Abstract
Background
Avian influenza virus (AIV) surveillance in birds is important for public health. Faecal droppings from wild-birds are more readily available for such studies, but the inability to identify the species-origin of faecal samples limits their value.
Objectives
Develop, optimise, and field-test a method to simultaneously detect AIV and identify the species-origin from faecal samples.
Study Design
Analytical sensitivity of the species-identification RT-PCR was assessed on serial dilutions of faecal droppings. Overall sensitivity of the methods for species-identification and AIV detection was assessed on 92 faecal and cloacal samples collected from wildlife, poultry markets, and experimentally H5N1-infected birds.
Results
All 92 samples were correctly identified to 24 different species, with a detection limit of 2.8μg of faecal material. All 20 specimens previously shown by virus culture to be positive for influenza virus were correctly identified by RT-PCR for influenza A using the same nucleic acid extracts used for species-identification.
Conclusions
We have optimised and evaluated a method for identifying the species of origin and detecting AIV from bird faecal droppings that can be applied to routine surveillance of influenza viruses in wild-birds.
Background
Aquatic wild-bird species are the natural reservoir for all 16 haemagglutinin (HA) and 9 neuraminidase subtypes of influenza virus.[1] Highly pathogenic avian influenza (HPAI) outbreaks in poultry arise from low pathogenic avian influenza(LPAI) viruses in wild-birds.[2] Some AIV also pose zoonotic and pandemic threats and are causes of concern for human health.[3,4,5] Past pandemics arose from LPAI viruses; thus, surveillance for pandemic preparedness must focus on both LPAI and HPAI viruses. An understanding of the ecology, evolution, and antigenic characteristics of LPAI and HPAI in wild-bird reservoirs is important for animal and public health.[6,7]
Surveillance is best carried out through oropharyngeal and cloacal swabs from trapped and identified wild-birds, but without ornithological expertise species-identification in wild-bird cloacal swabs and/or faeces may not be reliable. Many LPAI viruses are readily detected in cloacal swabs and from faeces.[1] While faeces are more accessible and often the only specimens available, they are difficult to determine species-origin. A method to accurately identify species-origin will allow these specimens to provide valuable information. Therefore, a reliable method to accurately identify species-origin of faecal samples without compromising the sensitivity to detect virus is beneficial for routine surveillance work.
Objectives
To develop, optimise, and field-test a technique to recover adequate DNA from low-level/quality host genomic DNA found in wild-bird faeces in order to identify the species-origin of faeces using DNA barcoding.[8] To detect AIV by RT-PCR using the same nucleic-acid extract.
Study design
Specimens
92 cloacal and faecal samples collected from wild-bird habitats and poultry markets, as well as experimentally H5N1-infected chicken (Gallus gallus) were used to evaluate our new methods. The swabs were collected into viral transport medium (VTM) prepared as previously mentioned[9] and stored at −80°C for up to 4.5 years. They had been tested for AIV by embryonated-egg inoculation and subtyped using standard methods.[9]
Host genomic DNA and viral RNA extraction
The faecal suspension in VTM was centrifuged to recover the faecal residue/pellet, which was then suspended in lysis buffer. Viral RNA and genomic DNA were extracted together using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) with modifications to manufacturer’s protocol. Samples were vortexed in a 5ml vial with lysis buffer and 1 tablet of InhibitorEX (Qiagen) to facilitate mixing. 2μg Carrier RNA was added to the lysis buffer-proteinase K-sample mixture to increase nucleic-acid yield. Purified DNA eluted from the column was concentrated with Microcon YM-100(Millipore, Billerica, MA) at 4°C to remove PCR inhibitors and concentrate the nucleic acid.
