The hatching enzyme of zebrafish, ZHE1, was expressed, purified and crystallized using the hanging-drop vapour-diffusion method. The crystal belonged to space group P212121 and diffracted X-rays to a resolution of 1.14 Å.
Keywords: ZHE1, hatching enzymes, zebrafish
Abstract
The hatching enzyme of the zebrafish, ZHE1 (29.3 kDa), is a zinc metalloprotease that catalyzes digestion of the egg envelope (chorion). ZHE1 was heterologously expressed in Escherichia coli, purified and crystallized by the hanging-drop vapour-diffusion method using PEG 3350 as the precipitant. Two diffraction data sets with resolution ranges 50.0–1.80 and 50.0–1.14 Å were independently collected from two crystals and were merged to give a highly complete data set over the full resolution range 50.0–1.14 Å. The space group was assigned as primitive orthorhombic P212121, with unit-cell parameters a = 32.9, b = 62.5, c = 87.4 Å. The crystal contained one ZHE1 molecule in the asymmetric unit.
1. Introduction
Fish hatching enzymes are metalloproteases that are used to digest the egg envelope (chorion) at the time of embryo hatching (Kawaguchi et al., 2006 ▶). In the case of medaka fish (Oryzias latipes), for which the hatching enzymes have been well studied, two metalloproteases, HCE1 (high choriolytic enzyme; EC 3.4.24.67) and LCE (low choriolytic enzyme; EC 3.4.24.66), with different target sequences have been reported to be required to completely digest the chorion (Kawaguchi et al., 2005 ▶). HCE1 swells the chorion by its proteolytic reaction and LCE then fully digests the swollen chorion (Yasumasu et al., 1992 ▶; Inohaya et al., 1997 ▶). In contrast, zebrafish hatching requires only one metalloprotease, ZHE1. ZHE1 swells and softens the zebrafish chorion, which is then ruptured by contractile movements of the embryo (Sano et al., 2008 ▶). ZHE1 is more similar to HCE1 than to LCE both in amino-acid sequence (63 versus 53% identity) and in substrate specificity (Sano et al., 2008 ▶). To compare the catalytic mechanisms and substrate-recognition modes among the fish hatching enzymes and astacin, a crustacean digestive metalloprotease with 43% sequence identity to ZHE1, we have been conducting crystallographic studies of fish hatching enzymes. The crystal structure of astacin has been reported (Bode et al., 1992 ▶). Crystallization of the fish hatching enzyme HCE1 has also been reported (Kudo et al., 2004 ▶), but its crystal structure has not yet been reported. Here, we report the crystallization and preliminary X-ray analysis of the zebrafish hatching enzyme ZHE1.
2. Materials and methods
2.1. Purification and crystallization of ZHE1
ZHE1 was expressed as described previously (Sano et al., 2008 ▶) with slight modifications. Recombinant ZHE1 without any tags was expressed using pET-3c expression vector (Novagen) in Escherichia coli strain BL21 (DE3) (Novagen) grown in 1 l LB medium at 310 K. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM when the OD600 reached ∼0.6 and the culture was grown for another 4 h at 310 K. The cells were harvested by centrifugation at 5800g at 277 K for 10 min, resuspended in 10 ml 50 mM Tris–HCl pH 8.0 and 1 mM EDTA (buffer A) and frozen at 253 K. The frozen cells were then thawed, supplemented with 20 ml buffer A and a final concentration of 1.7 mg ml−1 lysozyme and disrupted by sonication. After centrifugation at 5800g and 277 K for 10 min, the resulting pellet was washed three times with 20 ml 5%(v/v) Triton X-100 in buffer A by sonication, incubation at 310 K for 30 min and centrifugation at 5800g and 277 K for 10 min. A quarter of the resulting pellet, which predominantly consisted of inclusion bodies, was dissolved in 500 µl 50 mM Tris–HCl pH 8.0, 8 M urea and 0.1 M 2-mercaptoethanol and incubated at 310 K for 1 h. After centrifugation at 5800g and 277 K for 10 min, the solubilized materials were poured into 1 l of 50 mM Tris–HCl pH 8.0 containing 1 mM reduced glutathione, 0.1 mM oxidized glutathione, 0.8 M l-arginine and 5 µM ZnSO4 and incubated at 277 K for more than 48 h. The refolding mixture was then dialyzed against 50 mM Tris–HCl pH 8.0 at 277 K. After filtration, the dialysate was loaded onto an SP Sepharose Fast Flow 5 ml column (GE Healthcare) and a stepwise elution was achieved with 50 mM Tris–HCl pH 8.0 containing 0.4 M NaCl. After dialysis against 50 mM Tris–HCl pH 8.0, the eluate was applied onto a Source 15S PE 4.6/100 column (GE Healthcare) in the HPLC system and an elution program with a linear gradient of 0–0.5 M NaCl in 25 mM Tris–HCl pH 8.0 was used. ZHE1 eluted as a sharp peak at ∼0.25 M NaCl. The peak component was collected, dialyzed against 50 mM Tris–HCl pH 8.0 and concentrated to 5 mg ml−1 using Amicon Ultra 15 Ultracel-10k (Millipore); the ZHE1 concentration was estimated from the absorbance at 280 nm (A 280) with the calculated molar extinction coefficient of ZHE1 (∊280 = 27 765 M −1 cm−1), which contained two Trp, 11 Tyr and six half-cystine residues. The crystallization experiments were performed by the hanging-drop vapour-diffusion method with VDX crystallization plates (Hampton Research). A crystallization drop was prepared by mixing 1.0 µl ZHE1 solution (5 mg ml−1 in 50 mM Tris–HCl pH 8.0) and 0.5 µl reservoir solution [0.1 M MES buffer pH 6.0, 0.2 M potassium sulfate and 20%(w/v) PEG 3350] and placed in vapour equilibration with 300 µl reservoir solution at 278 K.
