Abstract
Cross-linking analysis of protein complexes and structures by tandem mass spectrometry (MS/MS) has advantages in speed, sensitivity, specificity, and the capability of handling complicated protein assemblies. However, detection and accurate assignment of the cross-linked peptides are often challenging due to their low abundance and complicated fragmentation behavior in collision-induced dissociation (CID). To simplify the MS analysis and improve the signal-to-noise ratio of the cross-linked peptides, we developed a novel peptide enrichment strategy that utilizes a cross-linker with a cryptic thiol group and using beads modified with a photocleavable cross-linker. The functional cross-linkers were designed to react with the primary amino groups in proteins. Human serum albumin was used as a model protein to detect intra- and intermolecular cross-linkages. Use of this protein-free selective retrieval method eliminates the contamination that can result from avidin–biotin based retrieval systems and simplifies data analysis. These features may make the method suitable to investigate protein–protein interactions in biological samples.
Formation of protein assemblies is a characteristic feature in biological pathways.1 Identification of protein–protein interactions within multiprotein complexes is essential for understanding biological mechanisms.2 Chemical modification of proteins within a complex, followed by identification of the modified residues, provides information on the surface accessibility of amino acid side chains and yields insight into those residues that are involved in interprotein contacts. More importantly, cross-linking can capture noncovalent protein–protein interactions and freeze transient complexes, facilitating subsequent purification, enrichment, and analysis.3,4
Many analytical approaches have emphasized the role of chemistry in the field of proteomics.5 Combining proteolytic digestion of cross-linked protein complexes and structure analysis by tandem mass spectrometry (MS/MS) provides advantages in speed and sensitivity and in the capability of handling large protein assemblies. However, the detection and accurate assignment of the cross-linked peptides are often challenging, complicated by the heterogeneity of the peptide mixture and low stoichiometry of the cross-linked products sought. Recently, Tang and co-workers reported a new type of cross-linker, the protein interaction reporter.6 It has specific fragmentation characteristics that can be exploited in low-energy collision-induced dissociation (CID)experiments (MS/MS), generating specific reporter ions that identify dead-end and intra- and intercross-linked species. In a subsequent article, they described the gas-phase fragmentation behavior of three similar cross-linkers.7 In low-energy CID experiments with Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS), these compounds showed selective fragmentation to generate specific product ions or reporter ions. In addition, photocleavable properties were incorporated in the reporter that allows release of cross-linked peptides from the biotin–avidin complex using UV light cleavage instead of solvent-based elution. Fluorescent affinity tags have also been used for enrichment and for subsequent analysis of peptides by MS.8
Unique affinity separation techniques coupled with cross-linking can directly minimize background contamination and, thus, improve MS sensitivity. Trester-Zedlitz et al. described a related approach employing avidin–biotin technology to retrieve cross-linked peptides.9 They introduced a modular solid-phase synthetic strategy for generating cross-linking reagents. This cross-linker has an affinity group and a cleavable isotope tag, reducing the complexity of the sample. Sinz and coauthors utilized high-resolution FTICR-MS to identify cross-linked peptides.10 Several other laboratories also reported experimental protocols to reduce the complexity of samples by constructing cleavable and isotope-labeled cross-linkers.11,12
The advantage of chemistry-based approaches is that they are robust and can selectively label macromolecular complexes in their functional native settings. The advancement of proteomics research requires the development of new chemical cross-linkers for global or large-scale identification of protein complexes.13 Additional groups can be added to the cross-linkers to provide functionalities for imaging, stabilizing, quantifying, and enrichment. Cleavable cross-linkers, in particular, can play a key role in selective enrichment of peptides. Cleavage can be achieved by chemical, enzymatic, thermal, or photolytic approaches. The use of photocleavable groups has greatly increased in recent years. Photocleavable, biotin-containing heterobifunctional reagents have also been developed. These reagents allowed easier detection and isolation of biomolecules, by first arresting the biomolecule of interest followed by the release of the reporter group. Lu and co-workers synthesized three new types of protein tagging reagents called visible isotope-coded affinity tags, which permit the absolute amount of a target protein or proteins to be quantified in a complex biological sample such as a eukaryotic cell lysate.14 A bead affinity chromatography system based on the photolytic elution method was integrated into a glass-silicon microchip to purify specific target proteins.15 In recent studies, magnetic beads were used to enrich peptides for MS analysis.16 For some special cases, specific chemicals/enzymes were required for selective bond cleavage. Most methods require the introduction of extra chemicals and severe cleavage conditions, either at an extreme pH, at a high salt concentration, and/or the extra addition of detergents or chaotropic agents, which often result in poor yield and a loss of protein function. Compared to these procedures, photocleavage is simple and no extra reagents are involved, hence, reducing the background contamination that is usually introduced by sample processing.17
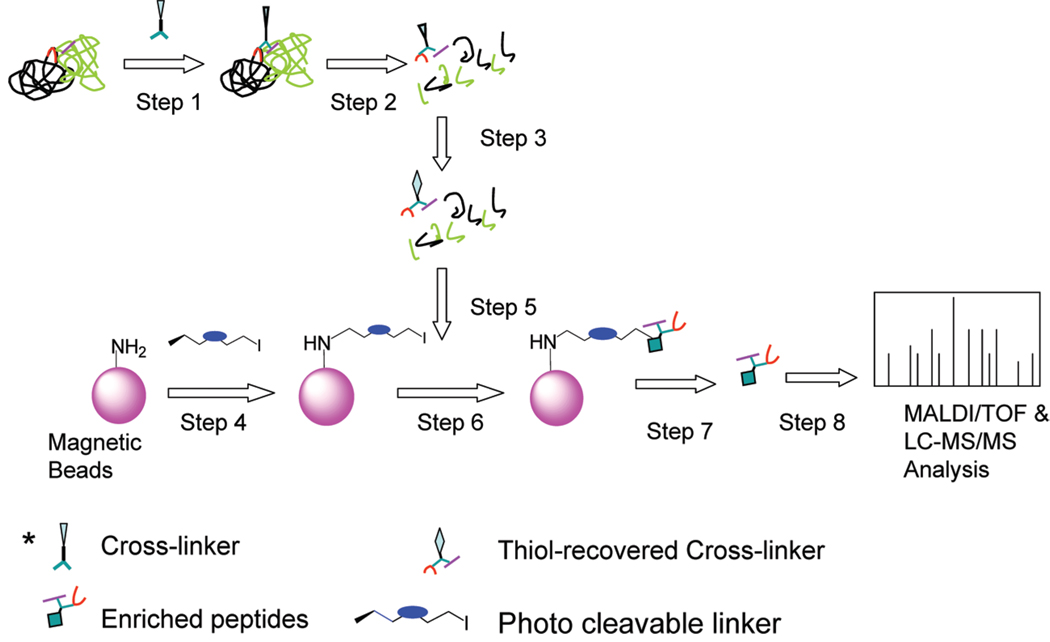
Here we report our proof-of-concept efforts to tackle this challenge by developing a selective retrieval methodology based on novel photocleavable cross-linking agents in order to directly enrich and identify cross-linked peptides obtained from protein complexes. Our approach consists of the following steps as illustrated in Figure 1: (1) generation of cross-links in the protein and protein complexes using a homobifunctional linker in which a cryptic group is embedded; (2) reduction and alkylation of Cys residues followed by digestion of proteins; (3) removal of the acetyl cryptic thiol protecting group with hydroxylamine; (4) modification of magnetic beads using a photocleavable cross-linker which supply iodo groups on the surface of magnetic beads; (5) binding of the cross-linked peptides to the magnetic beads, and the iodo group acts as a leaving group and generates a thiol ester bond with cross-linked peptides; (6) washing of the peptide resin to remove nonspecifically absorbed peptides; (7) enrichment of the cross-linked peptides via UV irradiation; (8) and identification of the sequences of the cross-linked peptide using tandem mass spectrometry. A major innovation of this work is the elimination of auxiliary enzymes and noncross-linked peptides from the procedure. This novel technique minimizes background contamination and improves MS analysis sensitivity. Consequently the information on protein complexes can be deciphered more effectively.
Figure 1.
Work flow for the selective retrieval of cross-linked peptides. Protein complexes are first reacted with the cross-linker (step 1) followed by proteolysis (step 2) and removal of the acetyl cryptic thiol protecting group with hydroxylamine (step 3). Modified magnetic beads, after incubating with the photocleavable cross-linker, (step 4) are mixed with the proteolytic peptides (step 5) followed by washing (step 6). The enriched anchored peptides are released using UV irradiation (step 7) and finally analyzed by tandem mass spectrometry (step 8).
