The crystal structures of truncated forms of the Streptococcus pyogenes phage-encoded hyaluronate lyases HylP2 and HylP3 have been determined by molecular replacement to 1.6 and 1.9 Å resolution, respectively.
Keywords: carbohydrates, lyases, streptococci, bacteriophages
Abstract
The crystal structures of truncated forms of the Streptococcus pyogenes phage-encoded hyaluronate lyases HylP2 and HylP3 were determined by molecular replacement to 1.6 and 1.9 Å resolution, respectively. The truncated forms crystallized in a hexagonal space group, forming a trimer around the threefold crystallographic axis. The arrangement of the fold is very similar to that observed in the structure of the related hyaluronate lyase HylP1. The structural elements putatively involved in substrate recognition are found to be conserved in both the HylP2 and HylP3 fragments.
1. Introduction
Hyaluronic acid (HA) is an important biological polymer that is found in the extracellular matrix and connective and epithelial tissues of higher animals (Necas et al., 2008 ▶). HA is composed of a linear repetition of β-1–4-linked N-acetylglucosamine (GlcNAc) and d-glucuronic acid (GlcUA) subunits. The bacterial enzymes involved in the degradation of HA, hyaluronidases, are thus important virulence factors and have gathered particular interest given their critical role during pathogenesis (Hynes et al., 2000 ▶). In terms of their enzymatic mechanisms, hyaluronidases are divided into ‘true’ hyaluronidases of eukaryotic origin that cleave the glycosidic β-1–4 linkage using a substrate-assisted acid–base catalytic mechanism (Markovic Housley et al., 2000 ▶) and hyaluronate lyases of bacterial origin that hydrolyze the same linkage via a β-elimination mechanism (Jedrzejas et al., 2002 ▶; Fig. 1 ▶ a). At the primary sequence level, the CAZy classification (Cantarel et al., 2009 ▶) places the eukaryotic hyaluronidases into glycoside hydrolase family 56 and the bacterial hyaluronate lyases into polysaccharide lyase families 6, 8 and 16, the last family being exclusively comprised of streptococcal bacteriophage and prophage members.
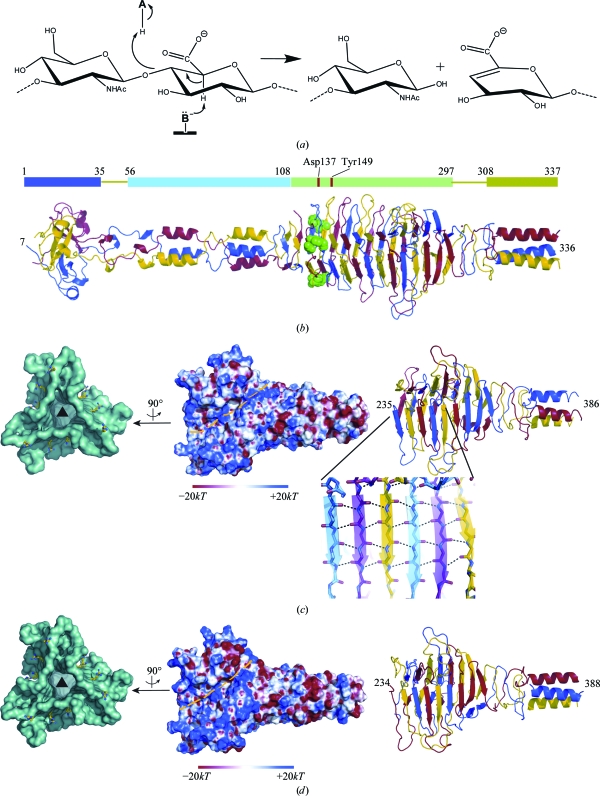
Figure 1.
