The first crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from an archaeon (P. horikoshii OT3) has been determined at 1.85 Å resolution. Comparative analyses of sequences and structures and modelling studies are presented.
Keywords: 4-methyl-5-β-hydroxyethylthiazole kinase, Pyrococcus horikoshii OT3, molecular modelling
Abstract
4-Methyl-5-β-hydroxyethylthiazole kinase (ThiK) catalyses the phosphorylation of the hydroxyl group of 4-methyl-5-β-hydroxyethylthiazole. This work reports the first crystal structure of an archaeal ThiK: that from Pyrococcus horikoshii OT3 (PhThiK) at 1.85 Å resolution with a phosphate ion occupying the position of the β-phosphate of the nucleotide. The topology of this enzyme shows the typical ribokinase fold of an α/β protein. The overall structure of PhThiK is similar to those of Bacillus subtilis ThiK (BsThiK) and Enterococcus faecalis V583 ThiK (EfThiK). Sequence analysis of ThiK enzymes from various sources indicated that three-quarters of the residues involved in interfacial regions are conserved. It also revealed that the amino-acid residues in the nucleotide-binding, magnesium ion-binding and substrate-binding sites are conserved. Binding of the nucleotide and substrate to the ThiK enzyme do not influence the quaternary association (trimer) as revealed by the crystal structure of PhThiK.
1. Introduction
Kinases are a ubiquitous group of enzymes that facilitate phosphoryl-transfer reactions from an ATP phosphate donor to an acceptor substrate. Thiamine is synthesized by most prokaryotes and by eukaryotes. In all cases, thiamine biosynthesis involves the separate formation of the pyrimidine and the thiazole. These are then coupled to form thiamine phosphate. A final phosphorylation catalyzed by 4-methyl-5-β-hydroxyethylthiazole kinase produces thiamine pyrophosphate (THI-PP), the biologically active form of the cofactor (Kawasaki, 1993 ▶). In bacteria, six gene products are involved in the formation of thiazole, whereas the formation of pyrimidine requires two. In contrast, only one gene product is required for biosynthesis of the thiazole and one for pyrimidine biosynthesis in yeast (Jurgenson et al., 2009 ▶). Thiamine can be transported into the cell and can be salvaged through several routes.
Labelling studies in Escherichi coli and Salmonella typhimurium have demonstrated that the thiazole is formed from tyrosine (Estramareix & Therisod, 1972 ▶; Bellion et al., 1976 ▶), deoxy-d-xylulose [without phosphate (David et al., 1982 ▶; Himmeldirk et al., 1996 ▶) or with phosphate (Nosaka et al., 1994 ▶)] and cysteine (Tazuya, Yamada et al., 1987 ▶). In Bacillus subtilis, the thiazole is formed from glycine rather than tyrosine (Tazuya, Morizaki et al., 1987 ▶). The de novo biosynthesis of this small molecule is complex and involves at least six gene products (ThiFSGH, ThiI and Dxs), with an additional gene product (ThiM) involved in salvage of the thiazole from the medium (Begley et al., 1999 ▶).
The 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) ring is generated through a complicated rearrangement reaction catalyzed by ThiC using 5-aminoimidazole ribotide (AIR) as the substrate. ThiD then phosphorylates HMP-P to give HMP-PP. Thiamine monophosphate (ThMP) is formed through the coupling reaction of 4-methyl-5-β-hydroxyethylthiazole phosphate (THZ-P) and HMP-PP using ThiE. Thiamine phosphate kinase adds the final phosphate group to ThMP to give thiamine disphosphate (ThDP), the active form of the cofactor. This reaction is carried out by ThiL in an ATP-dependent manner (McCulloch et al., 2008 ▶). ThDP can also be formed from thiamine in one step using thiamine pyrophosphokinase. In bacteria, this enzyme is called ThiN. In higher organisms, the thiamine pyrophosphokinase is THI80. As an alternative to the de novo pathway, thiamine or the components of thiamine can be salvaged. In bacteria, the thiazole alcohol can be converted to THZ-P by thiazole kinase (ThiM). In addition, HMP can be converted to HMP-P by HMP-P kinase (ThiD). ThiD is also the enzyme that converts HMP-P to HMP-PP in the de novo pathway. In bacteria, thiamine can be converted to ThMP by thiamine kinase (ThiK) or to ThDP by thiamine pyrophosphokinase (ThiN; Jurgenson et al., 2009 ▶).
Although sequence identity is not evident between ThiK enzymes from various sources (Fig. 1 ▶), they exhibit remarkable structural similarity and share a common nucleotide-binding and substrate-binding motif (Zhang et al., 1997 ▶; Petersen & Downs, 1997 ▶). This enzyme is a member of the ribokinase family of sugar kinases and has a structure similar to those of pyridoxal kinase (Newman et al., 2006 ▶), ribokinase (Sigrell et al., 1998 ▶; Andersson & Mowbray, 2002 ▶) and adenosine kinase (Mathews et al., 1998 ▶), as is evident from a DALI search (Holm & Sander, 1993 ▶). Thus, it is of interest to understand how phosphoryl transfer occurs in this particular group of enzymes and also to understand the variability and similarities between various ThiK enzymes.
Figure 1.