MATERIALS AND METHODS
Chemicals
Reagent grade solvents were used without any purification unless specified; chemicals and general organic solvents were purchased from the Sigma-Aldrich Chemical Co. Thin layer chromatography (TLC) was performed with Analtech silica gel F254 TLC plates (250 µm). Flash chromatography was carried out using Aldrich silica gel (60 mesh). 1H nuclear magnetic resonance (NMR) and 13C NMR spectra were collected using a Bruker 300 MHz DMX instrument. A UV–vis absorption spectrometer was used to follow the changes in absorbance of succinimidyl 5-(3-iodo-propoxy)-2-nitro-benzyl carbonic ester (SINB) fragmentation products during photolysis. The UV–vis spectra were measured on a Hewlett-Packard 8453A diode array spectrophotometer, with a wavelength range from 190 to 1100 nm and a 1 nm bandwidth. The steady-state emission-spectral measurements were carried out with 1 cm × 1 cm quartz cells with a Spectra Physics Lab series 150-30 Nd: YAG laser as the irradiation source.
Synthesis of 2-Acetylsulfanyl-Succinic Acid Bis-Succinimidyl Ester (SAMS, Scheme 1a, III)
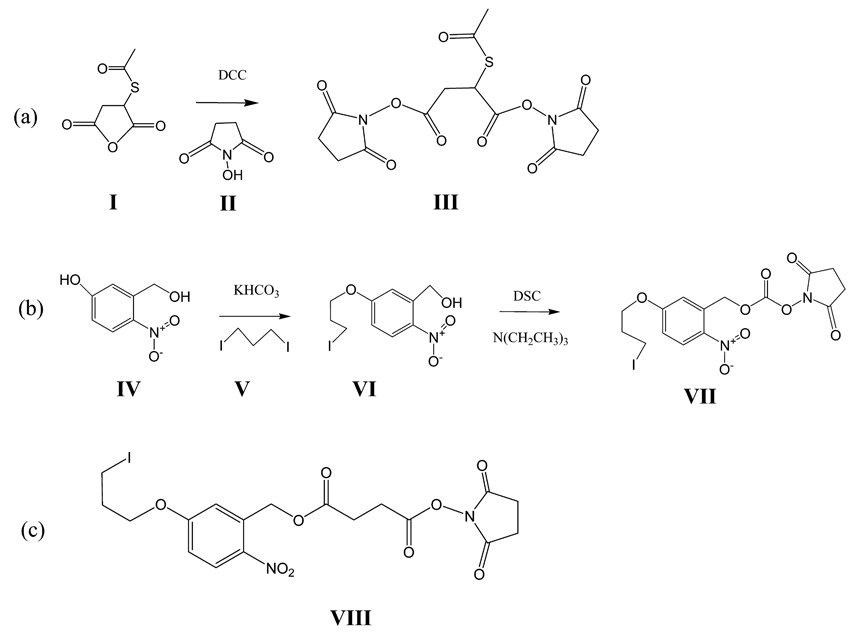
Scheme 1. Syntheses of Cross-Linkers a.
a (a) Synthesis of the selectively retrievable crosslinker, 2-acetylsulfanyl-succinic acid bis-succinimidyl ester (III, SAMS). Mercaptosuccinic anhydride (I) was reacted with N-hydroxysuccinic anhydride (II) forming SAMS (III). (b) Synthesis of the photocleavable cross-linker, succinimidyl 5-(3-iodo-propoxy)-2-nitro-benzyl carbonic ester (VII, SINB). 5-Hydroxy 2-nitro benzyl alcohol (IV) was reacted with di-iodo propane (V) to give 5-(3-iodopropoxy)-2-nitro benzyl alcohol (VI), which was react with disuccinimidyl carbonate (DSC) to give SINB (VII). (c) Structure of succinimidyl 5-(3-iodo-propoxy)-2-nitro-benzyl succinic ester (SSIN, VIII).
To a 10 mL acetonitrile solution of 345 mg (3 mmol) of N-hydroxysuccinimide (NHS) in a reaction vessel was added 1 mL of dimethylformaldehyde (DMF) solution of 174 mg (1 mmol) of S-acetyl succinimidyl anhydride dropwise. The reaction solution was stirred at room temperature for 30 min, and then 412 mg (2 mmol) of DCC was added. After 4 h of stirring, the dicyclohexylurea precipitate was removed by filtration and the solution was evaporated to near dryness. Product III was recrystallized in ethyl acetate/n-hexane (1:1 v/v) to give 290 mg of white crystal (yield 75%). 1H NMR (300 MHz, CDCl3): δ4.86(t, 1H), 3.35(m, 1H), 3.18(m, 1H), 2.71(s, 8H), 2.38(s, 3H).
Synthesis of 5-(3-Iodopropoxy)-2-Nitro Benzyl Alcohol (Scheme 1b, VI)
To a 20 mL DMF solution containing 360 mg (1.2 mmol) of compound V (1,3-di-iodo propane) and 120 mg (1.2 mmol) of KHCO3, 5 mL of a DMF solution containing 170 mg (1 mmol) of IV (5-hydroxy 2-nitro benzyl alcohol) was added dropwise. The reaction solution was stirred at 65 °C for 8 h. After the precipitate was removed by filtration, the solution was evaporated to near dryness. The product was then chromatographed on silica gel using ethyl acetate/n-hexane (1:4 v/v) to give 260 mg of VI, yellow powder (yield 77%). 1H NMR (300 MHz, CDCl3): δ8.15(d, 1H), 7.21(d, 1H), 6.90(d, 1H), 5.00(s, 2H), 4.16(t, 2H), 3.38(t, 2H), 2.20(m, 2H).
Synthesis of Succinimidyl 5-(3-Iodo-Propoxy)-2-Nitro-Benzyl Carbonic Ester (SINB, Scheme 1b, VII)
Disuccinimidyl carbonate (DSC), 250 mg (1 mmol), was dissolved in 25 mL of anhydrous acetonitrile. A total of 67 mg (0.2 mmol) of compound VI and 0.1 g (1 mmol) of N(CH2CH3)3 in 5 mL of acetonitrile were added dropwise under N2 protection in an ice bath. After 1 h of dropwise addition, the reaction was kept at room temperature and under N2 protection for 10 h. The solvent was then removed under vacuum, and the residue was dissolved in ethyl acetate and washed with 1% acetic acid and 1% NaHCO3, subsequently followed with deionized water washes, three times, and dried using Na2SO4. After rotary evaporation, the residue was then chromatographed on silica gel using ethyl acetate/n-hexane (1:1 v/v) to give 66 mg of SINB (yield 69%). 1H NMR (300 MHz, CDCl3): δ8.20(d, 1H), 7.11(d, 1H), 6.91(d, 1H), 5.80(s, 2H), 4.18(t, 2H), 3.32(t, 2H), 2.85(s, 4H), 2.20(m, 2H). The overall yield of the synthesis of SAMS and SINB are 75% and 53%, respectively.
Photolysis of SINB
Photolysis was performed with a 3.6 × 10−4 M SINB solution in CH3CN using a quartz cuvette. The irradiation was carried out at room temperature using Nd:YAG laser (Spectra Physics Lab series 150-30) with 355 nm UV irradiation. Irradiation time-dependent photolysis was monitored by periodically taking the absorption spectra of SINB (250–500 nm) to quantify the reaction quantum yield. Potassium ferrioxalate actinometry was used as a reference to determine the incident light intensity. Potassium ferrioxalate was recrystallized from warm water in the dark and dried in a vacuum oven. The concentration of the ferrioxalate was 0.006 M in water. The photoreaction of potassium ferrioxalate was monitored using UV–visible absorption.18 The amount of the product can be directly determined and then converted to the amount of photons delivered (in Einstein). The quantum yield (φ) of the photolysis of SINB in acetonitrile can then be calculated with the following equation using the above lamp flux value:
Thus, the amount of product can be determined by the absorbance change from UV–vis. C is a value that corrects for the incomplete light absorption at the excitation wavelength. C) = 1/(1 – T), where T is the transmittance of SINB before irradiation.
Cross-Linking of Standard Peptides
A peptide, CGQR (sequence CGQRSLQHPHKHAG, synthesized in our laboratory with average MH+ 1556.76) in PBS at 1.7 × 10−5 M, pH 8.5, was chosen to confirm the function of SAMS and to optimize cross-linking conditions. An acetonitrile solution of SAMS (2 µL) was added to 200 µL of CGQR solution. The mixture was kept at room temperature for 4 h. The mass of the CGQR peptide was monitored with matrix assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS).