2.2. X-ray data collection and processing
A single crystal of ZHE1 was prised apart and picked up in a nylon loop (Hampton Research), transferred to a cryoprotectant solution [0.08 M MES buffer pH 6.0, 0.16 M potassium sulfate, 16%(w/v) PEG 3350 and 20%(v/v) ethylene glycol] and then mounted for flash-cooling at 100 K using a nitrogen stream. X-ray diffraction data were collected from two different single crystals individually at the Photon Factory (Tsukuba, Japan). Data set 1 (the low-resolution data set), consisting of 180 images, was collected using an ADSC Quantum 210r detector system on beamline AR-NW12A with a wavelength of 1.000 Å, a crystal-to-detector distance of 117.1 mm, a rotation angle of 1.0°, a slit size of 100 × 100 µm and an exposure time of 5 s per image. Data set 2 (the high-resolution data set), consisting of 360 images, was collected using an ADSC Quantum 315r detector system on beamline BL-5A with a wavelength of 1.000 Å, a crystal-to-detector distance of 92.2 mm, a rotation angle of 0.5°, a slit size of 200 × 200 µm and an exposure time of 2 s per image. These two data sets were indexed and integrated separately in the resolution ranges 50.0–1.80 and 2.20–1.14 Å for data set 1 and data set 2, respectively. These data sets were then scaled together and merged, with an overlapping resolution range of 2.20–1.80 Å. The indexing, integration, scaling and merging of the diffraction data were performed with the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results
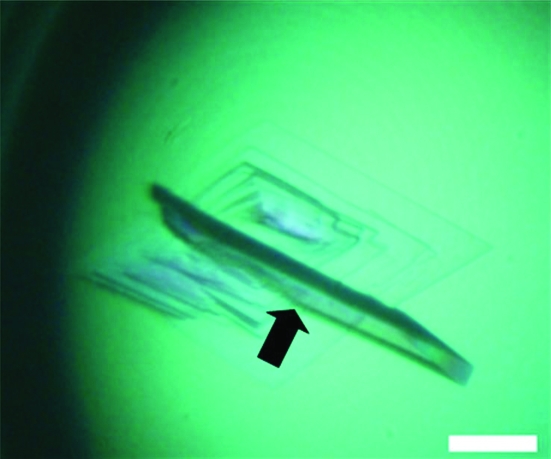
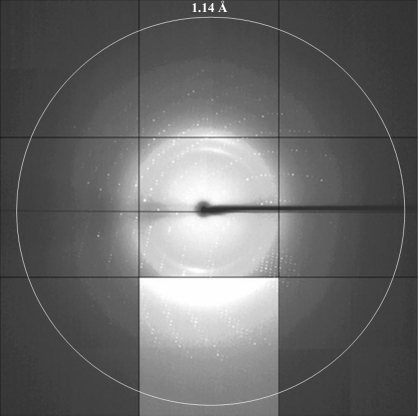
Typically, 0.6 mg purified ZHE1 was obtained from 1 l E. coli culture. Rod-shaped and thin-plate crystals of ZHE1 grew in a week under the conditions described in §2.1. X-ray diffraction data were obtained from these crystals at Photon Factory (Tsukuba, Japan). A rod-shaped crystal with approximate dimensions of 100 × 50 × 25 µm diffracted X-rays to 1.80 Å resolution and a data set was collected from this crystal (data set 1) in the resolution range 50.0–1.80 Å. The crystal belonged to space group P212121, with unit-cell parameters a = 32.9, b = 62.6, c = 87.5 Å. A thin-plate crystal with approximate dimensions of 250 × 200 × 25 µm (Fig. 1 ▶) diffracted X-rays to 1.14 Å resolution (Fig. 2 ▶) and a data set was collected from this crystal (data set 2) in the resolution range 50.0–1.14 Å. The crystal belonged to space group P212121, with unit-cell parameters a = 33.0, b = 62.7, c = 87.4 Å. Since the completeness in the low-resolution range (50.0–2.46 Å) of data set 2 was low (81.0%), the two data sets were merged to give a data set which was highly complete over the full resolution range 50.0–1.14 Å. The resolution ranges used to obtain the merged data set were 50.0–1.80 Å for data set 1 and 2.20–1.14 Å for data set 2. The statistics of the merged data set are summarized in Table 1 ▶. The Matthews coefficient (V M = 2.0 Å3 Da−1; Matthews, 1968 ▶) indicated that the crystal contained one ZHE1 molecule per asymmetric unit. Structure determination of ZHE1 by molecular replacement using the coordinates of astacin (42% sequence identity; PDB code 1ast; Bode et al., 1992 ▶), bone morphogenetic protein 1 (BMP-1) protease domain (37% sequence identity; PDB codes 3edg and 3edh; Mac Sweeney et al., 2008 ▶) and tolloid-like protease 1 (TLL-1) protease domain (38% sequence identity; PDB code 3edi; Mac Sweeney et al., 2008 ▶) as search models is currently under way.