(a) Scheme of the reaction catalyzed by bacterial hyaluronate lyases such as HylP1, HylP2 and HylP3. (b) Structure of the full-length HylP1 trimer with catalytic residues Asp137 and Tyr149 highlighted as green spheres and associated delineations of the various domains of HylP1, i.e. the N-terminal mixed globular α/β region (1–35, dark blue), the segmented α-helical coiled-coils region (56–108, light blue), the extended right-handed triple-stranded β-helix to residue 297 (light green) and the C-terminal α-helical region (308–337, dark green). Traces of the different monomers that conform the trimer are represented in blue, red and yellow. (c, d) Cartoon and electrostatic surface representations of the truncated forms of (c) HylP2 and (d) HylP3. The dashed yellow path on the electrostatic surfaces represents the putative binding groove for the HA polymer. The front-end view of the trimer surface (coloured in cyan) displays conserved arginine ladders (shown in ball-and-stick representation) that are likely to be involved in binding the negatively charged HA substrate. The electrostatic surfaces were calculated with APBS (Baker, 2004 ▶) and represented with PyMOL (DeLano, 2002 ▶).
Pathogenic bacteria such as Streptococcus pyogenes (group A streptococcus) produce an outer HA capsule in order to help to evade the human immune system (Cunningham, 2000 ▶). Phages that have co-evolved with these bacteria have acquired a hyaluronate lyase within their tail fibres to enable them to traverse the outer capsule during the infection process. In the genome sequence of serotype M1 S. pyogenes strain SF370 three prophage sequences have been localized (Ferretti et al., 2001 ▶) and each codes for a hyaluronate lyase. We have previously reported the full-length structure of one of these hyaluronate lyases, HylP1 (Smith et al., 2005 ▶), and in this article we describe the crystal structures of truncated forms of the remaining two prophage-encoded hyaluronidases HylP2 and HylP3.
2. Materials and methods
2.1. Molecular biology and protein purification
The full-length coding sequences of HylP2 (AAK33900.1) and HylP3 (AAK34249.1) were amplified from S. pyogenes SF370 (ATCC 700294) genomic DNA. The PCR products were cloned into expression vector pET28a (Novagen) on an NdeI–XhoI fragment for HylP2 and an NdeI–BamHI fragment for HylP3. N-terminal His6-tagged versions of HylP2 and HylP3 were produced in Escherichia coli BL21 (DE3) cultures harbouring the expression vectors following the protocol described in Smith et al. (2005 ▶) and purified using Ni2+-affinity chromatography and gel filtration. Pure protein samples were concentrated to 20 mg ml−1 and buffer-exchanged into 10 mM HEPES pH 7.4, 50 mM CaCl2 prior to structural studies.
2.2. X-ray crystallography
HylP2 and HylP3 protein samples were initially screened at 291 K using Crystal Screens 1 and 2 (Hampton Research). Using the hanging-drop vapour-diffusion method, 1 µl screen solution was mixed with 1 µl protein solution and placed on a well containing 0.5 ml mother liquor. Initial hits were further optimized and good diffracting crystals were prepared in 2 M NaCl, 10%(w/v) PEG 6000 for HylP2 and 0.2 M MgCl2, 0.1 M Tris–HCl pH 8.5, 30%(w/v) PEG 4000 for HylP3. MPD (2-methyl-2,4-pentanediol) was incorporated into the mother liquor at a concentration of 15%(v/v) to improve crystal quality and as a cryoprotectant. In both cases, hexagonal plates were harvested in rayon-fibre loops and bathed in cryoprotectant solution prior to flash-freezing in liquid nitrogen. Diffraction data were collected on European Synchrotron Radiation Facility (ESRF) beamline ID14-1 at a wavelength of 0.934 Å and a temperature of 100 K using an ADSC CCD Q210 detector. The HylP2 data were integrated using DENZO (Minor et al., 2006 ▶) and reduced with SCALEPACK (Minor et al., 2006 ▶). The HylP3 data were integrated with MOSFLM (Leslie, 1992 ▶) and reduced with SCALA from the CCP4 suite of programs (Collaborative Computational Project Number 4, 1994 ▶). Both the HylP2 and HylP3 structures were solved by molecular replacement using Phaser (Storoni et al., 2004 ▶). The HylP1 structure (PDB code 2c3f; Smith et al., 2005 ▶) was identified as the closest available molecular-replacement search module using BLAST (Altschul et al., 1990 ▶). HylP2 was solved using this model. The refined HylP2 model (PDB code 2wh7) was then used to solve the HylP3 structure. Model building was carried out with ARP/wARP (Perrakis et al., 1999 ▶). Water molecules were added to the model automatically using Coot (Emsley & Cowtan, 2004 ▶) and checked manually. All manual corrections to the model were made with Coot (Emsley & Cowtan, 2004 ▶).
