Mannosyl-3-phosphoglycerate synthase (MpgS) is a key enzyme in the biosynthesis of MG. Here, the purification, crystallization and preliminary crystallographic characterization of apo MpgS from Thermus thermophilus HB27 are reported.
Keywords: mannosyl-3-phosphoglycerate synthase, Thermus thermophilus HB27
Abstract
Mannosylglycerate (MG) is a compatible solute that is widespread in marine organisms that are adapted to hot environments, with its intracellular pool generally increasing in response to osmotic stress. These observations suggest that MG plays a relevant role in osmoadaptation and thermoadaptation. The pathways for the synthesis of MG have been characterized in a number of thermophilic and hyperthermophilic organisms. Mannosyl-3-phosphoglycerate synthase (MpgS) is a key enzyme in the biosynthesis of MG. Here, the purification, crystallization and preliminary crystallographic characterization of apo MpgS from Thermus thermophilus HB27 are reported. The addition of Zn2+ to the crystallization buffer was essential in order to obtain crystals. The crystals belonged to one of the enantiomorphic tetragonal space groups P41212 or P43212, with unit-cell parameters a = b = 113, c = 197 Å. Diffraction data were obtained to a resolution of 2.97 Å.
1. Introduction
Mannosylglycerate (MG) was first identified in red algae of the order Ceramiales (Bouveng et al., 1955 ▶). More recently, this solute has been found in thermophiles, such as the bacteria Rhodothermus marinus, Thermus thermophilus and Rubrobacter xylanophilus, and many hyperthermophilic archaea (Santos et al., 2007 ▶). The effect of heat stress on the solute pool of T. thermophilus has not been studied, but knockout mutants have been used to demonstrate the role of MG during osmoadaptation of this extreme thermophile (Alarico et al., 2007 ▶). Mannosylglycerate accumulates in R. marinus not only in response to osmotic stress, but also in response to above-optimal growth temperature (Silva et al., 1999 ▶). The finding that MG is widespread in marine hyperthermophiles and rarely found in mesophiles led to the view that this osmolyte is involved not only in osmoprotection but also in thermoprotection of cellular components. In fact, MG possesses a superior ability to protect enzymes against thermal denaturation in vitro and it has often been claimed to have potential utility in biotechnological and pharmaceutical applications (Borges et al., 2002 ▶; Faria et al., 2008 ▶).
The genes and enzymes involved in the synthesis of MG have been characterized in several (hyper)thermophilic bacteria and archaea (Martins et al., 1999 ▶; Borges et al., 2004 ▶; Empadinhas et al., 2001 ▶, 2003 ▶; Neves et al., 2005 ▶). In most organisms, MG synthesis proceeds via a two-reaction pathway. In the first step, GDP-α-d-mannose (GDP-man) and 3-phosphoglycerate are converted into mannosyl-3-phosphoglycerate (MPG) by the action of mannosyl-3-phosphoglycerate synthase (MpgS; EC 2.4.1.217). The phosphorylated intermediate is subsequently dephosphorylated by a specific phosphatase (MpgP; EC 3.1.5.70) to yield MG. Biochemical characterization of the enzyme led to the classification of mannosyl-3-phosphoglycerate synthase into the retaining glycosyl transferase family 55 (GT55; http://www.cazy.org). At present, this family only contains proteins that are homologous to T. thermophilus MpgS. In very rare cases, MG is synthesized in a single reaction catalysed by mannosylglycerate synthase (MgS; EC 2.1.4.–), which promotes transfer of the mannosyl group from GDP-man to d-glycerate (Martins et al., 1999 ▶). This enzyme has been classified into the retaining GT78 family and a front-face mechanism for glycosyl transfer with retention of anomeric configuration has been proposed (Flint et al., 2005 ▶). However, the catalytic mechanism of MpgS, the canonical enzyme for MG synthesis, has not been investigated.
Here, we describe the purification, crystallization and preliminary crystallographic analysis of apo MpgS from T. thermophilus HB27. Structural characterization of apo MpgS and its complexes with substrates or substrate analogues using X-ray crystallography will provide insights into the determinants of substrate specificity and catalysis.