Cross-Linking and Enzymatic Digestion of HAS
To 200 µL of freshly prepared human serum albumin (HSA, 1.67 × 10−5 M in 1% NaHCO3), 10 µL of SAMS was added (50 mM in acetonitrile), 2 µL aliquots every 5 min. After addition, the mixture was kept at room temperature for 2 h followed by the addition of 10 µL of 250 mM Tris-HCl buffer, pH 8.0, to quench any unreacted SAMS. To 50 µL of cross-linked HSA solution, dithiothreitol (DTT) (25 µL of 10 mM in 25 mM NH4HCO3) was added and kept at 56 °C for 1 h. Afterward, iodoacetamide (25 µL of 55 mM in 25 mM NH4HCO3) was added and kept in the dark at room temperature for 1 h. The mixture was desalted with a G-25 Sephadex quick spin column (Roche). Protein digestion is carried out by adding trypsin and Glu-C (1%, w/w) in 25 mM NH4HCO3, pH 8.4 and kept overnight at 37 °C. Formic acid is added to adjust the pH to 3–4, and the the sample was stored at −20 °C.
Selective Retrieval of Cross-Linked Peptides
To prepare deacetylated peptides, 5 µL of 100 mM freshly prepared N-hydroxylamine aqueous solution was added to 50 µL of digested peptides (pH adjusted to 8.0) and kept on a rotating platform for 1 h. At the same time, the magnetic beads (Dynabeads M-270 amine) were pretreated with PBS (pH 8.5) following the washing procedure in the manufacturer’s protocol. The beads were resuspended (2 × 108 beads, 0.01 µmol, 100 µL from stock solution) into 100 µL of freshly prepared SINB solution (5 × 10−4 M in acetonitrile/methanol, 1/9, v/v) with continuous shaking in the dark at room temperature. After 4 h, the SINB modified beads were removed and resuspended in the deacetylated peptides solution. The supernatant was removed after shaking in the dark for 1 h, and the beads were washed three times with 0.01% Tween20 with centrifugation at 3 rpm for 2 min between washes. A final wash with 100 µL of 50% acetonitrile/water (v/v) was performed on the beads, and the suspension was irradiated under UV laser light for 5 min. The output of the laser at 355 nm UV was 250 mJ at 30 Hz pulses. After irradiation, the supernatant was collected and concentrated to 50 µL using vacuum centrifugation. The samples were stored at −20 °C until MALDI-TOF and liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. Prior to mass spectrometric analysis, the samples were desalted/concentrated with a C18 ZipTip (Millipore Corporation) according to the manufacturer’s instructions.
MALDI-TOF Mass Spectrometry
MALDI-TOF mass spectrometry analysis of the cross-linked peptides was performed on a Voyager Biospectrometry DE-STR Workstation (Applied Bio-systems, Foster City, CA) equipped with a nitrogen laser (330 nm). The instrument was operated in positive ionization mode, and measurements were performed in the linear mode (mass range m/z 800–6000) using α-cyano-4-hydroxycinnamic acid as the matrix. A saturated matrix solution was prepared in 80% acetonitrile, 19.9% water, and 0.1% trifluoroacetic acid (TFA) (v/v/v). Samples were prepared using the dried droplet method by spotting 1 µL of matrix solution and 1 µL of sample solution onto the target. Spectra from 100 to 300 laser shots were accumulated to produce one spectrum. The instrument was calibrated using the monoisotopic masses of standard peptides: KPQQFFDLM (MH+1(mi) 1095.56), CQDSETRTFY (MH+1(mi) 1249.52), and YGGFMTSEKSQTPLVT (MH+1(mi) 1745.84).
LC–MS/MS Analysis
Proteolytic products of the cross-linked HSA and control reactions were analyzed by LC–MS/MS. The peptide mixtures were separated using C18-reverse phase (RP) chromatography on an Ultimate plus nano-high-performance liquid chromatography (HPLC) system (Dionex Corporation, Sunny-vale, CA). A C18 PepMap, 150 mm × 75 µm i.d., 3 µm, 100 Å (Dionex Corporation) column was used for separation with solvent A composed of 2% acetonitrile, 0.1% formic acid in water and solvent B of 80% acetonitrile, 0.1% formic acid in water. A gradient from 0% to 60% B in 90 min followed by isocratic elution with 90% B for 10 min was used after desalting at 100% A for X minutes. A LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA), equipped with a TriVersa NanoMate nanoelectrospray source (Advion BioSciences, Ithaca, NY), was coupled online to the nano-HPLC system. The electrospray ionization (ESI) voltage was set at 1600 V. The four most intense ions from an initial survey scan from m/z 300–1800 were selected for zoom scan and MS/MS. MS/MS was acquired using an isolation width of 2 m/z and a normalized collision energy of 35%.
Data Analysis
The cross-linked peptides were identified by manually screening the LC–MS/MS data. First, the cross-linker-modified and nonmodified HSA samples were digested separately under the same conditions. After collection of LC–MS/MS data, ions unique to the cross-linker-modified sample were found by comparing LC retention times and m/z values of ions detected in the LC–MS/MS runs from the two samples. These unique ions included cross-linked peptide along with dead-end peptides. Second, a de novo peptide sequence strategy to generate partial peptide sequences is implemented on the MS/MS spectra of the unique ions determined from as follows: After a partial sequence to the full sequence of HSA is matched, a predicted cross-linked peptide is located. Then, on the basis of the observed molecular mass and the MS/MS spectra, the other peptide chain of the cross-linked peptide pair is predicted. At this point, a list of predicted cross-linked peptide pairs is proposed. Finally, the cross-linked peptide pairs are confirmed by matching the product ions (e.g., b+, y+ ion series and/or internal product ions) observed in the MS/MS spectra to the theoretical product ions.
RESULTS AND DISCUSSION
Strategy for Enrichment of Cross-Linked Peptides and Design of Cross-Linkers
Development of new cross-linking strategies will allow researchers to study protein interactions at a higher resolution. To achieve nonprotein based enrichment, new functional cross-linkers were synthesized and utilized in processing peptides and proteins in the model system. Our approach is described in Figure 1. It is straightforward and can be summarized as follows: protein and protein complexes were cross-linked and then digested after reduction and alkylation; cryptic thiol groups were deprotected and then selectively react with photocleavable cross-linkers modified magnetic beads; finally, enriched peptides were collected after UV irradiation and then analyzed by tandem mass spectrometry in order to identify the sequences. The major innovation of this work is the elimination of protein elements, such as streptavidin, and noncross-linked peptides from the affinity capture procedure. This minimizes background contamination and improves MS sensitivity. As a result, the contact sites in the protein complex can be deciphered more easily. The cross-linkers and the synthetic schema are shown in Scheme 1.
When protein complexes are formed, proteins are closely associated to each other. Therefore, a shorter chain linker can provide more detailed information about complex formation. We chose succinic acid as the main bone of the cross-linker. The most common reactive groups in protein used for modification are primary amines or sulfhydryl groups of cysteine residues. Most linkers are primary amine reactive because of the relatively lower abundance of cysteine residues in proteins. N-Hydroxysuccinimide ester is a common group in popular linkers. Our strategy also utilizes the N-hydroxysuccinimide ester group along with a cryptic thiol group as the third functional group to provide further enrichment. The thiol is protected with an acetyl group that is easily removable with N-hydroxylamine, as shown in Scheme 2a. In order to anchor/release peptides for enrichment, we designed and synthesized a photocleavable cross-linker based on the o-nitrobenzyl group. Among the photocleavable moieties used to attach proteins to a substrate surface, the o-nitrobenzyl group19 is especially popular due to its unique properties, such as stability under ambient light, efficient cleavage upon exposure to UV irradiation, and nanosecond fast fragmentation reaction times upon photoexcitation.20 In this work, the photocleavable cross-linkers were designed to have a succinimidyl ester at one end and an iodo group at the other end (compound VII, Scheme 1b), thus allowing it to react with the amine modified beads. After the succinimidyl ester reacts with the amino group in the beads, the iodo group reacts with the thiol group of the beads for anchoring the cross-linked peptides.