Refinement cycles were performed with REFMAC (Murshudov et al., 1997 ▶). Diffraction data statistics and refined model parameters are shown in Table 1 ▶. Structural figures were drawn with PyMOL (DeLano, 2002 ▶). Coordinates and diffraction data have been deposited in the Protein Data Bank under codes 2wh7 and 2wb3 for HylP2 and HylP3, respectively.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the high-resolution outer shell.
| HylP2 | HylP3 | |
|---|---|---|
| Data processing | ||
| Space group | P6322 | P6322 |
| Unit-cell parameters (Å, °) | a = b = 50.8, c = 219.0, α = β = 90, γ = 120 | a = b = 49.5, c = 241.2, α = β = 90, γ = 120 |
| Molecules in ASU | 1 | 1 |
| Resolution range | 30.0–1.60 (1.66–1.60) | 30.0–1.90 (1.97–1.90) |
| Rmerge | 0.069 (0.172) | 0.028 (0.261) |
| 〈I/σ(I)〉 | 41.9 (13.1) | 36.7 (2.9) |
| Completeness | 91.2 (93.2) | 90.0 (51.0) |
| Redundancy | 4.4 (4.9) | 16.3 (11.8) |
| Refinement statistics | ||
| Resolution range (Å) | 109.76–1.60 | 19.58–1.90 |
| Rcryst | 0.212 | 0.189 |
| Rfree | 0.258 | 0.241 |
| No. of protein atoms | 1169 | 1283 |
| No. of solvent atoms | 200 | 101 |
| R.m.s.d. bonds (Å) | 0.021 | 0.020 |
| R.m.s.d. angles (°) | 1.717 | 1.846 |
| Mean B value (Å2) | ||
| Protein atoms | 17.1 | 33.9 |
| Solvent atoms | 29.8 | 36.7 |
| Ramachandran statistics† (%) | ||
| Preferred regions | 96.5 | 97.9 |
| Allowed regions | 2.8 | 2.1 |
| Outliers | 0.7 | 0.0 |
| PDB code | 2wh7 | 2wb3 |
Calculated using validation options in Coot.
3. Results and discussion
Samples of full-length HylP2 and HylP3 produced crystals suitable for X-ray analysis after one month of crystallization time. These crystals belonged to space group P6322, with a c-axis dimension (HylP2, c = 219 Å; HylP3, c = 241 Å) that was less than half of that observed for the HylP1 crystals (c = 586 Å). This length reduction along the c axis indicated a significant truncation in the crystallized proteins. HylP1 (Smith et al., 2005 ▶) models containing the N- and C-terminal portions of the structure were then used to solve the structure of HylP2 by molecular replacement (Fig. 1 ▶ b). Crystals of HylP2 were found to contain the C-terminal domain of the protein comprising residues Asn235–Leu386. The model of HylP2 was refined to an R cryst of 0.21 (R free = 0.26) using data to 1.6 Å resolution and was subsequently used to solve the structure of HylP3. HylP3 was also found to crystallize as a C-terminally truncated protein, in this case containing residues Thr234–Leu388. The HylP3 model was refined to an R cryst of 0.19 (R free = 0.24) using data to 1.9 Å resolution. As anticipated from the sequence similarity to HylP1 (HylP1, HylP2 and HylP3 share about 80% sequence identity with each other), the final refined models display an overall r.m.s.d. of 0.4 Å between each other. Statistics for data collection and refinement are given in Table 1 ▶.