2. Materials and methods
2.1. Expression and purification of MpgS from T. thermophilus HB27
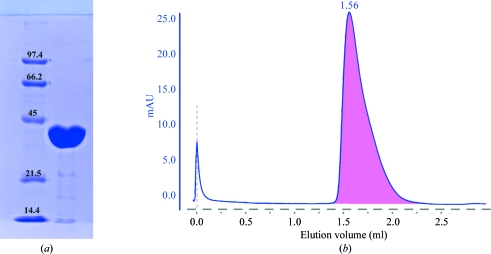
A plasmid containing the mpgS gene (accession No. YP_004563) cloned in pKK223-3 was kindly provided by M. S. da Costa, Coimbra, Portugal. Transformation, expression and purification of MgpS were performed as described previously (Empadinhas et al., 2003 ▶) with some modifications. Escherichia coli BL21 Rosetta strain (Novagen) cells bearing the construction were grown at 310 K in YT medium supplemented with ampicillin (100 µg ml−1) and chloramphenicol (40 µg ml−1) to an OD600 of 0.6–0.7 and induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) for 6 h. The cells were harvested by centrifugation (7000g, 15 min, 277 K) and suspended in 20 mM Tris–HCl buffer pH 7.6 containing 5 mM MgCl2 and a cocktail of protease inhibitors (Roche). Cells were disrupted in a French press and DNA was digested with benzonase (2 U ml−1; Novagen). Cell debris was removed by centrifugation (18 000g, 40 min, 277 K) and the cell extract was heated for 10 min at 353 K to precipitate the thermolabile proteins. After centrifugation (25 000g, 45 min, 277 K), the supernatant was applied onto a 70 ml Q-Sepharose column (GE Healthcare) equilibrated with buffer A (20 mM Tris–HCl pH 7.6). Elution was carried out with a linear gradient from buffer A to buffer B (20 mM Tris–HCl pH 7.6, 1 M NaCl). Fractions containing MpgS were eluted between 300 and 350 mM NaCl. The fractions were pooled and dialyzed against buffer A before loading them onto a 6 ml Resource Q column (GE Healthcare). MpgS eluted between 300 and 350 mM NaCl and the purest fractions were pooled and dialyzed against buffer A before loading them onto a 1 ml Mono Q column (GE Healthcare). The fractions eluted at 350 mM NaCl were pooled and loaded onto a 24 ml Superdex 75 10/300 GL column (GE Healthcare) equilibrated with buffer A containing 350 mM NaCl. In all chromatographic steps the MpgS activity was detected by visualizing the formation of MG derived from MPG after treatment with alkaline phosphatase by thin-layer chromatography (Empadinhas et al., 2003 ▶). The purified protein was judged to be pure by SDS–PAGE (12%), showing a single band with an apparent molecular mass of 43.6 kDa (Fig. 1 ▶ a). The protein concentration was determined using the Bradford assay (Bradford, 1976 ▶).
Figure 1.
(a) 12% SDS–PAGE of pure recombinant MpgS from T. thermophilus HB27. Lane 1, low-range molecular-weight markers (Bio-Rad): from the highest to the lowest weight, phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa) and lysozyme (14.4 kDa). Lane 2, MpgS monomer migrating according to its molecular weight (43.5 kDa). (b) Elution profile of MpgS loaded onto an analytical 2.4 ml Superdex 200 3.2/30 PC column. The elution volume suggests that the protein is most likely to be in the dimeric state. mAU stands for milliunits of absorption.
The molecular mass of MpgS was estimated by gel filtration (Fig. 1 ▶ b) using a 2.4 ml Superdex 200 3.2/30 PC column (GE Healthcare) equilibrated with 10 mM potassium phosphate buffer pH 7.2 containing 350 mM NaCl. Ribonuclease (13.7 kDa), chymotrypsinogen (25 kDa), ovalbumin (43 kDa), albumin (66 kDa), aldolase (158 kDa) and ferritin (440 kDa) were used as standards (GE Healthcare). Blue dextran 2000 (GE Healthcare) was used to determine the void volume of the column. MpgS eluted at the same volume (1.56 ml) as albumin, suggesting that a homodimeric structure is the prevalent oligomeric state under the running conditions.
