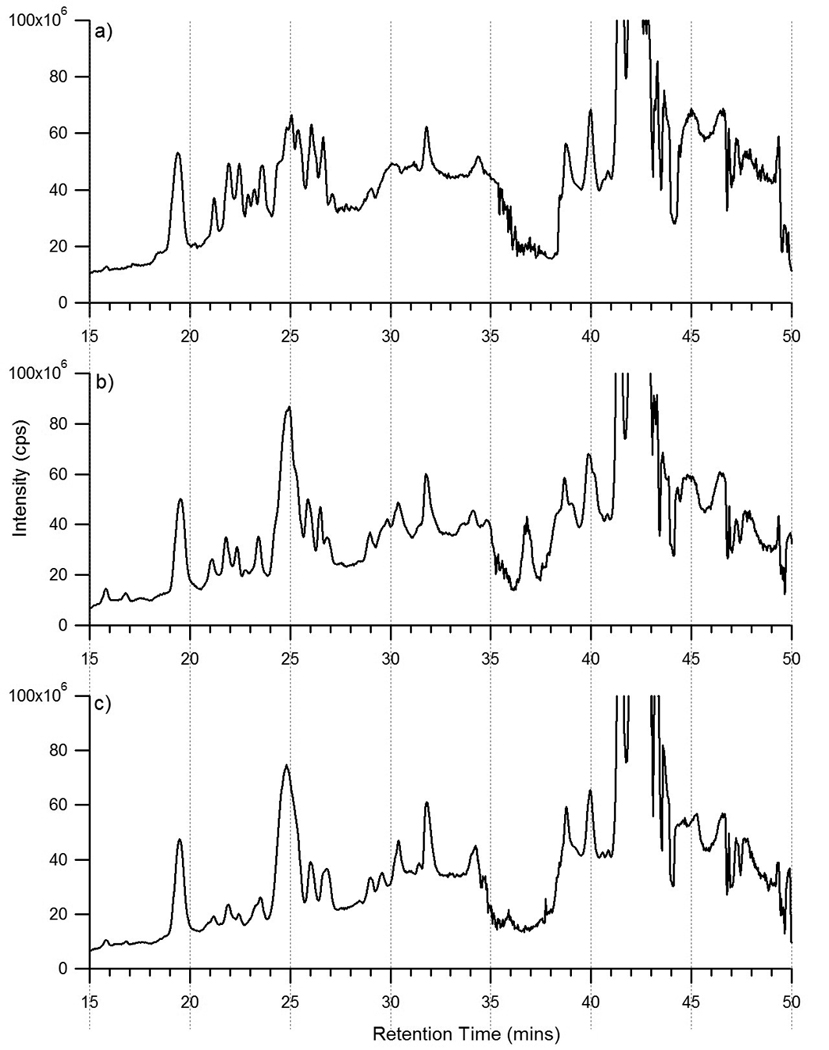
Total ion chromatograms of the combined fractions of the N-linked oligosaccharides released from a) commercially purchased human serum, b) donor 1 serum, and c) donor 2 serum obtained using the high capacity chip.
Pie charts based upon intensities of the identified glycans were generated to approximate the proportions of the different types of N-linked oligosaccharide for each individual (Figure 8, left). As shown, the N-linked glycans in human serum is mostly complex type oligosaccharides, accounting for ~96% of the total N-linked glycans present. High-mannose and hybrid type oligosaccharides are lower in abundance making up the remaining ~4% of the total N-linked glycans. These findings are consistent among the three individual sera samples and between the two chips used in this study. The abundances of the saccharides present were also compared. The abundances of the following saccharides present in each serum were determined, “Hex + HexNAc”, “dHex”, “NeuAc”, and “dHex + NeuAc”. The pie charts (Figure 8, right) show the amounts of each of the monosaccharide or disaccharide combinations.
Figure 8.