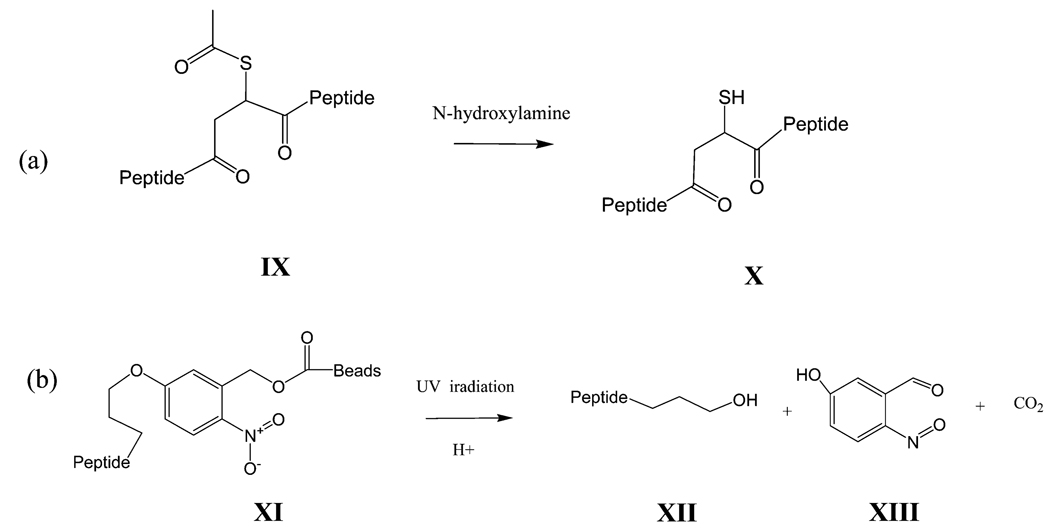
Scheme 2. Characterization of Cross-Linkers a.
a (a) Thiol recovery in a SAMS-crosslinked peptide. The thiol is protected with an acetyl group in crosslinked peptides (IX) that is removed by N-hydroxylamine to form thiol recovered cross-linked peptides (X). (b) UV cleavage of the photo cleavable crosslinker after modification. Crosslinked peptides were anchored on magnetic beads using a photocleavable spacer (XI). After UV cleavage, the enriched peptides (XII) were released and the photocleavable cross-linked formed nitrosobenzyldehyde (XIII).
Selection of Beads
Bead based affinity chromatography is a powerful method for purifying and preconcentrating specific proteins from crude biological samples. The target samples are accumulated and purified by an adsorption and elution process with ligand-containing beads.21 Several groups have reported on the enrichment method that utilizes biotin as the affinity tag.7,22 However, streptavidin must then be used for enrichment, which contributes noise during analysis from nonspecifically bound proteins. To achieve protein-free selective retrieval, SINB was used to conjugate the tagged/cross-linked peptides with amino-modified beads. The cross-linked peptides were chemically bound to the surface of the beads and detached using photoirradiation. Several commercially available beads were compared and among them, polystyrene magnetic beads were found to be sufficiently robust under the conjugating/irradiating conditions. Agarose beads (Sigma) were too fragile, and the beads fell apart during the rinse/irradiation step. Cleaved oligomers also yielded unacceptable levels of noise in MS analysis. Iron beads (Pierce) are sufficiently robust but so opaque that the UV light is blocked, and consequently less photolysis occurs on the surface of the beads. Polystyrene beads are also rather opaque, producing a dark yellow suspension. To address this issue, the bead suspension is diluted in a larger quartz cuvette that is used as the reaction container. Each irradiation reaction volume is 100 µL in a 1 mm thick, 1 cm × 1 cm quartz vial. After removal of unmodified peptides, the anchored peptides were released from the beads upon UV irradiation using laser pulses at 355 nm.
Characterization and Actinometry of SAMS and SINB
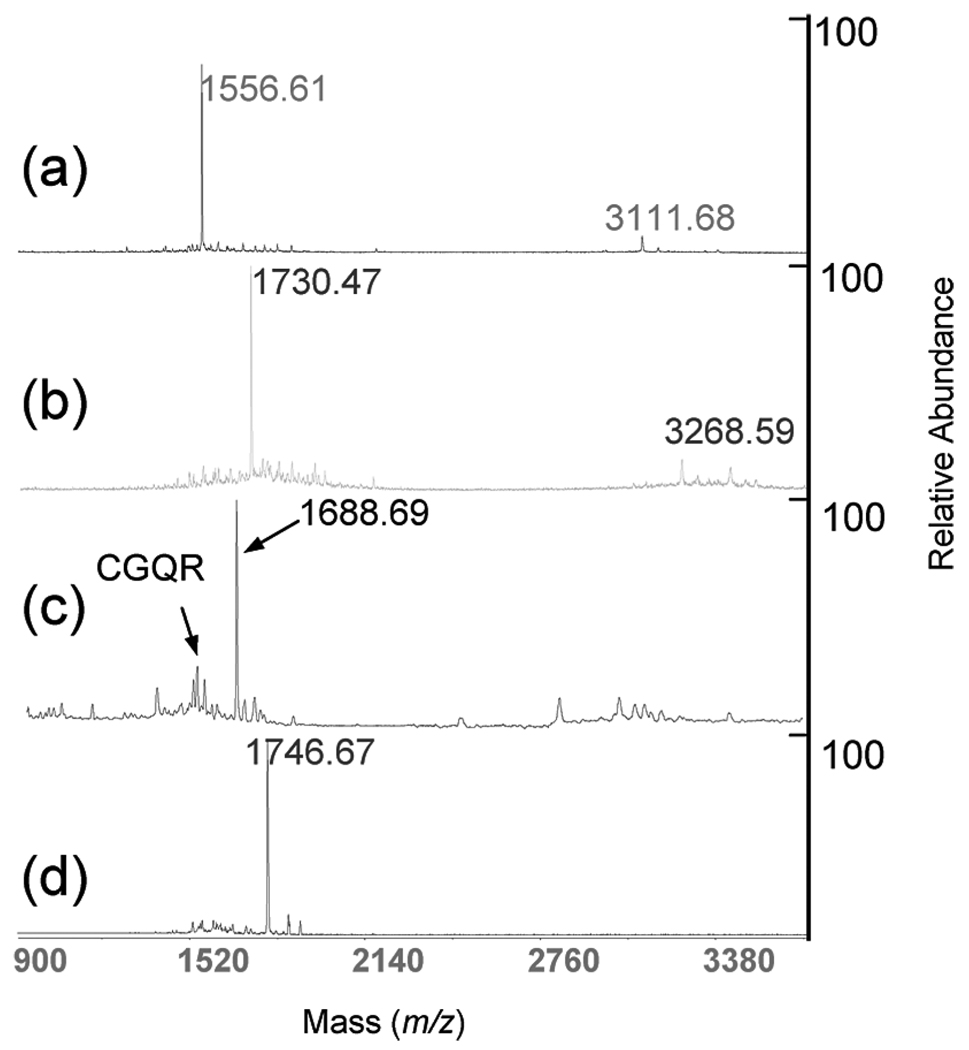
The mass of CGQR was measured using MALDI-TOF MS (Figure 2a). The ion at m/z 1730.47 in Figure 2b suggested that the lysine on CGQR was dead-end cross-linked (with another succinimidyl ester group hydrolyzed into a carboxylic group) by one SAMS. The S-acetyl group was removed with N-hydroxylamine and produced an ion at m/z 1688.69 (Figure 2c), showing the successful recovery of the thiol group in SAMS. The optimal ratio of lysine/SAMS for cross-linking was found to be 1:5.
Figure 2.
Characterization of the cross-linker, SAMS, by MALDI-TOF MS and a synthetic peptide. (a) Starting CGQR peptide average MH+ 1556.76 (sequence CGQRSLQHPHKHAG); (b) SAMS modified peptide, MH+ 1730.94; (c) SAMS modified CGQR peptide undergoing deacetylation, MH+ 1688.90. (d) Selective retrieval of the deacetylated modified peptide, 1746.98 m/z.