2.2. MpgS crystallization and cryoconditions
Preliminary crystallization trials were performed on a nanolitre scale with the commercially available kits Crystal Magic I and II (BioGenova) using the Cartesian Crystallization Robot Dispensing System (Genomics Solutions) with round-bottom 96-well Greiner CrystalQuick plates (Greiner Bio-One). Three crystallization drops were prepared per condition screened, using 100 nl reservoir solution and 100 nl protein solution at three different concentrations: 5, 10 and 17 mg ml−1. The drops were equilibrated against 100 µl reservoir solution. Crystals with flat faces and sharp edges were often found in the presence of several kinds of PEG (polyethylene glycol) in a buffering system with pH between 7 and 9 using all three concentrations, but with the highest concentration yielding larger crystals. Although nicely shaped, these crystals were hard to handle and very little diffraction was observed. When studying the effect of divalent metals on crystallization, a new crystal form was found that was only obtained when ZnCl2 was added within a narrow concentration interval of 400–600 µM to the crystallization solution of Crystal Magic II condition No. 5, consisting of 0.2 M magnesium acetate tetrahydrate, 0.1 M sodium cacodylate pH 6.5, 30% 2-methyl-2,4-pentanediol (MPD). This result was reproducible and was used during crystal growth by scaling-up. The native protein was concentrated to 17 mg ml−1 and 2 µl drops (with a 1:1 ratio of protein to reservoir solution) were set up at 293 K in 24-well crystallization plates using the hanging-drop vapour-diffusion technique. The drops were equilibrated against 500 µl reservoir solution composed of 0.2 M magnesium acetate, 0.1 M sodium cacodylate pH 6.5, 30–35% MPD with 600 µM ZnCl2 as an additive. Prismatic crystals, as illustrated in Fig. 2 ▶, developed within 5 d in both cases, with dimensions varying between 100 and 400 µm in the longest axis.
Figure 2.
Crystals of T. thermophilus HB27 MpgS.
For data collection, the crystals were initially cryoprotected prior to flash-cooling in liquid nitrogen by transferring them directly into reservoir solution without Zn2+ and supplemented with 12.5% glycerol. The highest resolution observed was about 2.9 Å. The diffraction limit was independent of both crystal age and cryo-treatment, as room-temperature in-house data collection (Bruker AXS Proteum diffractometer and μStar rotating-anode generator, Montel mirrors, Cu Kα radiation) showed no improvement of the diffraction resolution. However, the cryo-treatment affected the internal crystal order, as revealed by spot mosaic spread and by the higher background level of the diffraction pattern. After several tests, an improved flash-cooling procedure consisting of two steps was used in which the crystallization buffer was replaced by sodium citrate. The crystals were first transferred into a solution containing 0.2 M magnesium acetate, 0.2 M sodium citrate pH 6.5, 32.5% MPD and left to equilibrate for about 3 min; they were then dipped into a cryosolution with the same composition as in the first step but supplemented with 5% glycerol and immediately flash-cooled in liquid nitrogen. The spot quality improved significantly and a lower background was observed in the diffraction pattern, but no increase in the resolution limit was achieved.
2.3. Data collection and processing
An X-ray diffraction data set was collected at 100 K from an apo MpgS crystal using an ADSC Q315r CCD detector on ESRF beamline ID14-4 (ESRF, Grenoble). The diffraction images were processed with the XDS program package (Kabsch, 1993 ▶) and further processing was carried out with the CCP4 program package (Collaborative Computational Project, Number 4, 1994 ▶). A summary of the data-processing statistics is presented in Table 1 ▶. The diffraction pattern was seen to suffer from anisotropy, which resulted in the unusually large values of R merge and R meas in Table 1 ▶, despite a reasonable value for 〈I/σ(I)〉 of around 2. The crystal belonged to one of the enantiomorphic tetragonal space groups P41212 or P43212, with unit-cell parameters a = b = 113.3, c = 197.1 Å. Matthews coefficient calculations (Matthews, 1968 ▶) indicated the presence of two or three molecules in the asymmetric unit, with corresponding values of V M = 3.58 Å3 Da−1 and a predicted solvent content of 65.7%, or V M = 2.39 Å3 Da−1 and an estimated solvent content of 48.5%, respectively. A self-rotation Patterson function calculation did not reveal any strong peaks corresponding to either 180° or 120° noncrystallographic rotation axes.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.3050 |
| Space group | P41212 or P43212 |
| Unit-cell parameters (Å) | a = 113.3, c = 197.1 |
| Resolution (Å) | 45.2–2.97 (3.10–2.97) |
| No. of observations | 507641 (75956) |
| Unique reflections | 26864 (4114) |
| Completeness (%) | 99.4 (96.7) |
| Rmerge† (%) | 10.8 (127.2) |
| 〈I/σ(I)〉 | 19.48 (2.18) |
| Rmeas‡ (%) | 11.1 (130.8) |
R
merge = 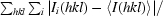
 × 100.