PCR amplification of mitochondrial cytochrome oxidase I (COI) gene for host-species identification and AIV M gene and H5 HA detection
PCR for the COI gene was performed using single and nested PCR methods (Table 1). Standard precautions were taken to minimize PCR cross-contamination. The nested PCR amplicons were purified directly using QIAgen PCR Purification Kit and used for sequencing. The amplified ~700bp PCR fragment of the COI gene was sequenced using 3730xl DNA Analyzer(Applied Biosystems, Foster City, CA), and analyzed by the barcoding software BOLD, which provides a taxonomic assignment to the query sequence by employing a linear search to collect nearest neighbours(lowest % divergence) from a global alignment of all reference sequences[11]. The BOLD database has 18,206 specimens with barcodes collected from 117 countries. 5779 bird-species have been deposited into the database for species-identification.[11] AIV M gene and H5 HA were detected by RT-PCR using previously described methods.[12,13] The forward and reverse primers for detection of M gene were 170801F-GGCATTTTGGACAAAKCGTCT and 170801R-CTTCTAACCGAGGTCGAAACG and for H5 HA were H5F-GCCATTCCACAACATACACCC and H5R-CTCCCCTGCTCATTGCTATG.
Table 1.
Experimental conditions and methodological details of the single PCR and nested PCR methods
| Single PCR | Nested PCR | |||||
|---|---|---|---|---|---|---|
| 1st Round | 2 Roundb | |||||
| Primersa | BirdF1(8) | BirdR1(8) | ExternalF1 (10) | ExternalR1 (10) | InternalF1 (10) | InternalR1 (10) |
| Sequence | 5′-TTCTCCAACCACAAAGACATTGGCAC-3′ | 5′-ACGTGGGAGATAATTCCAAATCCTG-3′ | 5′-TGTAAAAAGGWCTACAGCCTAACGC-3′ | 5′-GTRGCNGAYGTRAARTATGCTCG-3′ | 5′-AACAAACCACAAAGATATCGG3′ | 5′-TGGGARATAATTCCRAAGCCTGG-3′ |
| Pre-incubation | 95°C 10 min | 95°C 10 min | 95°C 10 min | |||
| Cycles | 45 | 45 | 35 | |||
| Denaturation | 95°C 10 sec | 95°C 10 sec | 95°C 10 sec | |||
| Amplification | ||||||
| Annealing | 58°C 10 sec | 58°C 10 sec | 55°C 10 sec | |||
| Elongation | 72°C 30 sec | 72°C 30 sec | 72°C 30 sec | |||
| Final Extension | 72°C 5 min | 72°C 5 min | 72°C 5 min | |||
| Size of Product | ~700bpc | ~700bp | ~670bpd | |||
For both methods, the master mix was prepared as follows: each tube had 9.8uL water, 2.4uL 25uM MgCl2, 0.4uL 25uM forward and reverse primers. 5uL of DNA sample was used for 20uL PCR reaction.
A 1000-fold dilution was performed after the first (external) round of PCR before adding 5uL of the PCR reaction to the second (internal) round RT-PCR reaction mix.
Final PCR product from the single PCR method was run in a 2% gel and the 700-base pair product was subsequently extracted using Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
The final PCR product from the nested PCR method was purified with the Qiagen PCR Purification Kit.
Determination of PCR sensitivity
Dilutions of the faecal swab pellet using lysis buffer were done before DNA extraction. Extraction and PCR was then performed as described above. To analyse the detection limit of the PCR methods in terms of copy number, serial dilution was performed on the DNA extract, and real-time PCR was performed using LightCycler480 according manufacturer’s protocol (Roche, Basal, Switzerland). Copy numbers determined from two different standards (746bp Gallus gallus COI PCR fragment cloned in a plasmid and 990bp PCR fragment of the cloned plasmid spanning the 746bp COI insert) were averaged.
Results
The single COI barcoding PCR method successfully amplified 57% of 47 faecal swab and 76% of 45 cloacal swab samples while the nested PCR successfully amplified all the specimens (Table 2). All PCR amplicons were successfully sequenced and led to identification of the correct species. In contrast, all 47 faecal and 45 cloacal samples were successfully PCR amplified and identified to the correct species-level by barcoding. Successfully identified specimens include 24 bird-species from 8 different Orders. Furthermore, all 20 of the 92 faecal and cloacal samples known to be AIV positive in egg culture were successfully detected by RT-PCR for AIV M gene or H5 HA. There was no evidence of PCR-cross contamination in the negative controls, which in some runs were added flanking each faecal specimen.