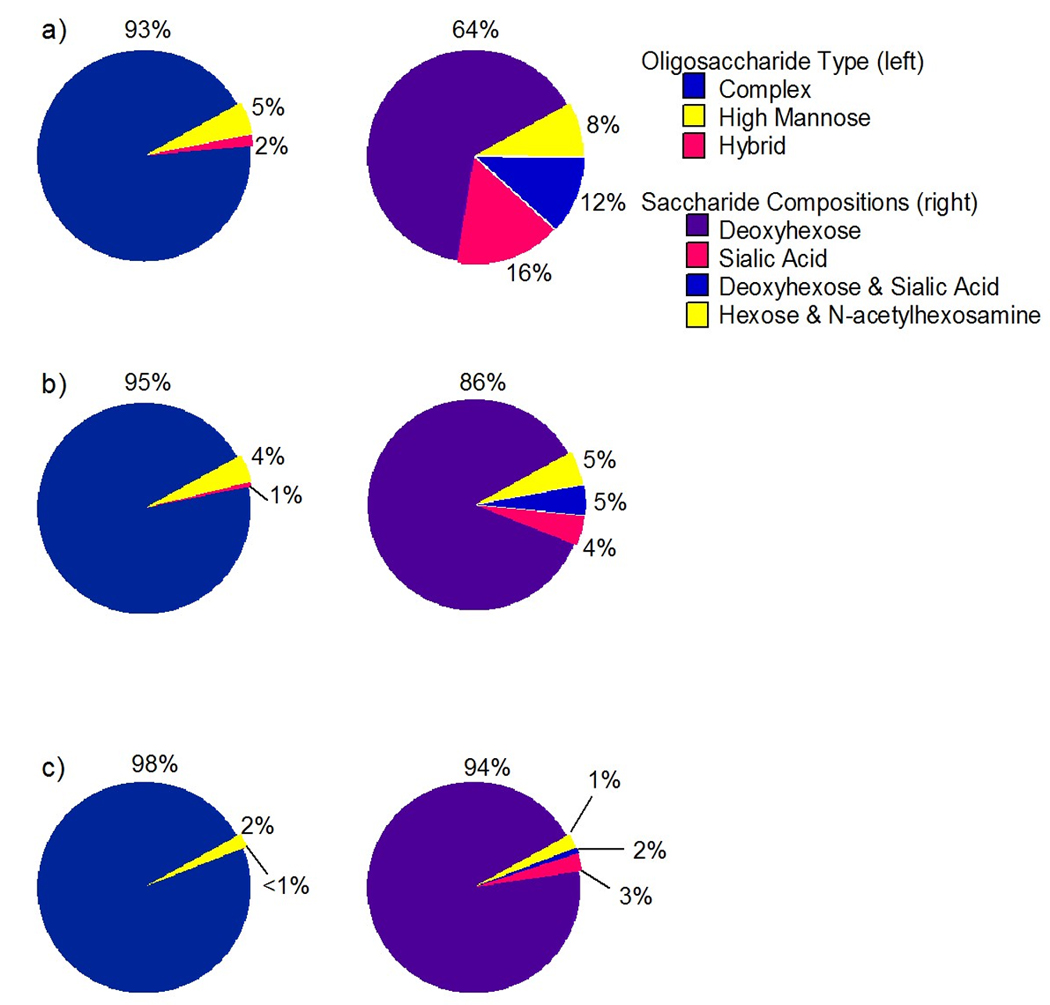
Pie charts illustrating the N-linked oligosaccharide types and the saccharide compositions for a) commercially purchased human serum, b) donor 1 serum, and c) donor 2 serum obtained using the high capacity chip.
4 CONCLUDING REMARKS
Native, underivatized N-linked oligosaccharides released from sera samples were separated on a microfluidic chip using graphitized carbon and analyzed using an integrated HPLC-Chip/TOF MS system. Nearly 200 glycans have been identified based on exact mass measurements using the glycan chip. A list of the N-linked oligosaccharides observed was generated including high mannose, fucosylated complex, sialylated complex, and fucosylated-sialylated complex type glycans. Additionally, profiles of three individual serum samples were analyzed and the glycan profiles were compared. Based upon the sera analysis, human serum mainly consists of complex type N-linked oligosaccharides that are fucosylated. Further studies, including tandem MS and exoglycosidase digestions, are necessary to determine the exact structures, linkage information, and monosaccharide type of each possible isomer observed.
Two different chips were used in this study, the glycan chip and the high capacity chip, both offered good sensitivity and reproducibility in separating a heterogeneous mixture of neutral and anionic oligosaccharides. For comparison, samples were also run on the high capacity chip, which had an analytical column that was 3.5 times longer in length than the glycan chip. Results from the high capacity chip were equivalent to those from the glycan chip both having less than 1% standard deviation in retention time reproducibility. Increasing the analytical column length improved resolution, however reproducibility in peak height and area were not comparable to the glycan chip.
Supplementary Material
Total ion chromatograms of a) the neutral oligosaccharides, b) the neutral and anionic oligosaccharides, and c) the anionic oligosaccharides released from human serum.
Table 3.
List of the N-linked oligosaccharides released from in human serum.