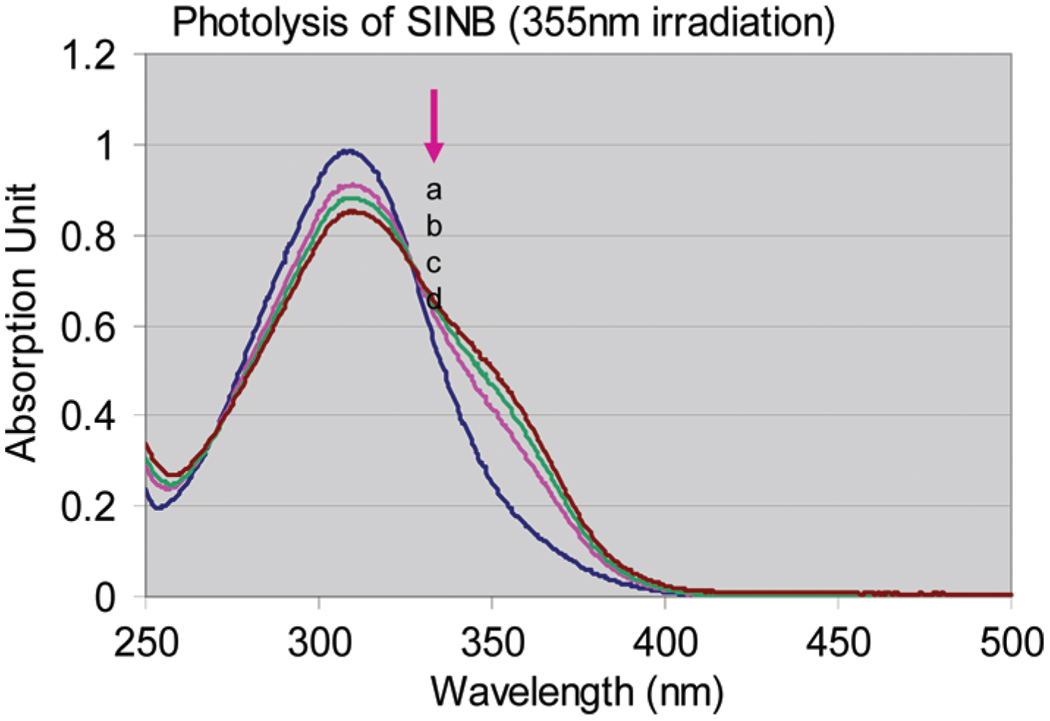
The photocleavable cross-linker, SINB, was cleavable under UV irradiation using 2 s intervals. The UV absorption spectra were monitored with a UV–vis spectrophotometer. Figure 3 shows the UV–vis spectrum of SINB as the blue curve. After UV irradiation, the maximum absorption peak (at 312 nm) decreased gradually. The isosbestic point at 325 nm confirmed that the photolysis occurred as a clean cleavage reaction. The quantum efficiency was 5%. To confirm the selective retrieval of cross-linked peptide, the magnetic beads were pretreated with SINB solution. At the same time, CGQR was modified with SAMS followed by S-acetyl deprotection. The recovered thiol-containing peptides were immediately incubated with the SINB modified beads. The beads were washed three times with 0.01% Tween20 followed by UV irradiation at 355 nm. The supernatant was collected for MALDI-TOF MS analysis (Figure 2d). The new major ion at m/z 1746.67 demonstrated the presence of the enriched peptides. General photolysis occurs on the benzyl alcohol group and gives rise to the nitrosobenzyldehyde (XIII in Scheme 2b). As shown in Scheme 2b, the photolysis was carried out in acidic aqueous solution. Therefore, the p-phenol ether was hydrolyzed, leaving a hydropropyl residue linked to the SH group of SAMS (XII in Scheme 2b). As a result, the starting acetyl group (C2H3O) in SAMS was converted to a 3-hydroxypropyl (C3H7O) group. The MS data revealed that the ion’s m/z value shift between the two spectra is 16 Da, which is the mass change of a CH4 group (elemental composition difference) after selective retrieval.
Figure 3.
Photolysis of SINB under UV 355 nm irradiation. The blue curve (a) is the UV spectrum of the original SINB solution without UV irradiation. Pink (b), green (c), and brown (d) curves are UV spectra of SINB under laser (355 nm) irradiation with a 2 s interval.
Cross-Linking of HSA and Selective Retrieval
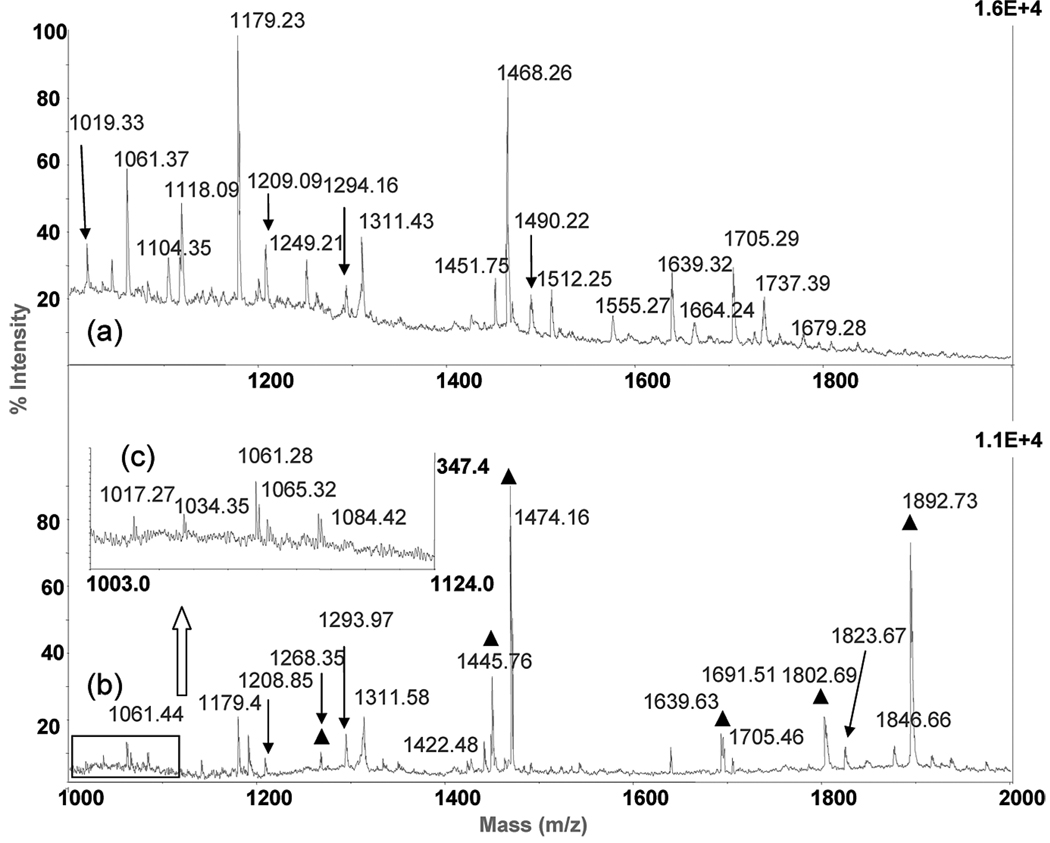
HSA (sequence shown in Table 1) is known to exist in monomeric as well as in oligomeric forms and was used as a model protein in the present study. Cross-linking was performed as described above. Both intra- and intermolecular cross-links were observed after reaction. The abundance of cross-linked peptides was very low and almost unobservable with MALDI-TOF MS (Figure 4a). The dominant observable peptides were noncross-linked. For example, the ions at m/z 1061.37, 1118.09, 1179.23, and 1468.26 are KLCTVATLR, VFLGMFLYE, RHPYFYAPE, and RHPDYSVVLLLR, respectively. However, the cross-linked peptides were enriched and easily observed after selective retrieval. For example, before enrichment the MH+ ion of RYK̃(SKDVCK)AAFTE (m/z 1876.64) appears as a barely observable minor peak embedded in the noise, (Figure 4a) [the italic K in the sequence represents a cross-linked lysine hereafter]. After enrichment, the MH+ ion of RYK̃(SKDVCK)AAFTE is m/z 1892.73, marked with a black triangle in Figure 4b, and becomes easily detectable. The observed mass is very close to the theoretical value of m/z 1892.84 (identified cross-linked peptides are listed in Table 2). The insert in Figure 4b is a zoomed in region of the boxed spectrum (Figure 4c). These newly observed ions are dead-ended cross-linked peptides, which were enriched. Ions at m/z 1017.27, 1034.35, and 1084.42 are peptides LTKVHTE, KSHCIAE, and LLFFAKR, respectively. The ion at m/z 1065.32 is the intracross-linked SISSKLKE peptide. The substantial increase in peptide signal is due to the enrichment procedure. More importantly, the new spectra obtained after selective retrieval provided more information about the cross-linked peptides.
Table 1.