Table 2.
Comparison the two PCR methods in identifying the host species through DNA barcoding using fecal and cloacal samples
| Single PCR | Nested PCR | |||||
|---|---|---|---|---|---|---|
| Species | Order | No. tested | Successful PCR and Correct Species Identification | Mean Specimen Similarity ± Standard Deviation (%)e | Successful PCR and Correct Species Identification | Mean Specimen Similarity ± Standard Deviation (%) |
| FECAL SAMPLES | ||||||
| Anas clypeata | Anseriformes | 3 | 3 | 100±0 | 3 | 100.0±0.1 |
| Anas platyrhynchos | Anseriformes | 1 | 1 | 95.9±0 | 1 | 100±0 |
| Ardea cinerea | Ciconiiformes | 1 | 1 | 100±0 | 1 | 100±0 |
| Columba livia | Columbiformes | 2 | 1 | 100±0 | 2 | 100±0 |
| Gallus gallus | Galliformes | 29 | 18 | 97.9±4.4 | 29 | 99.9±0.3 |
| Numenius phaeopus | Charadriiformes | 1 | 1 | 100±0 | 1 | 100±0 |
| Phalacrocorax carbo | Pelecaniformes | 2 | 1 | 100±0 | 2 | 100±0 |
| Phasianus colchicus | Galliformes | 3 | 1 | 97.7±0 | 3 | 100±0 |
| Charadrius leschenaultii | Charadriiformes | 1 | 0 | 1 | 99.9±0 | |
| Platalea minor | Pelecaniformes | 3 | 0 | 3 | 99.7±0.1 | |
| Rostratula benghalensis | Charadriiformes | 1 | 0 | 1 | 99.9±0 | |
| Total | 47 | 27 | 47 | |||
| CLOACAL SAMPLES | ||||||
| Anas acuta | Anseriformes | 4 | 4 | 100±0 | 4 | 100±0.1 |
| Anas penelope | Anseriformes | 1 | 1 | 100±0 | 1 | 97.8±0.0 |
| Anas platyrhynchos | Anseriformes | 1 | 1 | 99.7±0 | 1 | 100±0 |
| Gallus gallus | Galliformes | 13 | 13 | 100±0 | 13 | 98.8±2.9 |
| Muscicapa dauurica | Passeriformes | 1 | 1 | 100±0 | 1 | 95.9±0 |
| Prinia gracilis | Passeriformes | 1 | 1 | 100±0 | 1 | 100±0 |
| Sturnus sericeus | Passeriformes | 8 | 8 | 99.9±0.3 | 8 | 99.9±0.1 |
| Tringa nebularia | Charadriiformes | 1 | 1 | 100±0 | 1 | 100±0 |
| Tringa totanus | Charadriiformes | 3 | 3 | 100±0 | 3 | 100±0 |
| Zosterops japonicus | Passeriformes | 1 | 1 | 100±0 | 1 | 100±0 |
| Acrocephalus orientalis | Passeriformes | 1 | 0 | 1 | 100±0 | |
| Halcyon smyrnensis | Coraciiformes | 1 | 0 | 1 | 94.9±0 | |
| Hirundo rustica | Passeriformes | 1 | 0 | 1 | 100±0 | |
| Luscinia cyane | Passeriformes | 1 | 0 | 1 | 99.9±0.0 | |
| Phalacrocorax carbo | Pelecaniformes | 1 | 0 | 1 | 100±0 | |
| Pycnonotus jocosus | Passeriformes | 1 | 0 | 1 | 99.9±0 | |
| Rostratula benghalensis | Charadriiformes | 5 | 0 | 5 | 99.9±0.0 | |
| Total | 45f | 34 | 45 | |||
Mean and standard deviation of specimen similarity in one species group: Similarity scores (specimen similarity) are calculated from the alignment of query sequence to that of known species in the barcoding database.