| Mass | Oligosaccharide Composition | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Observed | Calculated | Error (ppm) |
Hex | dHex | HexNAc | NeuAc | Intensity (cps) |
Retention Time (mins) |
|
| 1 | 910.329 | 910.328 | 1.208 | 3 | 2 | 45782 | 22.4 | ||
| 2 | 1056.385 | 1056.386 | −0.284 | 3 | 1 | 2 | 16677 | 21.8 | |
| 3 | 1072.381 | 1072.381 | 0.000 | 4 | 2 | 9810 | 23.3 | ||
| 4 | 1072.383 | 1072.381 | 1.772 | 4 | 2 | 54997 | 20.7 | ||
| 5 | 1113.404 | 1113.407 | −3.144 | 3 | 3 | 10884 | 17.3 | ||
| 6 | 1113.404 | 1113.407 | −2.515 | 3 | 3 | 14570 | 18.8 | ||
| 7 | 1113.408 | 1113.407 | 0.359 | 3 | 3 | 26239 | 15.5 | ||
| 8 | 1234.433 | 1234.433 | −0.243 | 5 | 2 | 299942 | 22.5 | ||
| 9 | 1234.434 | 1234.433 | 0.405 | 5 | 2 | 392678 | 20.7 | ||
| 10 | 1259.468 | 1259.465 | 2.144 | 3 | 1 | 3 | 61619 | 21.7 | |
| 11 | 1259.469 | 1259.465 | 2.700 | 3 | 1 | 3 | 24353 | 20.8 | |
| 12 | 1259.469 | 1259.465 | 2.938 | 3 | 1 | 3 | 9908 | 19.3 | |
| 13 | 1275.461 | 1275.460 | 0.549 | 4 | 3 | 10307 | 24.0 | ||
| 14 | 1275.462 | 1275.460 | 1.882 | 4 | 3 | 9130 | 13.2 | ||
| 15 | 1275.463 | 1275.460 | 2.509 | 4 | 3 | 20804 | 18.7 | ||
| 16 | 1316.482 | 1316.487 | −3.494 | 3 | 4 | 10597 | 16.3 | ||
| 17 | 1316.488 | 1316.487 | 0.987 | 3 | 4 | 266473 | 19.4 | ||
| 18 | 1316.490 | 1316.487 | 2.583 | 3 | 4 | 81634 | 18.0 | ||
| 19 | 1396.483 | 1396.486 | −2.005 | 6 | 2 | 46505 | 23.3 | ||
| 20 | 1396.488 | 1396.486 | 0.931 | 6 | 2 | 66140 | 21.5 | ||
| 21 | 1396.489 | 1396.486 | 1.862 | 6 | 2 | 74100 | 23.4 | ||
| 22 | 1421.517 | 1421.518 | −0.563 | 4 | 1 | 3 | 31248 | 24.0 | |
| 23 | 1421.521 | 1421.518 | 2.181 | 4 | 1 | 3 | 48054 | 22.9 | |
| 24 | 1421.523 | 1421.518 | 3.517 | 4 | 1 | 3 | 141118 | 25.4 | |
| 25 | 1421.524 | 1421.518 | 4.010 | 4 | 1 | 3 | 68607 | 21.2 | |
| 26 | 1437.515 | 1437.513 | 1.600 | 5 | 3 | 62272 | 19.9 | ||
| 27 | 1462.547 | 1462.545 | 1.778 | 3 | 1 | 4 | 75349 | 18.6 | |
| 28 | 1462.547 | 1462.545 | 1.778 | 3 | 1 | 4 | 6067514 | 21.8 | |
| 29 | 1462.547 | 1462.545 | 1.846 | 3 | 1 | 4 | 12027 | 25.6 | |
| 30 | 1462.548 | 1462.545 | 2.393 | 3 | 1 | 4 | 3159838 | 20.2 | |
| 31 | 1478.537 | 1478.539 | −1.353 | 4 | 4 | 29186 | 18.9 | ||
| 32 | 1478.543 | 1478.539 | 2.097 | 4 | 4 | 46425 | 21.8 | ||
| 33 | 1478.545 | 1478.539 | 3.923 | 4 | 4 | 118891 | 18.8 | ||
| 34 | 1519.571 | 1519.566 | 2.961 | 3 | 5 | 167677 | 16.3 | ||
| 35 | 1519.574 | 1519.566 | 5.133 | 3 | 5 | 75554 | 15.3 | ||
| 36 | 1558.540 | 1558.539 | 0.706 | 7 | 2 | 18981 | 19.9 | ||
| 37 | 1558.541 | 1558.539 | 1.476 | 7 | 2 | 13601 | 18.8 | ||
| 38 | 1599.569 | 1599.566 | 2.126 | 6 | 3 | 27898 | 20.7 | ||
| 39 | 1624.594 | 1624.597 | −2.093 | 4 | 1 | 4 | 27115 | 19.6 | |
| 40 | 1624.596 | 1624.597 | −1.046 | 4 | 1 | 4 | 9343 | 23.5 | |
| 41 | 1624.599 | 1624.597 | 0.985 | 4 | 1 | 4 | 9127308 | 22.5 | |
| 42 | 1624.599 | 1624.597 | 1.293 | 4 | 1 | 4 | 38509 | 22.7 | |
| 43 | 1624.606 | 1624.597 | 5.171 | 4 | 1 | 4 | 65512 | 21.1 | |
| 44 | 1640.592 | 1640.592 | −0.244 | 5 | 4 | 11181 | 19.9 | ||
| 45 | 1640.594 | 1640.592 | 1.036 | 5 | 4 | 68461 | 22.1 | ||
| 46 | 1640.596 | 1640.592 | 2.316 | 5 | 4 | 10784 | 18.3 | ||
| 47 | 1640.599 | 1640.592 | 4.023 | 5 | 4 | 188542 | 21.4 | ||
| 48 | 1640.599 | 1640.592 | 4.084 | 5 | 4 | 156495 | 20.6 | ||
| 49 | 1665.620 | 1665.624 | −2.101 | 3 | 1 | 5 | 115568 | 17.9 | |
| 50 | 1665.630 | 1665.624 | 3.362 | 3 | 1 | 5 | 1716568 | 18.6 | |