× 100.
Redundancy-independent R factor R
meas = 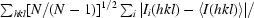
 × 100 (Diederichs & Karplus, 1997 ▶). For each unique Bragg reflection with indices hkl, I
i(hkl) is the ith observation of its intensity and N is its multiplicity.
× 100 (Diederichs & Karplus, 1997 ▶). For each unique Bragg reflection with indices hkl, I
i(hkl) is the ith observation of its intensity and N is its multiplicity.
3. Concluding remarks
The use of ZnCl2 as an additive was found to be indispensable in obtaining T. thermophilus MpgS crystals. It is hoped that the presence of this metal will afford a means of solving the phase problem via a MAD experiment at the Zn K absorption edge.
While this work was in progress, a report on the crystallization of MpgS from R. xylanophilus appeared in the literature (Sá-Moura et al., 2008 ▶). Despite catalysing the same reaction, this enzyme is highly unrelated to all MpgSs known to date and catalyzes the synthesis of both mannosyl-3-phosphoglycerate and glucosyl-3-phosphoglycerate. Accordingly, it has been classified into a different family (GT81) and is closely related to enzymes involved in the synthesis of glucosylglycerate, i.e. glucosyl-3-phosphoglycerate synthases. Despite their differences, comparison of the T. thermophilus and R. xylanophilus MpgSs is expected to shed light on the structural basis for the distinct substrate specificities of the two enzymes.
Acknowledgments
We would like to thank the ESRF for support during data collection and in particular the ID14-4 beamline staff. This work was funded by FCT grant PTDC/QUI/71142/2006. SG acknowledges FCT for grant SFRH/BD/23222/2005.
References
- Alarico, S., Empadinhas, N., Mingote, A., Simões, C., Santos, M. S. & da Costa, M. S. (2007). Extremophiles, 11, 833–840. [DOI] [PubMed]
- Borges, N., Marugg, J. D., Empadinhas, N., da Costa, M. S. & Santos, H. (2004). J. Biol. Chem.279, 9892–9898. [DOI] [PubMed]
- Borges, N., Ramos, A., Raven, N. D., Sharp, R. J. & Santos, H. (2002). Extremophiles, 6, 209–216. [DOI] [PubMed]
- Bouveng, H., Lindberg, B. & Wickberg, B. (1955). Acta Chem. Scand.9, 807–809.
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Empadinhas, N., Albuquerque, L., Henne, A., Santos, H. & da Costa, M. S. (2003). Appl. Environ. Microbiol.69, 3272–3279. [DOI] [PMC free article] [PubMed]
- Empadinhas, N., Marugg, J. D., Borges, N., Santos, H. & da Costa, M. S. (2001). J. Biol. Chem.276, 43580–43588. [DOI] [PubMed]
- Faria, T. Q., Mingote, A., Siopa, F., Ventura, R., Maycock, C. & Santos, H. (2008). Carbohydr. Res.343, 3025–3033. [DOI] [PubMed]
- Flint, J., Taylor, E., Yang, M., Bolam, D. N., Tailford, L. E., Martinez-Fleites, C., Dodson, E. J., Davis, B. G., Gilbert, H. J. & Davies, G. J. (2005). Nature Struct. Mol. Biol.12, 608–614. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Martins, L. O., Empadinhas, N., Marugg, J. D., Miguel, C., Ferreira, C., da Costa, M. S. & Santos, H. (1999). J. Biol. Chem.274, 35407–35414. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Neves, C., da Costa, M. S. & Santos, H. (2005). Appl. Environ. Microbiol.71, 8091–8098. [DOI] [PMC free article] [PubMed]
- Sá-Moura, B., Albuquerque, L., Empadinhas, N., da Costa, M. S., Pereira, P. J. B. & Macedo-Ribeiro, S. (2008). Acta Cryst. F64, 760–763. [DOI] [PMC free article] [PubMed]
- Santos, H., Lamosa, P., Faria, T. Q., Borges, N. & Neves, C. (2007). Physiology and Biochemistry of Extremophiles, edited by C. Gerday & N. Glansdorff, pp. 86–103. Washington: ASM Press.
- Silva, Z., Borges, N., Martins, L. O., Wait, R., da Costa, M. S. & Santos, H. (1999). Extremophiles, 3, 163–172. [DOI] [PubMed]