Sequence of Human Serum Albumin
| 1 | MKWVTFISLLFLFSSAYSRGVFRRDAHKSEVAHRFKDLGEENFKALVLIAFAQYLQQCPF |
| 61 | EDHVKLVNEVTEFAKTCVADESAENCDKSLHTLFGDKLCTVATLRETYGEMADCCAKQEP |
| 121 | ERNECFLQHKDDNPNLPRLVRPEVDVMCTAFHDNEETFLKKYLYEIARRHPYFYAPELLF |
| 181 | FAKRYKAAFTECCQAADKAACLLPKLDELRDEGKASSAKQRLKCASLQKFGERAFKAWAV |
| 241 | ARLSQRFPKAEFAEVSKLVTDLTKVHTECCHGDLLECADDRADLAKYICENQDSISSKLK |
| 301 | ECCEKPLLEKSHCIAEVENDEMPADLPSLAADFVESKDVCKNYAEAKDVFLGMFLYEYAR |
| 361 | RHPDYSVVLLLRLAKTYETTLEKCCAAADPHECYAKVFDEFKPLVEEPQNLIKQNCELFE |
| 421 | QLGEYKFQNALLVRYTKKVPQVSTPTLVEVSRNLGKVGSKCCKHPEAKRMPCAEDYLSVV |
| 481 | LNQLCVLHEKTPVSDRVTKCCTESLVNRRPCFSALEVDETYVPKEFNAETFTFHADICTL |
| 541 | SEKERQIKKQTALVELVKHKPKATKEQLKAVMDDFAAFVEKCCKADDKETCFAEEGKKLV |
| 601 | AASQAALGL |
Figure 4.
MALDI-TOF spectra with or without selective retrieval of cross-linked peptides: (a) original tryptic/Glu-C digested HSA peptides, (b) HSA digested peptides after selective retrieval, and (c) enlarged spectrum of boxed area in the spectrum in part b. The ▲ indicates the selectively retrieved peptides.
Table 2.
Identified Cross-Linked Peptides of HSAa
| sequence | sequence | selectively retrieved |
mass (Mi) theoretical |
mass (Mi) experimental |
cross-linked residue |
distance (Å) in monomer |
|
|---|---|---|---|---|---|---|---|
| Dead-End Cross-Linking | |||||||
| 1 | LTKVHTE | 1001.46 | 1001.40 | K267 | |||
| Yb | 1017.49 | 1017.27 | |||||
| 2 | YKFQNALLVRc | 1425.72 | 1425.56 | K426 | |||
| 3 | KSHCIAEc | 1018.40 | 1018.28 | K310 | |||
| Yb | 1034.43 | 1034.35 | |||||
| 4 | LLFFAKRc | 1068.55 | 1068.28 | K183 | |||
| Yb | 1084.58 | 1084.42 | |||||
| Intramolecular Cross Linking | |||||||
| 1 | LKE | FKDLGEc | 1252.57 | 1252.44 | K300, K36 | 19.6 | |
| Y | 1268.60 | 1268.36 | |||||
| 2 | HKPK | KQTALVE | 1452.75 | 1452.56 | K549, K560 | 16.1 | |
| 3 | KSHCIAE | RYKAAFTEc | 1984.89 | 1984.76 | K186, K310 | 20.7 | |
| Intrapeptide Cross-Linking | |||||||
| 1 | VGSKCCKHPEc | 1357.56 | 1358.22 | K460, 463 | 5.75 | ||
| Y | 1373.59 | 1373.88 | |||||
| 2 | SISSKLKEc | 1049.51 | 1049.01 | K298, 300 | 6.24 | ||
| Yb | 1065.54 | 1065.32 | |||||
| Intermolecular Cross-Linking | |||||||
| 1 | SKDVCK | RYKAAFTE | 1876.86 | 1876.64 | K337, K186 | 61.6 | |
| Y | 1892.89 | 1892.73 | |||||
| 2 | KTPVSD | RYKAAFTE | 1786.83 | 1786.82 | K490, K186 | 34.5 | |
| Y | 1802.86 | 1802.69 | |||||
| 3 | DKE | VCKNYAE | 1429.56 | 1429.62 | K588, K341 | 80.5 | |
| Y | 1445.58 | 1445.76 | |||||
| 4 | AHKSE | EFKPLVE | 1587.74 | 1587.66 | K28, K402 | 42.6 | |
| FKPLVE | Y | 1474.72 | 1474.16 | ||||
| 5 | AKR | YKAAFTE | Y | 1374.66 | 1374.30 | K468, K186 | 30.3 |
| 6 | FKPLVE | KQTALVE | Y | 1691.89 | 1691.51 | K402, K549 | 41.5 |
| 7 | KYLYE | KQTALVE | Y | 1674.83 | 1674.34 | K161, K549 | 33.0 |
| 8 | HKPK | FKPLVE | 1396.73 | 1396.64 | K560, K402 | 39.1 | |
| 9 | LKE | KTPVSD | 1190.56 | 1190.44 | K300, K490 | 46.3 | |
| 10 | LKE | YKAAFTE | 1373.63 | 1373.48 | K300, K186 | 26.8 | |
| 11 | CCKAD | (YTKKVPQVSTPTLVE) | (trimer) | 3306.38 | 3306.52 | K438–K584 | 36.6 |
| CCKAD | K437–K584 | 33.5 | |||||
These peptides were first measured using MALDI-TOF analysis and their sequence confirmed by LC-ESI-MS/MS. Mi, monoisotopic protonated mass.
Peptides observed by MALDI-TOF MS analysis but not sequenced by MS/MS.
Observed in the mono band in the SDS-PAGE experiment.
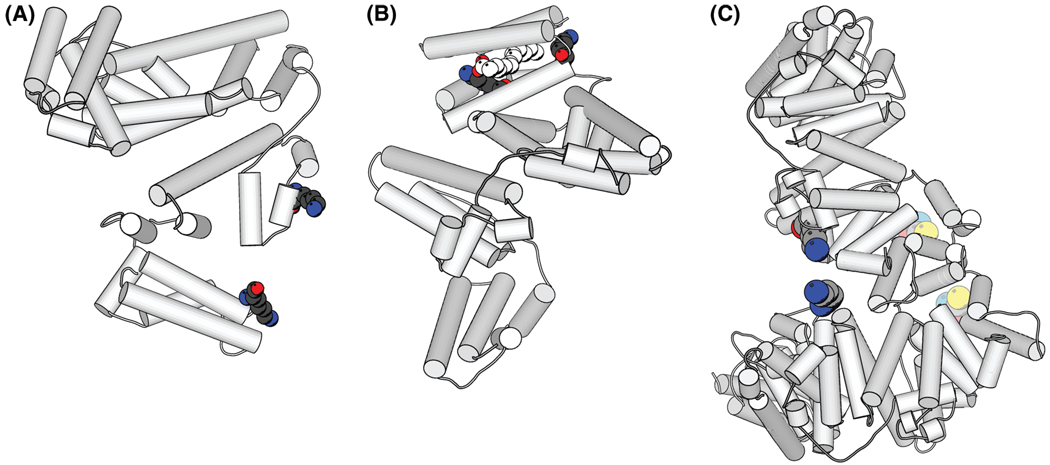
A total of 20 cross-linked peptides were observed and are listed in Table 2. Four dead-ended peptides (YKFQNALLVR, KSHCIAE, LLFFAKR, and LTKVHTE) were found with one end of the succinimidyl group hydrolyzed. This low number of dead-end cross-linking suggests that the reaction is optimized to produce double sided cross-links. The production of these dead-end cross-link peptides provides information on the contact site of lysine within the complex. To determine intra/intermolecular cross-linking, we utilized the HSA crystal structure determined by Zhu et. al23 to measure the shortest distance on the surface between the Cβ atoms of cross-linked lysines (Rasmol). The measurement of the Cβ–Cβ distance is more adequate than that of the Cα–Cα distance. The orientation of the Cα–Cβ bond is determined by the backbone atoms irrespective of side chain dynamics because it cannot be rotated in order to satisfy cross-link cross-link length requirements. In comparison, a Cα atom alone does not provide information on the directionality of the side chain, therefore determination of a maximum possible cross-link distance between Cα atoms is more ambiguous. Since our maximal link length, including the length of SAMS and two side chains of lysines, is about 19.5 Å, the intramolecular cross-links can be identified by comparing the distance between lysines with this cutoff value. From this comparison, two intrapeptide cross-linked peptides and three intramolecular cross-linked peptides were observed. These are VGSKCCK-HPE, SISSKLKE, FK̃(LKE)DLGE, K̃(HKPK)QTALVE, and YK̃ (KSHCIAE)FTE. Figure 5a shows the cross-linked lysine residues in peptides FK̃ (LKE)DLGE on the structure of HSA. Figure 5b shows lysines in the cross-linked peptides K̃ (HKPK) QTALVE, which suggests that a link is formed within this hydrophobic pocket. This pocket is normally occupied with the natural ligand, myristic acid,25 shown in white in the spacefilling model in Figure 5b. The cross-linker SAMS apparently occupies the same pocket of this ligand. Additional intermolecular cross-linked peptides were observed both in the preselective retrieval and in the enriched fraction (Table 2). As an example, Figure 5c shows the hypothetical structure of the HSA dimer with the cross-linked peptide RYK̃ (SKDVCK)AAFTE. The location of this cross-link is in agreement with the dimer formation reported by Komatsu,26 in which the cysteine residue (34) formed a disulfide bond producing the dimer. Their circular dichroism (CD) and NMR studies also showed that the structure of HSA remained the same. Figure 5c suggests that the possible arrangement of protein chains in the dimer is consistent with both with our cross-linked experimental data and that reported in the literature.26 To differentiate between intra- and intermolecular cross-linking, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or size exclusion chromatography can be used. Our preliminary SDS PAGE experiment provided two additional intrapeptide cross-links (Table 2). Intramolecular cross-links can provide valuable information to characterize protein structures for which no three-dimensional experimental data is available.