| 51 | 1665.630 | 1665.624 | 3.542 | 3 | 1 | 5 | 853982 | 17.9 | |
| 52 | 1681.625 | 1681.619 | 3.746 | 4 | 5 | 375906 | 16.9 | ||
| 53 | 1681.627 | 1681.619 | 4.817 | 4 | 5 | 182486 | 19.1 | ||
| 54 | 1712.618 | 1712.613 | 2.861 | 4 | 1 | 3 | 1 | 241490 | 27.3 |
| 55 | 1712.620 | 1712.613 | 3.679 | 4 | 1 | 3 | 1 | 15618 | 28.7 |
| 56 | 1720.591 | 1720.592 | −0.407 | 8 | 2 | 33826 | 18.7 | ||
| 57 | 1720.594 | 1720.592 | 1.395 | 8 | 2 | 25184 | 20.9 | ||
| 58 | 1720.595 | 1720.592 | 1.569 | 8 | 2 | 11130 | 19.4 | ||
| 59 | 1728.611 | 1728.608 | 1.446 | 5 | 3 | 1 | 99234 | 25.2 | |
| 60 | 1728.616 | 1728.608 | 4.744 | 5 | 3 | 1 | 8690 | 27.7 | |
| 61 | 1728.624 | 1728.608 | 8.851 | 5 | 3 | 1 | 19353 | 23.8 | |
| 62 | 1769.632 | 1769.635 | −1.695 | 4 | 4 | 1 | 34710 | 26.3 | |
| 63 | 1769.635 | 1769.635 | −0.170 | 4 | 4 | 1 | 130393 | 26.9 | |
| 64 | 1769.638 | 1769.635 | 1.695 | 4 | 4 | 1 | 14512 | 25.9 | |
| 65 | 1769.639 | 1769.635 | 2.317 | 4 | 4 | 1 | 11667 | 24.1 | |
| 66 | 1769.640 | 1769.635 | 2.995 | 4 | 4 | 1 | 184545 | 25.8 | |
| 67 | 1769.643 | 1769.635 | 4.351 | 4 | 4 | 1 | 57992 | 24.7 | |
| 68 | 1769.646 | 1769.635 | 6.046 | 4 | 4 | 1 | 8277 | 28.0 | |
| 69 | 1786.646 | 1786.650 | −2.071 | 5 | 1 | 4 | 23046 | 22.1 | |
| 70 | 1786.653 | 1786.650 | 1.455 | 5 | 1 | 4 | 1193131 | 22.0 | |
| 71 | 1786.653 | 1786.650 | 1.791 | 5 | 1 | 4 | 4797501 | 23.5 | |
| 72 | 1802.645 | 1802.645 | 0.222 | 6 | 4 | 10358 | 19.4 | ||
| 73 | 1802.649 | 1802.645 | 1.997 | 6 | 4 | 123948 | 23.7 | ||
| 74 | 1802.651 | 1802.645 | 3.107 | 6 | 4 | 30881 | 21.5 | ||
| 75 | 1827.683 | 1827.677 | 3.283 | 4 | 1 | 5 | 764636 | 18.4 | |
| 76 | 1827.683 | 1827.677 | 3.666 | 4 | 1 | 5 | 13463 | 18.5 | |
| 77 | 1827.684 | 1827.677 | 4.158 | 4 | 1 | 5 | 2288931 | 19.1 | |
| 78 | 1827.686 | 1827.677 | 4.979 | 4 | 1 | 5 | 10005 | 25.8 | |
| 79 | 1843.678 | 1843.672 | 3.526 | 5 | 5 | 13256 | 19.6 | ||
| 80 | 1843.679 | 1843.672 | 3.797 | 5 | 5 | 28793 | 18.3 | ||
| 81 | 1843.679 | 1843.672 | 3.959 | 5 | 5 | 23215 | 16.5 | ||
| 82 | 1874.670 | 1874.666 | 1.920 | 5 | 1 | 3 | 1 | 28985 | 26.4 |
| 83 | 1874.671 | 1874.666 | 2.774 | 5 | 1 | 3 | 1 | 8173 | 25.3 |
| 84 | 1882.638 | 1882.645 | −3.612 | 9 | 2 | 10876 | 19.8 | ||
| 85 | 1882.647 | 1882.645 | 1.009 | 9 | 2 | 58204 | 18.8 | ||
| 86 | 1882.652 | 1882.645 | 3.718 | 9 | 2 | 37797 | 17.5 | ||
| 87 | 1890.662 | 1890.661 | 0.317 | 6 | 3 | 1 | 11949 | 24.8 | |
| 88 | 1890.665 | 1890.661 | 2.169 | 6 | 3 | 1 | 8860 | 25.5 | |
| 89 | 1890.669 | 1890.661 | 4.126 | 6 | 3 | 1 | 38465 | 25.9 | |
| 90 | 1915.688 | 1915.693 | −2.662 | 4 | 1 | 4 | 1 | 9906 | 28.7 |
| 91 | 1915.692 | 1915.693 | −0.626 | 4 | 1 | 4 | 1 | 1200614 | 26.8 |
| 92 | 1915.693 | 1915.693 | 0.365 | 4 | 1 | 4 | 1 | 334390 | 26.9 |
| 93 | 1915.697 | 1915.693 | 2.140 | 4 | 1 | 4 | 1 | 342642 | 25.9 |
| 94 | 1916.699 | 1916.713 | −7.617 | 4 | 3 | 4 | 31288 | 24.7 | |
| 95 | 1931.688 | 1931.688 | 0.362 | 5 | 4 | 1 | 42212 | 27.2 | |
| 96 | 1931.691 | 1931.688 | 1.708 | 5 | 4 | 1 | 337158 | 27.1 | |
| 97 | 1931.692 | 1931.688 | 2.019 | 5 | 4 | 1 | 2127370 | 25.1 | |
| 98 | 1931.693 | 1931.688 | 2.537 | 5 | 4 | 1 | 254681 | 28.2 | |
| 99 | 1931.694 | 1931.688 | 3.313 | 5 | 4 | 1 | 847587 | 26.2 | |
| 100 | 1932.691 | 1932.708 | −8.589 | 5 | 2 | 4 | 7924 | 22.5 | |
| 101 | 1932.696 | 1932.708 | −6.468 | 5 | 2 | 4 | 2009310 | 26.2 | |
| 102 | 1932.696 | 1932.708 | −6.157 | 5 | 2 | 4 | 20298 | 26.5 | |