The HylP2 and HylP3 fragments crystallized with one molecule in the asymmetric unit, forming a trimer around the threefold crystallographic axis. The trimers are constructed using an extensive inter-chain hydrogen-bond network that involves approximately 80% of the protein residues. Each monomer contributes about 2100 Å2 of accessible surface area (15% of the total surface area) to the formation of the trimer interface (Figs. 1 ▶ c and 1 ▶ d). The largest section of this interface (HylP2 residues Asn235–Ser322, HylP3 residues Thr234–Leu329) is formed by the intertwined interaction of β-strands contributed from each monomer in a sequential way. This interaction results in large and concave β-sheets that cover each face of the triangular-shaped tube. The fold is further stabilized by the packing of hydrophobic residues (mainly leucine and isoleucine) inside the central core. The structures then continue with a short stretch of antiparallel β-sheets (HylP2 residues Gly326–Lys350, HylP3 residues Leu329–Ala351) that each monomer forms on its own. This region is followed by a loosely structured region at the C-terminal side that ends with a triple α-helical bundle (HylP2 residues Lys373–Leu386, HylP3 residues Ala373–Leu388).
The catalytic residues in HylP1 are likely to be Asp137 and Tyr149 and these residues are located at the N-terminal end of the central β-sheet (Fig. 1 ▶ b). In the truncated forms of HylP2 and HylP3 the residues forming the catalytic dyad are not present but the rest of the structural features putatively involved in the recognition of HA are reiterated in the structures of HylP2 and HylP3. Thus, in both fragments a ladder of arginine side chains spaced about 14 Å was observed that may be involved in binding the carboxylate moieties of the glucuronic acid subunits of HA (Figs. 1 ▶ c and 1 ▶ d). The central groove containing the arginine ladder is edged by several loops that display residues with double conformations, indicating local disorder that might in turn be connected to substrate binding. The length of these central cavities in HylP2 and HylP3 and the superposition with the full-length fold of HylP1 also suggest the accommodation of HA substrates with up to eight glucuronic acid–N-acetyl glucosamine repeats; this is in agreement with activity studies on HylP1, in which the enzyme cleaves HA in an endo fashion, releasing mainly Δ4,5-unsaturated hyalurono-hexasaccharides and Δ4,5-unsaturated hyalurono-octasaccharides (Smith et al., 2005 ▶).
The structural solution of the HylP2 and HylP3 proteins sheds light on a class of hyalutonate lyase which has evolved both a structural role as part of a phage-tail fibre, as well as an enzymatic role, to aid the transition of a bacteriophage through the outer HA capsule of S. pyogenes.
Supplementary Material
PDB reference: HylP2, 2wh7, r2wh7sf
PDB reference: HylP3, 2wb3, r2wb3sf
Acknowledgments
We gratefully acknowledge financial support from the Royal Society and the Biotechnology and Biological Sciences Research Council (BBSRC). EJT is a Royal Society University Research Fellow.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol.215, 403–410. [DOI] [PubMed]
- Baker, N. A. (2004). Methods Enzymol.383, 94–118. [DOI] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res.37, D233–D238. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cunningham, M. W. (2000). Clin. Microbiol. Rev.13, 470–511. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Ferretti, J. J. et al. (2001). Proc. Natl Acad. Sci. USA, 98, 4658–4663.
- Hynes, W. L., Dixon, A. R., Walton, S. L. & Aridgides, L. J. (2000). FEMS Microbiol. Lett.184, 109–112. [DOI] [PubMed]
- Jedrzejas, M. J., Mello, L. V., de Groot, B. L. & Li, S. (2002). J. Biol. Chem.277, 28287–28297. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Markovic Housley, Z., Miglierini, G., Soldatova, L., Rizkallah, P. J., Muller, U. & Schirmer, T. (2000). Structure, 8, 1025–1035. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Necas, J., Brauner, P. & Kolar, J. (2008). Vet. Med. (Praha), 53, 397–411.
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Smith, N. L., Taylor, E. J., Lindsay, A. M., Charnock, S. J., Turkenburg, J. P., Dodson, E. J., Davies, G. J. & Black, G. W. (2005). Proc. Natl Acad. Sci. USA, 102, 17652–17657. [DOI] [PMC free article] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HylP2, 2wh7, r2wh7sf
PDB reference: HylP3, 2wb3, r2wb3sf
PDB reference: HylP2, 2wh7, r2wh7sf
PDB reference: HylP3, 2wb3, r2wb3sf