Sequence alignment of PhThiK with BsThiK (B. subtilis ThiK) and EfThiK (En. faecalis V583 ThiK) was performed using ClustalW (Thompson et al., 1994 ▶) and the figure was produced using ESPript (Gouet et al., 1999 ▶). The secondary-structural elements of PhThiK and residue numbering are shown above the sequence alignment. Sequence identities are highlighted in red and similarities are shown as red letters. Both identical and similar sequences are shown in blue boxes. The residues involved in the ATP, THZ-P and Mg2+ active sites of BsThiK, PhThiK and EfThiK are indicated by green stars, cyan triangles and orange circles, respectively. The conserved residues in 131 unique sequences of various ThiKs are indicated by blue circles.
The first detailed picture of the binding mode and catalytic reaction was obtained from the X-ray crystallograpic structure determination of complexes of the ThiK enzyme from B. subtilis (BsThiK) with the substrate THZ and ATP/THZ-P (Campobasso et al., 2000 ▶). Recently, the coordinates of the structure of ThiK from the pathogenic bacterium Enterococcus faecalis V583 (EfThiK) complexed with ADP (PDB code 3dzv) have been deposited in the Protein Data Bank (http://www.rcsb.org) by the Joint Center for Structural Genomics (JCSG). Although several archaeal ThiK proteins have been identified, none of them have been structurally and/or functionally characterized. The present paper is the first crystal structure report of an archaeal ThiK protein: that from Pyrococcus horikoshii OT3 at 1.85 Å resolution.
2. Materials and methods
2.1. Expression and purification
The 4-methyl-5-β-hydroxyethylthiazole kinase (ThiK) protein from the archaeon P. horikoshii OT3 (PhThiK) used in this study has a molecular weight of 28.95 kDa and consists of 265 amino-acid residues. The plasmid encoding PhThiK was digested with NdeI and BglII and the fragment was inserted into the expression vector pET-11a (Novagen) linearized with NdeI and BamHI. Escherichia coli BL21-CodonPlus(DE3)-RIL cells were transformed with the recombinant plasmid and grown at 310 K in Luria–Bertani medium containing 100 µg ml−1 ampicillin for 20 h. The transformants were cultured at 310 K overnight in a medium containing 1% polypeptone, 0.5% yeast extract, 0.5% NaCl, 100 µg ml−1 ampicillin pH 7.0 and 25 µg ml−1 chloramphenicol. The cells were lysed by sonication in 20 mM Tris–HCl pH 8.0 containing 500 mM NaCl, 5 mM β-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysate was incubated at 363 K for 13 min and centrifuged at 15 000 rev min−1 for 30 min at 277 K.
After buffer-exchange by loading it onto a HiPrep 26/10 desalting column (GE Healthcare Biosciences) pre-equilibrated with 20 mM Tris–HCl pH 8.0, the protein sample was loaded onto a Toyopearl Super Q-650 M column (Tosoh) pre-equilibrated with 20 mM Tris–HCl pH 8.0. The protein bound to the column and was eluted with a linear gradient from 0 to 400 mM NaCl. The protein-containing fraction was subjected to buffer-exchange to 20 mM Tris–HCl pH 8.0 and was loaded onto a Resource Q column (GE Healthcare Biosciences) pre-equilibrated with the same buffer. The protein was bound to the column and eluted in a linear gradient from 0 to 400 mM NaCl. The protein was subjected to buffer-exchange to 10 mM potassium phosphate buffer pH 7.0 and loaded onto a Bio-Scale CHT10-I column (Bio-Rad) pre-equilibrated with the same buffer. The protein was bound to the column and eluted with a linear gradient from 10 to 450 mM potassium phosphate buffer pH 7.0. The protein was subjected to gel filtration on a HiLoad 16/60 Superdex 200 pg column (GE Healthcare Biosciences) pre-equilibrated with 20 mM Tris–HCl and 200 mM NaCl pH 8.0. The protein-containing fraction was collected and the purified PhThiK protein was concentrated using Vivaspin 10 kDa cutoff (Sartorius). The final preparation was dissolved in 20 mM Tris–HCl containing 200 mM NaCl and 1 mM DTT pH 8.0. The purity was greater than 95% as analyzed by SDS–PAGE. The protein concentration was determined by measuring the absorbance at 280 nm (Kuramitsu et al., 1990 ▶). The yield of the purified protein was 1.9 mg per litre of culture.
2.2. Crystallization
Initial crystallization trials were attempted for the purified PhThiK protein (1.9 mg ml−1) in 20 mM Tris–HCl pH 8.0 containing 200 mM NaCl and 1 mM DTT using commercially available kits from Hampton Research. Crystals were obtained employing the microbatch-under-oil method with a 7:3 ratio of silicon and paraffin oils in Nunc HLA plates (Nalge Nunc International). Crystals suitable for X-ray diffraction studies grew in a few weeks at 293 K using 1 µl protein solution mixed with 1 µl of a solution containing 0.1 M CHESS buffer pH 9.3 and 0.88 M sodium citrate as the precipitant. After one month, crystals reached final dimensions of 0.20 × 0.20 × 0.35 mm. Prior to immersion in liquid nitrogen for storage and data collection, the crystals were transferred to mother liquor containing 25%(v/v) glycerol for cryoprotection.