| 103 | 1948.695 | 1948.703 | −4.259 | 6 | 1 | 4 | 11377 | 25.8 | |
| 104 | 1948.706 | 1948.703 | 1.386 | 6 | 1 | 4 | 20296 | 25.1 | |
| 105 | 1973.724 | 1973.735 | −5.168 | 4 | 2 | 5 | 15638 | 25.6 | |
| 106 | 1989.733 | 1989.730 | 1.910 | 5 | 1 | 5 | 930833 | 19.6 | |
| 107 | 1989.734 | 1989.730 | 2.412 | 5 | 1 | 5 | 160677 | 18.9 | |
| 108 | 2005.725 | 2005.724 | 0.199 | 6 | 5 | 7255 | 26.2 | ||
| 109 | 2005.730 | 2005.724 | 2.991 | 6 | 5 | 28419 | 21.7 | ||
| 110 | 2005.735 | 2005.724 | 5.285 | 6 | 5 | 9211 | 20.4 | ||
| 111 | 2005.739 | 2005.724 | 7.429 | 6 | 5 | 8333 | 23.3 | ||
| 112 | 2036.717 | 2036.719 | −0.835 | 6 | 1 | 3 | 1 | 8490 | 25.5 |
| 113 | 2036.725 | 2036.719 | 2.897 | 6 | 1 | 3 | 1 | 37708 | 26.5 |
| 114 | 2046.758 | 2046.751 | 3.322 | 5 | 6 | 14074 | 25.9 | ||
| 115 | 2077.744 | 2077.746 | −0.626 | 5 | 1 | 4 | 1 | 14685 | 24.7 |
| 116 | 2077.745 | 2077.746 | −0.433 | 5 | 1 | 4 | 1 | 9275 | 25.8 |
| 117 | 2077.746 | 2077.746 | 0.241 | 5 | 1 | 4 | 1 | 20910 | 25.8 |
| 118 | 2077.750 | 2077.746 | 2.021 | 5 | 1 | 4 | 1 | 6334387 | 27.1 |
| 119 | 2077.750 | 2077.746 | 2.262 | 5 | 1 | 4 | 1 | 1291595 | 26.3 |
| 120 | 2077.752 | 2077.746 | 2.888 | 5 | 1 | 4 | 1 | 462553 | 28.9 |
| 121 | 2093.737 | 2093.740 | −1.433 | 6 | 4 | 1 | 58304 | 26.6 | |
| 122 | 2093.741 | 2093.740 | 0.143 | 6 | 4 | 1 | 23809 | 25.7 | |
| 123 | 2093.748 | 2093.740 | 3.391 | 6 | 4 | 1 | 16774 | 27.3 | |
| 124 | 2093.752 | 2093.740 | 5.302 | 6 | 4 | 1 | 25235 | 28.1 | |
| 125 | 2094.757 | 2094.761 | −1.957 | 6 | 2 | 4 | 32869 | 27.2 | |
| 126 | 2119.772 | 2119.793 | −9.718 | 4 | 3 | 5 | 9140 | 26.1 | |
| 127 | 2119.777 | 2119.793 | −7.265 | 4 | 3 | 5 | 9017 | 23.7 | |
| 128 | 2134.769 | 2134.767 | 1.077 | 5 | 5 | 1 | 37770 | 21.7 | |
| 129 | 2134.772 | 2134.767 | 2.530 | 5 | 5 | 1 | 84649 | 22.8 | |
| 130 | 2134.773 | 2134.767 | 2.764 | 5 | 5 | 1 | 79351 | 24.8 | |
| 131 | 2135.769 | 2135.787 | −8.662 | 5 | 2 | 5 | 10507 | 25.7 | |
| 132 | 2135.781 | 2135.787 | −3.137 | 5 | 2 | 5 | 137517 | 25.3 | |
| 133 | 2214.776 | 2214.767 | 4.018 | 8 | 3 | 1 | 9436 | 25.8 | |
| 134 | 2223.808 | 2223.803 | 2.024 | 5 | 2 | 4 | 1 | 15432 | 26.8 |
| 135 | 2223.810 | 2223.803 | 3.148 | 5 | 2 | 4 | 1 | 40849 | 27.6 |
| 136 | 2239.807 | 2239.798 | 3.884 | 6 | 1 | 4 | 1 | 39421 | 27.5 |
| 137 | 2255.793 | 2255.793 | 0.000 | 7 | 4 | 1 | 36630 | 26.7 | |
| 138 | 2255.795 | 2255.793 | 0.798 | 7 | 4 | 1 | 102109 | 28.4 | |
| 139 | 2255.796 | 2255.793 | 1.419 | 7 | 4 | 1 | 202445 | 27.6 | |
| 140 | 2280.823 | 2280.825 | −0.965 | 5 | 1 | 5 | 1 | 25359 | 26.6 |
| 141 | 2280.829 | 2280.825 | 1.710 | 5 | 1 | 5 | 1 | 98095 | 24.3 |
| 142 | 2280.834 | 2280.825 | 3.858 | 5 | 1 | 5 | 1 | 829591 | 24.7 |
| 143 | 2280.836 | 2280.825 | 4.691 | 5 | 1 | 5 | 1 | 30533 | 26.6 |
| 144 | 2281.833 | 2281.845 | −5.609 | 5 | 3 | 5 | 276493 | 24.8 | |
| 145 | 2296.822 | 2296.820 | 1.088 | 6 | 5 | 1 | 58964 | 26.1 | |
| 146 | 2296.823 | 2296.820 | 1.350 | 6 | 5 | 1 | 122398 | 27.0 | |
| 147 | 2296.826 | 2296.820 | 2.569 | 6 | 5 | 1 | 100740 | 26.2 | |
| 148 | 2296.826 | 2296.820 | 2.743 | 6 | 5 | 1 | 60532 | 28.5 | |
| 149 | 2377.855 | 2377.840 | 6.434 | 9 | 2 | 3 | 12240 | 27.1 | |
| 150 | 2442.878 | 2442.878 | 0.287 | 6 | 1 | 5 | 1 | 23093 | 30.4 |
| 151 | 2442.881 | 2442.878 | 1.310 | 6 | 1 | 5 | 1 | 79868 | 26.9 |
| 152 | 2442.883 | 2442.878 | 2.047 | 6 | 1 | 5 | 1 | 23088 | 25.5 |
| 153 | 2442.884 | 2442.878 | 2.374 | 6 | 1 | 5 | 1 | 16939 | 26.1 |