Figure 1.
Single crystals of ZHE1 grown at 278 K using a reservoir solution containing 0.1 M MES buffer pH 6.0, 0.2 M potassium sulfate and 20%(w/v) PEG 3350. The thickest plate crystal indicated by the arrow in this figure was used to collect the high-resolution data set (data set 2). The white bar corresponds to a length of 100 µm.
Figure 2.
A diffraction image of the ZHE1 crystal from data set 2. The edge of the detector corresponds to a resolution of 1.01 Å. The middle lower panel is shown with enhanced contrast.
Table 1. Crystal and diffraction parameters for ZHE1.
The statistics are for the merged data set. Values in parentheses are for the highest resolution shell.
| X-ray source | Photon Factory AR-NW12A and BL-5A |
| Wavelength (Å) | 1.000 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 32.9, b = 62.5, c = 87.4 |
| Resolution range (Å) | 50.0–1.14 (1.18–1.14) |
| Observed reflections | 467403 |
| Unique reflections | 66154 |
| Data completeness (%) | 99.1 (93.9) |
| Redundancy | 7.1 (5.4) |
| Rmerge† | 0.058 (0.363) |
| 〈I/σ(I)〉 | 24.7 (4.0) |
R
merge = 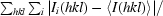
 , where I
i(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity of reflection hkl.
, where I
i(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity of reflection hkl.
Acknowledgments
The synchrotron-radiation experiments were performed on beamlines AR-NW12A and BL-5A at Photon Factory (Tsukuba, Japan) with the approval of Photon Factory, KEK (Proposal Nos. 2003S2-002 and 2006S2-006). This work was partly supported by the National Project on Protein Structural and Functional Analyses (NPPSFA), by the Targeted Proteins Research Program (TPRP) and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Bode, W., Gomis-Rüth, F. X., Huber, R., Zwilling, R. & Stöcker, W. (1992). Nature (London), 358, 164–167. [DOI] [PubMed]
- Inohaya, K., Yasumasu, S., Araki, K., Naruse, K., Yamazaki, K., Yasumasu, I., Iuchi, I. & Yamagami, K. (1997). Dev. Growth Differ.39, 191–197. [DOI] [PubMed]
- Kawaguchi, M., Yasumasu, S., Hiroi, J., Naruse, K., Inoue, M. & Iuchi, I. (2006). Dev. Genes Evol.216, 769–784. [DOI] [PubMed]
- Kawaguchi, M., Yasumasu, S., Shimizu, A., Hiroi, J., Yoshizaki, N., Nagata, K., Tanokura, M. & Iuchi, I. (2005). FEBS J.272, 4315–4326. [DOI] [PubMed]
- Kudo, N., Yasumasu, S., Iuchi, I. & Tanokura, M. (2004). Acta Cryst. D60, 725–726. [DOI] [PubMed]
- Mac Sweeney, A., Gil-Parrado, S., Vinzenz, D., Bernardi, A., Hein, A., Bodendorf, U., Erbel, P., Logel, C. & Gerhartz, B. (2008). J. Mol. Biol.384, 228–239. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sano, K., Inohaya, K., Kawaguchi, M., Yoshizaki, N., Iuchi, I. & Yasumasu, S. (2008). FEBS J.275, 5934–5946. [DOI] [PubMed]
- Yasumasu, S., Yamada, K., Akasaka, K., Mitsunaga, K., Iuchi, I., Shimada, H. & Yamagami, K. (1992). Dev. Biol.153, 250–258. [DOI] [PubMed]