2.3. X-ray data collection and analysis
Diffraction data were collected from a single crystal on the RIKEN Structural Genomics Beamline (BL26B1) at SPring-8 in Japan using a Rigaku R-AXIS VII image-plate detector (Rigaku MSC Co., Tokyo, Japan). The PhThiK crystal was briefly soaked in the mother liquor and was cooled in a nitrogen-gas stream at 100 K using the SPring-8 Precise Automatic Cryo-sample Exchanger (SPACE), which was controlled by BSS beamline-scheduling software (Ueno et al., 2004 ▶, 2005 ▶). The wavelength of the incident X-ray beam was 1 Å and a total of 180 frames were recorded. Each diffraction frame was taken with an oscillation angle of 1° and an exposure time of 10 s. The crystal-to-detector distance was kept at 200 mm. The crystal diffracted to beyond 1.85 Å resolution and the completeness of the final data was 99.8%. The data were processed using DENZO and SCALEPACK from the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶). Statistics related to data collection are given in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data statistics | |
| Space group | I23 |
| Resolution (Å) | 1.85 |
| Unit-cell parameters (Å, °) | a = b = c = 112.24, α = β = γ = 90 |
| VM (Å3 Da−1) | 2.1 |
| Solvent content (%) | 40.4 |
| Resolution range (Å) | 30.0–1.85 (1.92–1.85) |
| Measured reflections | 81063 |
| Unique reflections | 20162 (2004) |
| Rmerge† (%) | 6.7 (29.2) |
| Completeness (%) | 99.8 (99.5) |
| Average I/σ(I) | 19.5 (4.4) |
| Redundancy | 4.0 (3.9) |
| Refinement statistics | |
| Resolution (Å) | 20.0–1.85 |
| Rwork (%) | 16.4 |
| Rfree (%) | 18.4 |
| Protein atoms | 1950 |
| Water molecules | 208 |
| Phosphate ion | 1 |
| R.m.s.d. bond lengths (Å) | 0.005 |
| R.m.s.d. bond angles (°) | 1.30 |
| Estimated coordinate error (Å) | 0.16 |
| Overall B factor (Å2) | 18.90 |
| Ramachandran plot (%) | |
| Most favoured | 94.7 |
| Allowed | 5.3 |
| PDB code | 3hpd |
R
merge = 
 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl,  is the sum over all reflections and
is the sum over all reflections and  is the sum over i measurements of reflection h.
is the sum over i measurements of reflection h.
2.4. Structure determination and refinement
The structure was determined by the molecular-replacement technique using the AMoRe software package (Navaza, 1994 ▶). One subunit of the B. subtilis ThiK (BsThiK) structure crystallized in a monoclinic space group (PDB code 1c3q; Campobasso et al., 2000 ▶) was used as a search model. Calculation of rotation and translation functions using diffraction data between 10.0 and 4.0 Å resolution revealed a clear solution for a single subunit in the asymmetric unit with a correlation coefficient of 55% and an R factor of 46%. The initial model was subjected to rigid-body refinement. This was followed by positional refinement and simulated annealing using CNS (Brünger et al., 1998 ▶). 5% of reflections were used for the calculation of R free to monitor the progress of refinement. Both 2F o − F c and F o − F c maps were calculated from the phases based on the roughly refined model at this stage. The atomic model was built using the programs FRODO (Jones, 1985 ▶) and Coot (Emsley & Cowtan, 2004 ▶) in an iterative manner. The composite OMIT map was calculated at different stages of refinement in order to remove model bias. The R factor fell to below 30% using Coot (Emsley & Cowtan, 2004 ▶) and solvent molecules were gradually added to the model based on electron densities above 0.9σ and 2.8σ in 2F o − F c and F o − F c maps, respectively. Interestingly, we found a blob of positive electron density at the active site that could be interpreted as a phosphate ion and this was included in the model. At this stage, individual B-factor refinement was started and additional water molecules were included in the subsequent refinement cycle. The residues 132–143, which are part of a flexible loop, and the last residue (Leu265) of the C-terminal region could not be located in the electron-density map. The final R and R free values were 16.4 and 18.4%, respectively, and the stereochemical quality of the structure was validated using PROCHECK (Laskowski et al., 1993 ▶). Statistics of the structural refinement and validation of the model are provided in Table 1 ▶.
2.5. Molecular modelling
The refined model of PhThiK was used for modelling studies after removing water molecules and the phosphate ion. Since no structures are available for complexes with the reactants or the products released after catalysis, this prompted us to model ADP and phosphorylated hydroxyethylthiazole (THZ-P) complexed with the refined PhThiK model based on the structures of the complexes of BsThiK and EfThiK. The complex of the reactant ATP and the substrate hydroxyethylthiazole (THZ) with PhThiK was also modelled in order to compare it with the available structures of BsThiK complexes (Campobasso et al., 2000 ▶). The ternary PhThiK–ADP–THZ-P complex was constructed by superimposing the C198S mutant of the BsThiK complex (PDB code 1esq; Campobasso et al., 2000 ▶) for THZ-P and using the EfThiK complex for ADP (PDB code 3dzv; Joint Center for Structural Genomics, unpublished work). Subsequently, a 5 Å water shell was generated around the ADP and THZ-P of the complex using Insight II (Accelrys Inc, San Diego, California, USA). After generating H atoms, the modelled complex was subjected to 100 cycles of steepest-descent energy minimization followed by conjugate-gradient energy minimization. The CVFF force field was used with a distance-dependent dielectric constant of unity. The protein Cα-trace atoms were tethered harmonically with a force constant of 419 kJ Å−2. The remaining atoms in the model were allowed to move freely during minimization. The model of the ternary complex of PhThiK with the reactant ATP and THZ was minimized in a similar manner to the model of the ternary complex of PhThiK with ADP and THZ-P.