| 154 | 2442.886 | 2442.878 | 3.275 | 6 | 1 | 5 | 1 | 39494 | 27.7 |
| 155 | 2442.899 | 2442.878 | 8.637 | 6 | 1 | 5 | 1 | 7598 | 29.0 |
| 156 | 2442.899 | 2442.878 | 8.637 | 6 | 1 | 5 | 1 | 29203 | 30.4 |
| 157 | 2443.882 | 2443.898 | −6.424 | 6 | 3 | 5 | 20106 | 26.6 | |
| 158 | 2443.889 | 2443.898 | −3.601 | 6 | 3 | 5 | 13340 | 28.2 | |
| 159 | 2500.915 | 2500.920 | −1.759 | 6 | 2 | 6 | 20610 | 25.4 | |
| 160 | 2588.941 | 2588.936 | 1.931 | 6 | 2 | 5 | 1 | 7110 | 28.0 |
| 161 | 2645.940 | 2645.957 | −6.387 | 6 | 1 | 6 | 1 | 8623 | 25.4 |
| 162 | 2645.969 | 2645.957 | 4.535 | 6 | 1 | 6 | 1 | 32261 | 24.9 |
| 163 | 2661.956 | 2661.952 | 1.503 | 7 | 6 | 1 | 48468 | 27.0 | |
| 164 | 2661.961 | 2661.952 | 3.531 | 7 | 6 | 1 | 9259 | 27.5 | |
| 165 | 2661.962 | 2661.952 | 3.569 | 7 | 6 | 1 | 27192 | 26.5 | |
| 166 | 2661.970 | 2661.952 | 6.612 | 7 | 6 | 1 | 21375 | 28.0 | |
| 167 | 2808.017 | 2808.010 | 2.564 | 7 | 1 | 6 | 1 | 14398 | 32.1 |
| 168 | 2808.032 | 2808.010 | 8.013 | 7 | 1 | 6 | 1 | 6357 | 26.5 |
| 169 | 3757.327 | 3757.349 | −5.696 | 10 | 9 | 1 | 23101 | 23.0 | |
| 170 | 3831.386 | 3831.385 | 0.026 | 11 | 10 | 75673 | 26.8 | ||
ACKNOWLEDGMENTS
The authors thank Dr. Helen Chew and Dr. Kit Lam for the kind gifts of serum samples used in this study. The authors would also like to thank Prof. Xi Chen, Prof. Jerry Hedrick, Crystal Kirmiz, Eric D. Dodds, and Richard R. Seipert for their technical support and guidance. Funding was provided by the National Institutes of Health.
Abbreviations
- cps
counts per second
- dHex
deoxyhexose
- Hex
hexose
- HexNAc
N-acetylhexosamine
- NeuAc
sialic acid
- PGC
porous graphitized carbon
- SPE
solid-phase extraction
- XIC
extracted ion chromatogram
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Dwek RA. Glycobiology: Toward Understanding the Function of Sugars. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 2.Varki A, Cummings R, Esko J, Freeze H, et al. Essentials of Glycobiology. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- 3.Ratner DM, Adams EW, Disney MD, Seeberger PH. Tools for glycomics: mapping interactions of carbohydrates in biological systems. Chembiochem. 2004;5:1375–1383. doi: 10.1002/cbic.200400106. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XL. Roles of glycans and glycopeptides in immune system and immune-related diseases. Curr Med Chem. 2006;13:1141–1147. doi: 10.2174/092986706776360897. [DOI] [PubMed] [Google Scholar]
- 5.Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 6.Brooks SA, Dwek MV, Schumacher U. Functional and Molecular Glycobiology. BIOS Scientific Publ; 2002. [Google Scholar]
- 7.Butler M, Quelhas D, Critchley AJ, Carchon H, et al. Detailed glycan analysis of serum glycoproteins of patients with congenital disorders of glycosylation indicates the specific defective glycan processing step and provides an insight into pathogenesis. Glycobiology. 2003;13:601–622. doi: 10.1093/glycob/cwg079. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- 9.Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, et al. Frontiers in glycomics: bioinformatics and biomarkers in disease. An NIH white paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11–13, 2006) Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- 10.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 11.Sparks SE. Inherited disorders of glycosylation. Mol Genet Metab. 2006;87:1–7. doi: 10.1016/j.ymgme.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Morelle W, Flahaut C, Michalski JC, Louvet A, et al. Mass spectrometric approach for screening modifications of total serum N-glycome in human diseases: application to cirrhosis. Glycobiology. 2006;16:281–293. doi: 10.1093/glycob/cwj067. [DOI] [PubMed] [Google Scholar]