Figure 5.
Crystal structure of HSA (PDB code 1n5u). The location of the cross-linked peptides are highlighted in color in the ribbon and space fill diagram. (a) Intramolecular cross-linking on the surface of HSA. Balls represent the cross-linked peptide FK̃ (LKE)DLGE. (b) The intramolecular cross-linked peptides K̃ (HKPK)QTALVE within HSA. The white balls represent overlap with the ligand (myristic acid) observed in the crystal structure. The cross-linker SAMS apparently occupies the space of the ligand. (c) The cross-linked HSA dimer. Bright colored balls represent the cross-linked amino group in lysine in RYK̃ (SKDVCK)AAFTE. Pale colored balls represent the cross-linked thiol in Cys (34).26 This is a hypothetical structure of the HSA dimer (c) to illustrate the feasibility of the links. The plots were generated with Molscript.24 Atoms in the structure: C, light gray; O, red; N, sky blue; S, yellow.
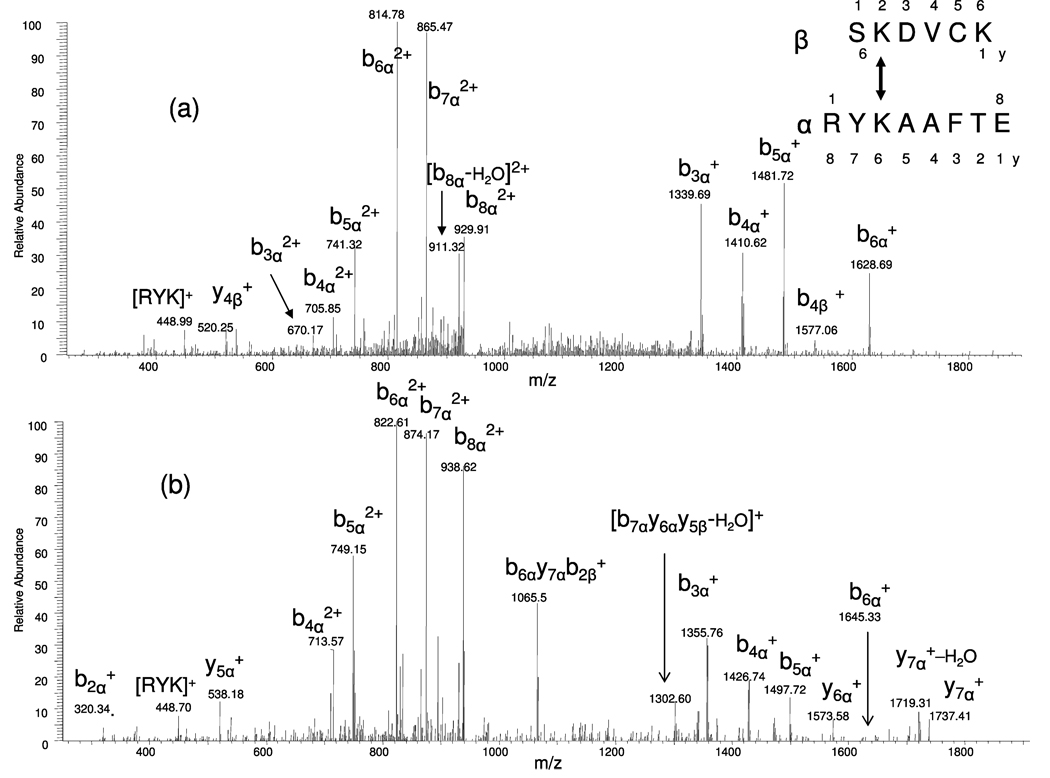
Figure 6 presents the MS/MS spectra of the cross-linked peptide RYKAAFTE(α) and SKDVCK(β). The MS/MS spectra of nonretrieved and selectively retrieved peptides are shown in parts a and b of Figure 6, respectively, from the doubly charged precursor ions at m/z 938.82 and at m/z 946.86. The nomenclature used in the MS/MS spectra is in accordance with that suggested by Schilling and coauthors.27 The product ions of the longer cross-linked peptide are noted as α and the shorter one as β. When the lysine residues are cross-linked, the total peptide mass of the other cross-linked peptide is included in the calculated mass. The major product ions in both mass spectra shown in Figure 6 are b3α+, b4α+, b5α+, b6α+, b7α+, and also the doubly charged ions as b3α2+, b4α2+, b5α2+, b6α2+, b7α2+, b8α2+. For example, the m/z b3α+ measured before selective retrieval is 1339.69. After selective retrieval, the m/z b3α+ is measured at 1355.76. Closer examination of the mass spectra (Figure 6b) shows that the product ions shifted by 16 Da from Figure 6a, which is due to the CH4 mass increase after the enrichment procedure, as described above.
Figure 6.
Electrospray MS/MS ionization spectra of the cross-linked peptide RYK̃ (SKDVCK)AAFTE. (a) Peptide before selective retrieval, precursor doubly charged ion m/z 938.82; (b) peptide after selective retrieval, precursor doubly charged ion m/z 946.86. The product ions of the shorter cross-linked peptide are noted as yβ and bβ ions. Those of longer cross-linked peptide are noted as bα and yα ions.
The efficiency of the described enrichment procedure, however, requires further improvement. Some concerns are that it is possible that the physical absorption of the peptide onto the beads is too strong and/or that the photocleavable spacer is not sufficiently stable. The surface of a bead contains many phenyl groups, from polystyrene and from the nitro-benzyl group of the photocleavable spacer. As a result, peptides containing tyrosine and phenylalanine residues may also be enriched due to nonspecific absorption. We have observed this type of enrichment, and this is shown in Figure 4b. The ion at m/z 1179.40 has the sequence of RHPYFYAPE, and the sequence of m/z 1311.58 is HPDYSVVLLLR (confirmed by MS/MS). As described in the Materials and Methods, the PBS Tween20 buffer was used to remove unbound peptides. However, we found that the anchored peptides were also washed out after extensive washes. We realized afterward that the carbonate ester in SINB can be easily hydrolyzed in water with subsequent release of CO2. Because of this instability, carbonate mono esters and some anchored peptides were lost during the washing step. This issue has been addressed by replacing SINB with an improved photocleavable spacer, SSIN (succinimidyl 5-(3-iodo-propoxy)-2-nitro-benzyl succinic ester), structure shown in Scheme 1c compound VIII, which is more robust during washing and more sensitive to UV light. Since SSIN is a benzyl succinic ester, it is also more stable than the benzyl carbonate ester during extensive washes. The SSIN immobilized beads remain intact even after overnight soaking. To efficiently remove physically absorbed peptides, the beads were soaked in the buffer (0.01% Tween20) for 2 h during each washing step. These adjustments allowed us to observe a dramatic increase of cross-linked peptides, as shown in Figure 4b. This method has proven to be capable of improving the signal-to-noise of cross-linked peptides of interest and minimize noise as well.