3. Results and discussion
3.1. Quality of the PhThiK model
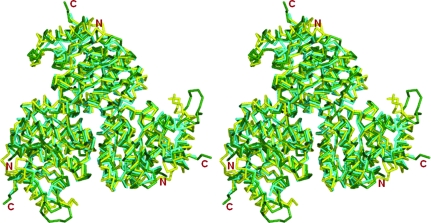
The crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from P. horikoshii OT3 (PhThiK) was solved by the molecular-replacement method. The asymmetric unit contained a single subunit with a phosphate ion at the binding pocket as represented in Fig. 2 ▶(a). The PhThiK structure was refined to an R factor of 16.4% (R free = 18.4%) at 1.85 Å resolution with standard deviations of bond lengths and bond angles of 0.005 Å and 1.3°, respectively. The final model of the enzyme consisted of 253 amino acids (residues 1–132 and 144–264), one phosphate ion and 208 water molecules. Residues 132–143, which are part of a flexible loop connecting helices α6 and α7, were disordered and the carboxy-terminal residue Leu265 was not visible in the electron-density maps. The quality of the refined protein model assessed using the program PROCHECK (Laskowski et al., 1993 ▶) showed that none of the residues were in the disallowed region of the Ramachandran plot. The structural superposition was carried out using ALIGN (Cohen, 1997 ▶) and surface areas were calculated using NACCESS (http://www.bioinf.manchester.ac.uk/naccess/). Figures were prepared using the program PyMOL (DeLano, 2002 ▶).
Figure 2.
Ribbon diagram showing (a) the PhThiK subunit in the asymmetric unit and (b) the quaternary assembly of the trimeric PhThiK structure. The phosphate ion is shown in ball-and-stick representation. Symmetry-related molecules are indicated A′ and A′′.
3.2. Trimeric structure of PhThiK
The PhThiK monomer in the asymmetric unit and its threefold-related partner constitute a trimer resembling those of the BsThiK (Campobasso et al., 2000 ▶) and EfThiK structures. The trimeric assembly of PhThiK can be described as a symmetrically triangular-shaped arrangement with dimensions of 75 × 66 × 45 Å (Fig. 2 ▶ b). In the trimeric crystal structure, intersubunit interfaces are formed with threefold symmetry-related partners by extensive hydrophobic interactions and hydrogen bonds including ion-pair interactions. The residues involved in hydrogen bonds between subunits are listed in Table 2 ▶. The hydrophobic interactions are formed by residues from helices (α2 and α10), η1 and various loops; these connect β1α2, β3α4, β4α5, η1α8 and α9α10 of molecule A with helices (α2, α3 and α10), β2 and residues forming part of loops (α1β1, β1α2, α2β2, β2α3) of the symmetry-equivalent molecule.
Table 2. Residues involved in hydrogen bonds between subunits in the PhThiK trimer.
| Subunits A and B | Subunits B and C | Subunits A and C | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | Distance (Å) | B | C | Distance (Å) | A | C | Distance (Å) | |
| 1 | Val25 O | Asn28 ND2 | 2.90 | Val25 O | Asn28 ND2 | 2.90 | Asn28 ND2 | Val25 O | 2.90 |
| 2 | Asp68 OD2 | His45 NE2 | 2.86 | Asp68 OD2 | His45 NE2 | 2.86 | Asn32 OD1 | Thr191 N | 2.66 |
| 3 | Thr99 OG1 | Glu49 OE1 | 2.78 | Thr99 OG1 | Glu49 OE1 | 2.78 | Leu35 O | Arg189 NH1 | 2.99 |
| 4 | Lys100 N | Glu52 OE1 | 2.79 | Lys100 N | Glu52 OE1 | 2.79 | His45 NE2 | Asp68 OD2 | 2.86 |
| 5 | Arg102 NE | Glu49 OE1 | 2.65 | Arg102 NE | Glu49 OE1 | 2.65 | Glu49 OE1 | Thr99 OG1 | 2.79 |
| 6 | Arg102 NH2 | Glu49 OE2 | 2.93 | Arg102 NH2 | Glu49 OE2 | 2.93 | Glu49 OE1 | Arg102 NE | 2.65 |
| 7 | Arg189 NH1 | Leu35 O | 2.99 | Arg189 NH1 | Leu35 O | 2.99 | Glu49 OE2 | Arg102 NH2 | 2.93 |
| 8 | Thr191 N | Asn32 OD1 | 2.66 | Thr191 N | Asn32 OD1 | 2.66 | Glu52 OE1 | Lys100 N | 2.79 |
| 9 | Gly236 N | Asp244 OD1 | 2.72 | Gly236 N | Asp244 OD1 | 2.72 | Asp244 OD1 | Gly236 N | 2.72 |
| 10 | Ser237 N | Asp244 OD1 | 2.82 | Ser237 N | Asp244 OD1 | 2.82 | Asp244 OD1 | Ser237 N | 2.82 |
| 11 | Ser237 OG | Asp244 OD2 | 2.53 | Ser237 OG | Asp244 OD2 | 2.53 | Asp244 OD2 | Ser237 OG | 2.53 |
3.3. Structure of PhThiK monomer
The monomeric structure of PhThiK in the asymmetric unit belongs to the ribokinase superfamily. The topology of the PhThiK structure shows the typical ribokinase fold (Campobasso et al., 2000 ▶; Sigrell et al., 1998 ▶; Mathews et al., 1998 ▶) of an α/β protein comprising of 11 α-helices, one 310-helix and a nine-stranded mixed β-sheet that has topology β2, β1, β3, β4, β5, β6, β7, β8, β9 (Fig. 2 ▶ a). As observed in other available three-dimensional ThiK structures, the first six strands (β2, β1, β3, β4, β5 and β6) are parallel to each other, whereas strands β6, β7, β8 and β9 are antiparallel. Helices α3, α4, α5, α6 and α7 flank one side of the central β-sheet and the remaining seven helices including a 310-helix (η1) form a bundle on the opposite side. Helices α2, α9 and α10 form a bundle adjoining the β-sheet.