- 13.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 14.Kirmiz C, Li B, An HJ, Clowers BH, et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, et al. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5:1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 16.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 17.Williams TI, Toups KL, Saggese DA, Kalli KR, et al. Epithelial ovarian cancer: disease etiology, treatment, detection, and investigational gene, metabolite, and protein biomarkers. J Proteome Res. 2007;6:2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 18.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 19.Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, et al. Alterations in the serum glycome due to metastatic prostate cancer. J Proteome Res. 2007;6:1822–1832. doi: 10.1021/pr060664t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards E, Thomas-Oates J. Hyphenating liquid phase separation techniques with mass spectrometry: on-line or off-line. Analyst. 2005;130:13–17. doi: 10.1039/b405130k. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Borges J, Aturki Z, Rocco A, Fanali S. Recent applications in nanoliquid chromatography. J Sep Sci. 2007;30:1589–1610. doi: 10.1002/jssc.200700061. [DOI] [PubMed] [Google Scholar]
- 22.Davies M, Smith KD, Harbin AM, Hounsell EF. High-performance liquid chromatography of oligosaccharide alditols and glycopeptides on a graphitized carbon column. J Chromatogr. 1992;609:125–131. doi: 10.1016/0021-9673(92)80155-n. [DOI] [PubMed] [Google Scholar]
- 23.Davies MJ, Smith KD, Carruthers RA, Chai W, et al. Use of a porous graphitised carbon column for the high-performance liquid chromatography of oligosaccharides, alditols and glycopeptides with subsequent mass spectrometry analysis. J Chromatogr. 1993;646:317–326. doi: 10.1016/0021-9673(93)83344-r. [DOI] [PubMed] [Google Scholar]
- 24.Knox JH, Kaur B. Structure and performance of porous graphitic carbon in liquid chromatography. Journal of Chromatography. 1986;352:3–25. [Google Scholar]
- 25.Koizumi K, Okada Y, Fukuda M. High-performance liquid chromatography of mono- and oligo-saccharides on a graphitized carbon column. Carbohydrate Research. 1991;215:67–80. [Google Scholar]
- 26.Mechref Y, Novotny MV. Miniaturized separation techniques in glycomic investigations. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;841:65–78. doi: 10.1016/j.jchromb.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 27.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 28.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal Biochem. 1996;240:210–226. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 29.Itoh S, Kawasaki N, Ohta M, Hyuga M, et al. Simultaneous microanalysis of N-linked oligosaccharides in a glycoprotein using microbore graphitized carbon column liquid chromatography-mass spectrometry. J Chromatogr A. 2002;968:89–100. doi: 10.1016/s0021-9673(02)00951-2. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki N, Ohta M, Hyuga S, Hashimoto O, Hayakawa T. Analysis of carbohydrate heterogeneity in a glycoprotein using liquid chromatography/mass spectrometry and liquid chromatography with tandem mass spectrometry. Anal Biochem. 