Four of the 45 cloacal samples did not yield a specific PCR product by the above methods. The DNA extracts from these 4 specimens, were re-extracted using the Qiagen Investigator Kit. Nested PCR was done for all 4 samples.
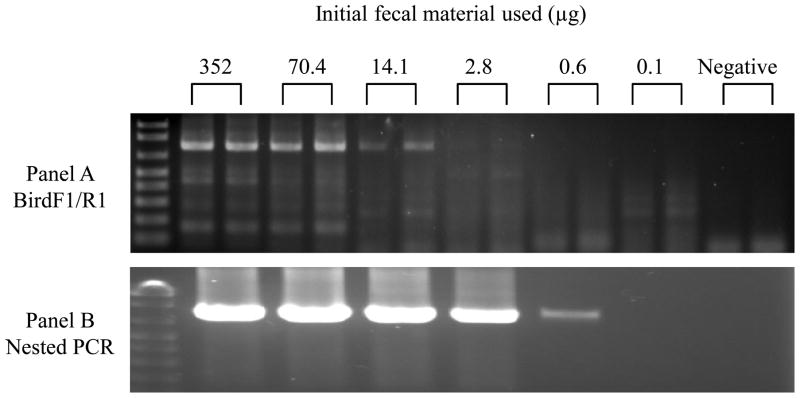
Typically, a swab collected from faeces picks up 45–75 mg of faecal material and contains on average 1.52 × 104 copies of COI(n=11 samples). Figure 1 shows the sensitivity of the single (panel A) and nested PCR (panel B) methods when applied to serial dilutions of a faecal specimen. The single PCR method (gel extraction required) allows for successful amplification and DNA barcoding of a 3,152-fold dilution of the initial faecal material (i.e. 14μg of the initial faecal sample). The nested PCR method can detect the equivalent of 2.8μg (15,625-fold dilution) of the initial faecal sample. Serial dilution of the DNA extract shows that the nested PCR method can successfully detect an average of as low as 3 to 4 genome copies of COI gene (n=4 samples). In all cases, the amount of faecal material required was well within what routine sampling provides.
Figure 1.
Comparison of the sensitivities of the single (panel A) and nested PCR (panel B) methods: Agarose gel electrophoresis of products from PCR amplification of partial fragments of mitochondrial COI from fecal samples. Serial dilutions were done to determine the minimal amount of initial fecal sample needed for successful species identification. Fecal sample in each reaction was respectively 352μg (representing a dilution of 1/625 of fecal swab material), 70.4μg, 14.1μg, 2.8μg, 0.6μg and 0.1μg. Negative control was distilled water.
Discussion
We demonstrate that DNA barcoding can be reliably applied to faecal and cloacal swabs to accurately identify species-origin, even in specimens stored for up to 4.5 years. The quantity of faecal material obtained on a swab is ample for this purpose. Furthermore, AIV detection by RT-PCR and DNA barcoding can be performed on the same nucleic-acid extract that contains both host DNA and viral RNA without loss of sensitivity. This method can be applied to field surveillance of AIV in wild-birds in two ways. Identification of species-origin of faeces by DNA barcoding can be done on the minority (usually <1%) of specimens found to be AIV positive by virus-culture or RT-PCR. Alternatively, all faecal surveillance samples can be processed with one nucleic acid extraction using PCR to detect species-origin of faeces and RT-PCR to detect the presence of AIV RNA. Other genes, mitochondrial (cytochrome b), ribosomal (16S), and nuclear (c-mos and glyceraldehyde-3-phosphodehydrogenase) can be amplified from these samples for a more detailed phylogenetic analysis of the host species (data not shown).