Inspection of the difference electron-density map during the final stage of refinement showed a positive density peak in the region of the expected nucleotide-binding site into which a phosphate ion could be unambiguously fitted. The average B factor of the phosphate ion was 45.4 Å2. The phosphate ion occupies the position of the β-phosphate of ATP/ADP when the BsThiK and EfThiK complexes are superimposed. There is a direct hydrogen bond between one of the phosphate O atoms (O2) and Gly194 N. Moreover, water molecules form a cluster occupying both the nucleotide-binding and substrate-binding site cleft. The phosphate ion also interacts with the protein through water molecules. The residues involved in the water-mediated interactions are Lys118, Thr165, Leu187, Val190, Gly192 and Thr193. Also noteworthy is the water molecule (165) that occupies the position of the thiazole S atom that interacts with the backbone O atom of Gly65. Among the interacting residues, Gly65, Thr165, Leu187, Gly192 and Gly194 are identical in PhThiK, BsThiK and EfThiK.
3.4. Docking of ternary complexes of PhThiK
In the ribokinase-like kinases, the highly conserved Asp residue (replaced by Cys195 in PhThiK and Cys198 in BsThiK) is believed to serve as a catalytic base during the phosphorylation process. Since there are no structural models of the complexes with the reactants as well as with the products that are released upon the catalytic reaction of ThiK enzymes, we modelled the binding modes for the products ADP/THZ-P and reactants ATP/THZ with PhThiK as described in §2. The residues involved in the active site are indicated in Fig. 1 ▶. The active site is located at the interface between adjacent subunits. Each active site contains a thiazole-binding site and a nucleotide-binding site.
3.4.1. Thiazole binding site
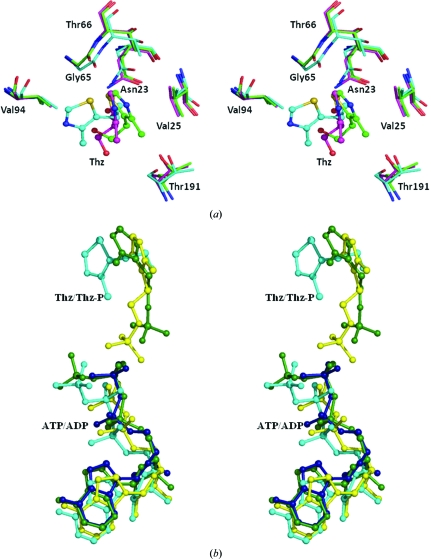
The phosphorylated hydroxyethylthiazole (THZ-P) and ADP conformations from the energy-minimized model do not substantially deviate from the crystallographically observed models. In the energy-minimized model of the ATP/THZ complex the orientation of the thiazole moiety of the substrate THZ is different, as can be seen in Figs. 3 ▶(a) and 3 ▶(b), when compared with other complexes (Campobasso et al., 2000 ▶). However, the interactions of THZ are nearly the same as those observed in other complexes. In the energy-minimized model, the THZ molecule interacts with the highly conserved residues Asn23, Gly65 and Val94. In the BsThiK complexes one face of the thiazole ring stacks against Val27 (Val25 in PhThiK), while the opposite face is more solvent-exposed. Interestingly, the thiazole ring in the PhThiK complex is moved towards Val94 and away from Val25. Interestingly, despite the difference in the relative orientation of the thiazole ring, the side chain of the catalytic Cys195 is also located at the bottom of the active site near to the hydroxyl group of the THZ in the energy-minimized model of PhThiK. The relative orientation of the residues involved in thiazole binding is nearly the same in both BsThiK and the PhThiK energy-minimized model (Fig. 3 ▶ a).
Figure 3.
(a) Stereoview showing the overlay of residues involved in the binding site of thiazole in the energy-minimized model of PhThiK and the crystal structures of BsThiK complexes. The ligand atoms are shown in ball-and-stick representation and the PhThiK (cyan) residue numbering is indicated. The R3 and P21 forms of BsThiK are depicted in green and magenta, respectively. (b) The relative orientation of nucleotide and substrate with respect to protein in energy-minimized models of PhThiK (cyan and yellow), the mutant complex of BsThiK (green) and EfThiK (blue).
The interactions between phosphorylated hydroxyethylthiazole and protein in the energy-minimized model of PhThiK are nearly the same as observed in the mutant complex of BsThiK, even though the THZ-P molecule points towards the β-phosphate of ADP. The phosphate O atoms of THZ-P are 3.2 and 3.5 Å from the β-phosphate of ADP. The thiazole S atom interacts with Gly65 O and the phosphate group of THZ-P is involved in hydrogen-bonding interactions with the backbone N atoms of Thr193 and Gly194 and with Cys195 SG. The side chain of Thr191 and the backbone atoms of Gly192 and Gly194 are only involved in van der Waals interactions.