1999;269:297–303. doi: 10.1006/abio.1999.4026. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki N, Ohta M, Hyuga S, Hyuga M, Hayakawa T. Application of liquid chromatography/mass spectrometry and liquid chromatography with tandem mass spectrometry to the analysis of the site-specific carbohydrate heterogeneity in erythropoietin. Anal Biochem. 2000;285:82–91. doi: 10.1006/abio.2000.4739. [DOI] [PubMed] [Google Scholar]
- 32.Pabst M, Bondili JS, Stadlmann J, Mach L, Altmann F. Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal Chem. 2007;79:5051–5057. doi: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y-G, Jang K-S, Joo H-S, Kim H-K, et al. Simultaneous profiling of N-glycans and proteins from human serum using a parallel-column system directly coupled to mass spectrometry. Journal of Chromatography B. 2007;850:109–119. doi: 10.1016/j.jchromb.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Ninonuevo M, An H, Yin H, Killeen K, et al. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 35.Ninonuevo MR, Park Y, Yin H, Zhang J, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 36.Costello CE, Contado-Miller JM, Cipollo JF. A glycomics platform for the analysis of permethylated oligosaccharide alditols. J Am Soc Mass Spectrom. 2007;18:1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson NG, Wilson NL, Wirth HJ, Dawes P, et al. Negative ion graphitised carbon nano-liquid chromatography/mass spectrometry increases sensitivity for glycoprotein oligosaccharide analysis. Rapid Commun Mass Spectrom. 2004;18:2282–2292. doi: 10.1002/rcm.1626. [DOI] [PubMed] [Google Scholar]
- 38.Wong JW, Durante C, Cartwright HM. Application of fast Fourier transform cross-correlation for the alignment of large chromatographic and spectral datasets. Anal Chem. 2005;77:5655–5661. doi: 10.1021/ac050619p. [DOI] [PubMed] [Google Scholar]
- 39.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 41.An HJ, Lebrilla CB. Supression of sialylated by sulfated oligosaccharides in negative MALDI-FTMS. Israel Journal of Chemistry. 2001;41:117–128. [Google Scholar]
- 42.Zhang J, Lamotte L, Dodds ED, Lebrilla CB. Atmospheric pressure MALDI Fourier transform mass spectrometry of labile oligosaccharides. Anal Chem. 2005;77:4429–4438. doi: 10.1021/ac050010o. [DOI] [PubMed] [Google Scholar]
- 43.Yin H, Killeen K. The fundamental aspects and applications of Agilent HPLC-Chip. J Sep Sci. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- 44.Yin H, Killeen K, Brennen R, Sobek D, et al. Microfluidic chip for peptide analysis with an integrated HPLC column, sample enrichment column, and nanoelectrospray tip. Anal Chem. 2005;77:527–533. doi: 10.1021/ac049068d. [DOI] [PubMed] [Google Scholar]
- 45.Royle L, Campbell MP, Radcliffe CM, White DM, et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376:1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Yamada E, Tsukamoto Y, Sasaki R, Yagyu K, Takahashi N. Structural changes of immunoglobulin G oligosaccharides with age in healthy human serum. Glycoconj J. 1997;14:401–405. doi: 10.1023/a:1018582930906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total ion chromatograms of a) the neutral oligosaccharides, b) the neutral and anionic oligosaccharides, and c) the anionic oligosaccharides released from human serum.