The use of faeces is less intrusive to sensitive ecological locations, and can provide information to generate environmental risk-maps for AIV transmission. In addition to AIV surveillance, this method is useful when investigating the role of wild-birds in AIV poultry outbreaks since lack of wildlife expertise often leads to the misidentification of wild-birds in the vicinity of outbreaks.[14] Identifying the species-origin of wild-bird faeces by our method in the vicinity of poultry outbreaks is therefore beneficial to AIV surveillance.
Available experimental studies suggest that HPAI H5N1 viruses are shed preferentially in the respiratory tract of birds[7,15] and if so faeces may not be the optimal specimen for HPAI H5N1 detection. However, many LPAI viruses are more readily detected in cloacal and faecal swabs,[1] and such viruses are equally, if not more relevant, to pandemic preparedness.
Acknowledgments
We acknowledge Paul Leader and his team in Asia Ecological Consultant Ltd. for bird trapping and identification. We also acknowledge research funding from the National Institutes of Health (NIAID Contract HHSN266200700005C) and the Area of Excellence Scheme of the University Grants Committee (Grant AoE/M-12/06) of the Hong Kong SAR Government. All experimental protocols, including those that involved H5N1 infection of chicken (Gallus gallus) followed the standard operating procedures of the approved bio-safety level-3 animal facilities and were approved by the Animal Ethics Committee of the University of Hong Kong with reference number CULATR 1226-06. The authors declare no conflict of interest.
Abbreviations
- AIV
Avian influenza virus
- HA
haemagglutinin
- HPAI
highly pathogenic avian influenza
- LPAI
low pathogenic avian influenza
- VTM
virus transport medium
- COI
cytochrome oxidase I
- RT-PCR
Reverse Transcriptase-Polymerase Chain Reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawaoka Y, Webster RG. Molecular mechanism of acquisition of virulence in influenza virus in nature. Microb Pathog. 1988;5:311–318. doi: 10.1016/0882-4010(88)90032-0. [DOI] [PubMed] [Google Scholar]
- 3.Webster RG, Laver WG. Studies on the origin of pandemic influenza. I. Antigenic analysis of A 2 influenza viruses isolated before and after the appearance of Hong Kong influenza using antisera to the isolated hemagglutinin subunits. Virology. 1972;48:433–444. doi: 10.1016/0042-6822(72)90054-2. [DOI] [PubMed] [Google Scholar]
- 4.Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avian Influenza technical Task Force, FAO. Should wild-birds now be considered a permanent reservoir of the virus? FAO AIDE News. 2006;40:1–13. [Google Scholar]
- 7.Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R, Osterhaus AD, Fouchier RA, Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of Birds through DNA Barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung YH, Zhang LJ, Chow CK, Tsang CL, Ng CF, Wong CK, Guan Y, Peiris JS. Poultry drinking water used for avian influenza surveillance. Emerg Infect Dis. 2007;13:1380–1382. doi: 10.3201/eid1309.070517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tavares ES, Baker AJ. Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol Biol. 2008;8:81. doi: 10.1186/1471-2148-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratnasingham S, Hebert PDN. bold: The Barcode of Life Data System. Mol Ecol Notes. 2007;1; 7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. ( http://www.barcodinglife.org) [DOI] [PMC free article] [PubMed]
- 12.Chan KH, Peiris JS, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J Clin Virol. 2008 May;42(1):65–9. doi: 10.1016/j.jcv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. Recommendations and laboratory procedures for detection of avian influenza A(H5N1) virus in specimens from suspected human cases. http://www.who.int/csr/disease/avian_influenza/guidelines/RecAIlabtestsAug07.pdf.
- 14.Whitworth D, Newman S, Mundkur T, Harris P. FAO Animal Production and Health Manual No. 5. Food and Agriculture Organization of the United Nations; Rome: 2007. Wild-birds and Avian Influenza: An Introduction to Applied Field Research and Disease Sampling Techniques; pp. 1–12. [Google Scholar]
- 15.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, Buranathai C, Nguyen TD, Chaisingh A, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79(17):11269–79. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]