3.4.2. Nucleotide binding site
In the BsThiK complex the ATP molecule stretches out along the C-terminal edge of the central β-sheet and is closest to strands β5, β6, β7 and β8. The adenine base is stacked between two loops. One of the loops connects β6 and β7 and the other loop is positioned before the 310-helix. There are no hydrogen bonds between the adenine base and the protein (Campobasso et al., 2000 ▶). The relative orientations of the substrate and the nucleotide with respect to the protein in the energy-minimized models of PhThiK and the crystal structures of BsThiK and EfThiK are shown in Fig. 3 ▶(b). Interestingly, in the energy-minimized model of the PhThiK complex the adenine N atoms are involved in hydrogen-bonding interactions with the backbone O atoms of the highly conserved Gly166 and Ala167. The backbone O atom of Asn182 also makes interactions with the adenine N atom. A similar interaction could also be inferred from the EfThiK complex. However, in the EfThiK complex the former two residues make contacts with sugar O atoms and the latter is similar to that of the PhThiK model. The adenine moiety of ADP makes hydrogen bonds to the backbone O atoms of Gly166 and Ala167 located in a loop between β6 and β7 on one side and the backbone O atoms of Asn182 and His184 on the other. The sugar ring of ADP mainly interacts with Asn182 O and the side chain of Asp169. Leu187 and Leu218 are only involved in van der Waals interactions with the sugar ring. The guanidinium moiety of the conserved Arg121 in the C198S-mutant BsThiK–ATP–THZ-P complex provides an additional electrostatic stabilization of the β-phosphate. The same interaction is also observed in the energy-minimized model of the PhThiK complex. However, this interaction is made via a water-molecule bridge in the EfThiK complex. In the energy-minimized model of PhThiK–ATP–THZ the ATP interactions with the enzyme are nearly similar to those in the C198S mutant of the BsThiK complex.
3.5. Structural comparison
A detailed comparative study between PhThiK and the two available ThiK structures from B. subtilis [wild type, PDB code 1ekq; wild-type complex with hydroxyethylthiazole (THZ) in R3 form, PDB code 1ekk; wild-type complex with THZ in P21 form, PDB code 1c3q; C198S mutant, PDB code 1esj; C198S mutant complex with phosphorylated THZ and ATP; PDB code 1esq] and E. faecalis V583 (wild-type complex with ADP; PDB code 3dzv) was carried out to explore the influence of the substrate and nucleotide binding on the tertiary and quaternary structure of ThiK.
3.5.1. Comparison of trimer
PhThiK has a sequence identity of 41% with BsThiK and of 25% with EfThiK, although the amino-acid sequences show considerable divergence outside of the binding-site and interfacial regions (Fig. 5). The overall structures from all three species closely resemble one another.
PhThiK exists as a monomer in the asymmetric unit and the trimer is formed with its symmetry partner, whereas the R3 form of BsThiK crystallized in two crystallographically independent subunits and each subunit further makes trimers with its symmetry equivalent. These two trimers are related by noncrystallographic translational symmetry. In the P21 form of BsThiK a single trimer exists in the asymmetric unit. All of the residues are only observed in the R3 and P21 forms of the wild type–THZ complexes. The residues in the loop that connects α6 and α7 are disordered in all remaining ThiK structures from all three species, including the wild-type and mutant BsThiK complexes. In the wild-type BsThiK structure this loop mainly interacts with some of the residues in α6 and also with the side chain (CB) of the conserved Asn123 (Asn120 in PhThiK and Asn121 in EfThiK). This hydrophobic interaction appears in both subunits. Interestingly, the side chain of this conserved Asn residue interacts with the γ-phosphate of the nucleotide in the mutant complex of BsThiK, whereas the β-phosphate is involved in this interaction in both EfThiK and the energy-minimized models of PhThiK. When the PhThiK trimer is superimposed on the trimers of different forms of wild-type as well as mutant BsThiK complexes, 735–749 pairs of Cα positions superpose with an r.m.s.d. ranging from 0.9 to 1.3 Å (Fig. 4 ▶).
Figure 4.
Stereo drawing of the structural superposition of the trimer of PhThiK (cyan) with trimers of BsThiK (green) and EfThiK (yellow).
In EfThiK, the two subunits are related by 21 screw symmetry in the asymmetric unit and each subunit further generates a trimer with its symmetry-related subunits. When the PhThiK trimer is superimposed on the trimers of the EfThiK complex, 687–705 pairs of Cα positions superpose with r.m.s.d. ranging from 1.3 to 1.4 Å (Fig. 4 ▶). Interestingly, a salt bridge is formed between the side chain of Arg49 of subunit A and Glu73 of subunit B in the asymmetric unit of EfThiK. The former positively charged residue is replaced by the negatively charged Glu48 in PhThiK and Glu50 in BsThiK, while the latter negatively charged residue is replaced by Arg72 in PhThiK and Val74 in BsThiK. Therefore, this ion-pair interaction is absent in both PhThiK and BsThiK. However, the corresponding structural equivalent of these residues is situated at the threefold interface in the PhThiK and BsThiK structures. It is worth mentioning that the residues involved in the twofold interface other than ion-pair interactions in EfThiK are not involved in the threefold interface in the ThiK structures from these three species.
The intersubunit interactions in the PhThiK structure are more extensive than those in the BsThiK complexes. The surface area buried on trimer formation in the former is larger by about 977–1104 Å2 than in the latter. It is only about 225 Å2 lower in the case of the EfThiK complex. The interfacial regions of the molecule are nearly the same in all three species and they are also reflected in the numbers of intersubunit hydrogen bonds, which are nearly the same in all three species, except for trimer I of the wild-type BsThiK structure. However, there are differences in the number of residues involved in the interfacial regions in the three species. There are 160 intersubunit nonbonded contacts in the structure of PhThiK as calculated using PDB-sum (http://www.ebi.ac.uk/pdbsum/), whereas there are 150 in the case of trimer I of EfThiK and 135 in trimer II. There are approximately 95 intersubunit nonbonded contacts in wild-type BsThiK and 82 contacts in the R3 form of the wild type THZ complex, whereas 106 nonbonded contacts are observed in the monoclinic form of the wild type THZ complex of BsThiK. Interestingly, the intersubunit nonbonded contacts in PhThiK and BsThiK appeared to be more nonpolar than in the EfThiK complex. This is because some of the charged residues in the interfacial regions in EfThiK are replaced by either hydrophobic or neutral charged residues in the PhThiK and BsThiK structures. In particular, Glu25, Glu29, Asp47, His67 and Asp245 are located at the interface in EfThiK. The corresponding equivalent residues in PhThiK (BsThiK) are Phe24 (Asn26), Asn28 (Asn30), Ala46 (Ala48), Thr66 (Thr68) and Ser237 (Ser243). Thus, the trimeric structures of both the PhThiK and EfThiK complexes can be expected to be somewhat stabler than those of the BsThiK complexes.
3.5.2. Comparison of monomers
When the PhThiK molecule was superimposed on the subunits of different forms of wild-type as well as mutant BsThiK complexes, 246–249 pairs of Cα positions superposed with r.m.s.d.s ranging from 0.9 to 1.0 Å. 238–244 pairs of Cα positions superposed with r.m.s.d.s ranging from 1.3 to 1.4 Å with substantial deviation in the N-terminal residues (some of which form the α1 helix) when EfThiK molecules were used for calculation. Thus, the tertiary and quaternary structures of ThiK are substantially unaffected by substrate and nucleotide binding.
3.6. Sequence analysis of ThiK
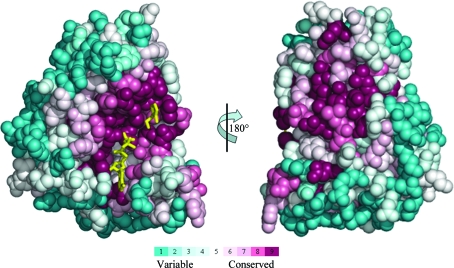
4-Methyl-5-β-hydroxyethylthiazole kinase sequences were extracted from the SWISS-PROT/TREMBL database with the PSI-BLAST algorithm using an E-value cutoff of 0.0001. The EfThiK sequence could not be detected when a PSI-BLAST E-value cutoff of 0.001 was used. A data set containing 131 unique sequences with less than 90% identity was constructed. The length of the sequences varied from 229 to 273 residues and the pairwise sequence identity varied from 25% to 82%. Multiple sequence alignment of these sequences was carried out using ClustalW (Thompson et al., 1994 ▶). The ThiK sequences were spread over three groups: 115 sequences from bacteria, 14 sequences from archaea and two sequences from yeast. The eukaryotic yeast sequences contained two domains (thiamine-phosphate pyrophosphorylase and hydroxyethylthiazole kinase). The conservation scores for each residue position in the 131 sequences were calculated using the program CONSURF v.3.0 (Landau et al., 2005 ▶) and were then mapped onto the three-dimensional structure of PhThiK (Fig. 5 ▶). The most highly conserved residues, indicated in maroon, are clustered around the substrate-binding and nucleotide-binding regions.
Figure 5.
The evolutionary conservation score of the amino-acid positions in the 131 ThiK sequences calculated by the CONSURF server and mapped onto the structure of the PhThiK monomer. The two pictures are related by a rotation of 180° about the vertical axis. The right face of the picture on the left and the left face of that on the right approximately correspond to the intersubunit interface. The nucleotide and substrate are also shown as yellow stick models.
As indicated in Fig. 1 ▶, 41 residues fall into the most conserved regions based on an analysis of 131 unique ThiK sequences from various sources as calculated using CONSURF v.3.0 (Landau et al., 2005 ▶). The residues involved in the nucleotide-binding and magnesium ion-binding sites fall into the most conserved regions. The residues involved in substrate binding, except for Leu187 in PhThiK, are also within the 41 most conserved residues. About 44 residues are involved in the intersubunit interfacial regions. Of these, 20 residues are highly conserved and 13 of them are conserved. The remaining 11 residues are variable. This indicates that 75% of the interfacial residues in the trimer are conserved. His45 and Asp68, which belong to a less conserved region, are involved in an intersubunit salt bridge. The residues involved in ATP/ADP binding and substrate binding in BsThiK and EfThiK are conserved. In ribokinase-like kinases (e.g. ribokinases, adenosine kinases and pyridoxal kinases), the highly conserved Asp residue (replaced by Cys195 in PhThiK) is believed to serve as a catalytic base during the phosphorylation process. It worth mentioning that only six sequences from Gram-positive bacteria, including EfThiK, have an Asp at this position; the other sequences contain Cys. This suggests that the majority of ThiKs utilize the Cys residue as an alternative catalytic base for the phosphorylation reaction. However, the specific activity of a mutant enzyme in which the Cys is replaced by Asp is ninefold higher than the native BsThiK enzyme (Campobasso et al., 2000 ▶).
4. Conclusions
The crystal structure of PhThiK from P. horikoshii OT3 was determined at 1.85 Å resolution. The PhThiK structure reported here is the first of an archaeal ThiK enzyme. The phosphate ion occupies the position of the β-phosphate of the nucleotide as observed in BsThiK and EfThiK complexes. Sequence analysis of 131 ThiK sequences from various sources revealed that 41 residues fall into the most conserved regions. The active-site residues are highly conserved, showing that they may have the same catalytic properties as in BsThiK. In the crystal, ThiK molecules from three different sources form a trimer with crystallographic or noncrystallographic threefold symmetry, irrespective of whether they are of the unbound form and/or of the bound nucleotide/substrate complex.
Supplementary Material
PDB reference: 4-methyl-5-β-hydroxyethylthiazole kinase, 3hpd, r3hpdsf
Acknowledgments
JJ thanks beamlines BL12B2 and BL26B1 of SPring-8 for excellent facilities and assistance and also thanks the Bioinformatics Centre, Pondicherry University, Pondicherry, India. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Andersson, C. E. & Mowbray, S. L. (2002). J. Mol. Biol.315, 409–419. [DOI] [PubMed]
- Begley, T. P., Downs, D. M., Ealick, S. E., McLafferty, F. W., Van Loon, A. P. G. M., Taylor, S., Campobasso, N., Chiu, H.-J., Kinsland, C., Reddick, J. J. & Xi, J. (1999). Arch. Microbiol.171, 293–300. [DOI] [PubMed]
- Bellion, E., Kirkley, D. & Faust, J. (1976). Biochim. Biophys. Acta, 437, 229–237. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Campobasso, N., Mathews, I. I., Begley, T. P. & Ealick, S. E. (2000). Biochemistry, 39, 7868–7877. [DOI] [PubMed]
- Cohen, G. E. (1997). J. Appl. Cryst.30, 1160–1161.
- David, S., Estramareix, B., Fischer, J.-C. & Therisod, M. (1982). J. Chem. Soc. Perkin Trans.1, 2131–2137.
- DeLano, W. L. (2002). The PyMol Molecular Graphics System. DeLano Scientific LLC, Palo Alto, California, USA.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Estramareix, B. & Therisod, M. (1972). Biochim. Biophys. Acta, 273, 275–282. [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Himmeldirk, K., Kennedy, I. A., Hill, R. E., Sayer, B. G. & Spenser, I. D. (1996). Chem. Commun., pp. 1187–1188. [DOI] [PubMed]
- Holm, L. & Sander, C. (1993). J. Mol. Biol.233, 123–138. [DOI] [PubMed]
- Jones, T. A. (1985). Methods Enzymol.115, 157–171. [DOI] [PubMed]
- Jurgenson, C. T., Begley, T. P. & Ealick, S. E. (2009). Annu. Rev. Biochem.78, 569–603. [DOI] [PMC free article] [PubMed]
- Kawasaki, Y. (1993). J. Bacteriol.175, 5153–5158. [DOI] [PMC free article] [PubMed]
- Kuramitsu, S., Hiromi, K., Hayashi, H., Morino, Y. & Kagamiyama, H. (1990). Biochemistry, 29, 5469–5476. [DOI] [PubMed]
- Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T. & Ben-Tal, N. (2005). Nucleic Acids Res.33, W299–W302. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Mathews, I. I., Erion, M. D. & Ealick, S. E. (1998). Biochemistry, 37, 15607–15620. [DOI] [PubMed]
- McCulloch, K. M., Kinsland, C., Begley, T. P. & Ealick, S. E. (2008). Biochemistry, 47, 3810–3821. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Newman, J. A., Das, S. K., Sedelnikova, S. E. & Rice, D. W. (2006). J. Mol. Biol.363, 520–530. [DOI] [PubMed]
- Nosaka, K., Nishimura, H., Kawasaki, Y., Tsujihara, T. & Iwashima, A. (1994). J. Biol. Chem.269, 30510–30516. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Petersen, L. & Downs, D. M. (1997). J. Bacteriol.179, 4894–4900. [DOI] [PMC free article] [PubMed]
- Sigrell, J. A., Cameron, A. D., Jones, T. A. & Mowbray, S. L. (1998). Structure, 6, 183–193. [DOI] [PubMed]
- Tazuya, K., Morisaki, M., Yamada, K., Kumaoka, H. & Saiki, K. (1987). Biochem. Int.14, 153–160. [PubMed]
- Tazuya, K., Yamada, K., Nakamura, K. & Kumaoka, H. (1987). Biochim. Biophys. Acta, 924, 210–215. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Ueno, G., Hirose, R., Ida, K., Kumasaka, T. & Yamamoto, M. (2004). J. Appl. Cryst.37, 867–873.
- Ueno, G., Kanda, H., Kumasaka, T. & Yamamoto, M. (2005). J. Synchrotron Rad.12, 380–384. [DOI] [PubMed]
- Zhang, Y., Taylor, S. V., Chiu, H.-J. & Begley, T. P. (1997). J. Bacteriol.179, 3030–3035. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: 4-methyl-5-β-hydroxyethylthiazole kinase, 3hpd, r3hpdsf
PDB reference: 4-methyl-5-β-hydroxyethylthiazole kinase, 3hpd, r3hpdsf