To assign the cross-linked peptides obtained from MS and MS/MS measurements, several software tools, such as Automated Peptide Assignment Program and MS2 assign were developed, but data analysis remains a challenge.27 Aebersold’s group recently described a method to identify cross-linked peptides from complex samples and large protein sequence databases by combining isotopically tagged cross-linkers, chromatographic enrichment, targeted proteomics, and a new search engine called xQuest. This software reduces the search space by an upstream candidate–peptide search before the recombination step.28 A cross search engine was also developed to provide the foundation for an over arching strategy to detect cross-linked peptides from digests of large (>170 kDa) cross-linked proteins.29
Fragmentation Behavior of Cross-Linked Peptides
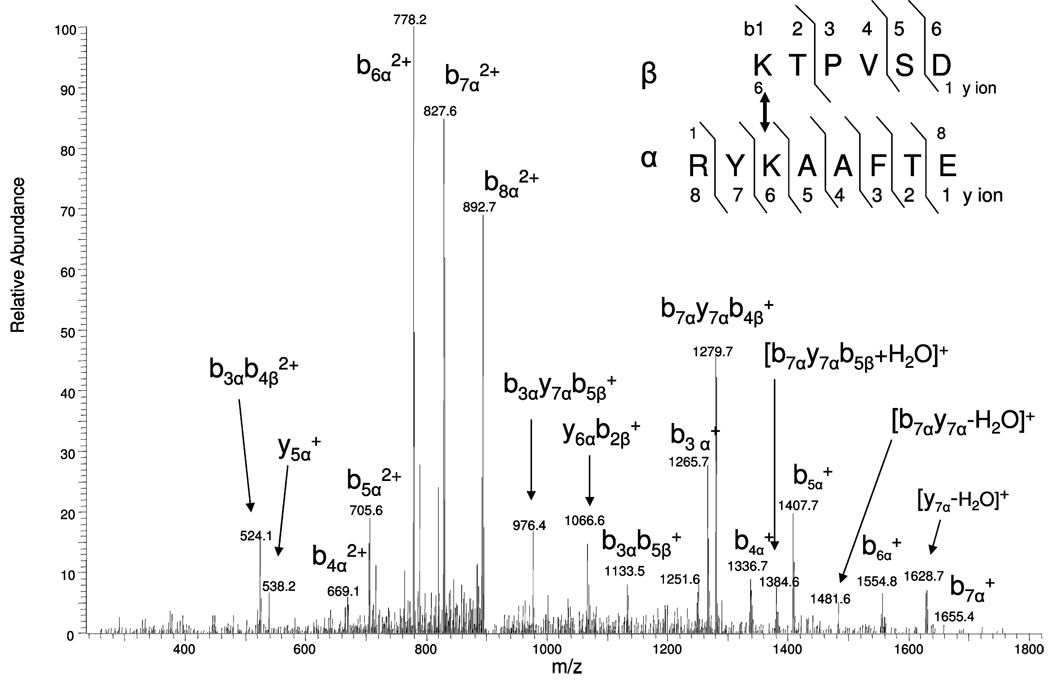
Peptides undergoing CID produce product ions that directly provide information on their amino acid sequence. After digestion, cross-linked peptides will contain two C-termini and two N-termini. Our initial examination of the MS/MS data obtained from cross-linked peptides revealed more b and y product ion series from the cross-linked peptide containing the larger number of amino acid residues. This was observed for both the singly and doubly protonated ions. Figure 7 shows the MS/MS for the cross-linked peptide RYKAAFTE(α) and KTPVSD(β); the precursor doubly charged ion is m/z 901.84. The predicted protonated monoisotopic mass of these peptides are m/z 646.34 and 985.51, respectively. The cross-linking group (mass of 174.03 m/z) will bind to the ε-amino group of lysine residues resulting in an addition of 172.03 Da (minus 1 Da for each peptide) for each cross-linked peptides. As shown in Figure 7, both precross-linked product ions, i.e., y5α+, [AAFTE]+ (m/z 538.24), and postcross-linked product ions are observed. The postcross-linked product ions are the major ions including ions of b3α+, [RYK(KTPVSD)]+ (m/z 1265.70); b4α+ [RYK(KTPVSD)A]+ (m/z 1336.74); b5α+ [RYK(KTPVSD)AA]+ (m/z 1407.73); b6α+, [RYK(KTPVSD)AAF]+ (m/z 1554.79); b7α+, [RYK(KTPVSD)AAFT]+ (m/z 1655.43); b4α2+ (m/z 669.05); b5α2+ (m/z 705.61); b6α2+ (m/z 778.24); b7α2+ (m/z 827.65); and b8α 2+ (m/z 892.70). Also, internal cleavages were observed during the MS/MS conditions used (including overlapping series), for example, b3αb4α2+, [RYK(KTPV)]2+, m/z 524.12; b3αy7αb5α+, [YK(KTPVS)]+, m/z 976.40; y6αb2α+, [K(KT) AAFTE]+, m/z 1066.6; b3α b5α+, [RYK(KTPVS)]+, m/z 1133.48; b7αy7αb4α+, [YK(KTPV)AAFT]+, m/z 1279.68; [b7αy7αb5α + H2O]+, [YK(KTPVS)AAFT + H2O]+, m/z 1384.6; and [b7αy7α − H2O]+, [YK(KTPVSD)AAFT − H2O]+, m/z 1481.60. The MS/MS data obtained from the b, y series and overlapping internal product ion series allows one to easily deduce the amino acid sequence of the cross-linked peptides. In Figure 7, we have observed more product ions from the bα series rather than yα; the presence of arginine near the amino terminus of the peptide may explain this observation. These observations should aid in developing software to automate the search of MS/MS data obtained from cross-linking studies. Although the signal of cross-linked peptides greatly increased, the MS/MS spectra still remain complicated due to both intra- and intermolecular cross-linked peptides. In our future work, we plan to apply isotopic or gas phase dissociable cross-linkers6 to improve our data analysis.
Figure 7.
Electrospray ionization MS/MS spectrum of the cross-linked peptide RYKAAFTE(α) and KTPVSD(β) (doubly charged precursor ion m/z 901.84) after enrichment.
CONCLUSIONS
A novel method has been established to selectively retrieve cross-linked peptides utilizing photocleavable technology. Thiol-generating cross-linkers were synthesized, and their structure and physical properties were characterized. The conjugation between the cross-linkers and standard peptides and model proteins were investigated. Human serum albumin was chosen as a model to interact with our cross-linker, producing protein–protein cross-links. The cross-linked peptides were anchored to the beads with a photocleavable spacer and then enriched with photoirradiation. MS/MS fragmentation behavior of cross-linked peptide was also studied. Use of protein-free selective retrieval eliminated the contamination that usually results from avidin–biotin based retrieval systems. The signal of MS/MS spectra was improved allowing for improved sequence assignment and, thus, protein searching. The sensitivity for detection also improved substantially after enrichment. The method described in this study enables identification of low amounts of cross-linked peptides in an abundance of digested peptide mixtures for protein–protein interaction studies. Future studies will employ other model systems with multiple protein components and tests its utility in biological systems.
ACKNOWLEDGMENT
This article is dedicated to the memory of Professor George Orr, Ph.D., deceased November 2005. Dr. George Orr was the founder of the Einstein Biodefense Proteomics Research Center and an inspiration for this project. This work was supported by NIH/NIAID Contract HHSN266200400054C.
References
- 1.Remaut H, Waksman G. Trends Biochem. Sci. 2006;31(8):436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Vasilescu J, Figeys D. Curr. Opin. Biotechnol. 2006;17:394–399. doi: 10.1016/j.copbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Sinz A. Mass Spectrom. Rev. 2006;25(4):663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 4.Gingras AC, Gstaiger M, Raught B, Aebersold R. Nat. Rev. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 5.Trakselis MA, Alley SC, Ishmael FT. Bioconjugate Chem. 2005;16(4):741–750. doi: 10.1021/bc050043a. [DOI] [PubMed] [Google Scholar]
- 6.Tang X, Munske GR, Siems WF, Bruce JE. Anal. Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury SM, Munske GR, Ronald RC, Bruce JE. J. Am. Soc. Mass Spectrom. 2007;18:493–497. doi: 10.1016/j.jasms.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brittain SM, Ficarro SB, Brock A, Peters EC. Nat. Biotechnol. 2005;23(4):463–468. doi: 10.1038/nbt1076. [DOI] [PubMed] [Google Scholar]
- 9.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. J. Am. Chem. Soc. 2003;125:2416–2425. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 10.Kalkhof S, Ihling C, Mechtler K, Sinz A. Anal. Chem. 2005;77:495–503. doi: 10.1021/ac0487294. [DOI] [PubMed] [Google Scholar]
- 11.Muller DR, Schindler P, Towbin H, Wirth U, Voshol H, Hoving S, Steinmetz MO. Anal. Chem. 2001;73:1927–1934. doi: 10.1021/ac001379a. [DOI] [PubMed] [Google Scholar]
- 12.Petrotchenko EV, Olkhovik VK, Borchers CH. Mol. Cell. Proteomics. 2005;4:1167–1179. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Soderblom EJ, Bobay BG, Cavanagh J, Goshe MB. Rapid Commun. Mass Spectrom. 2007;21:3395–3408. doi: 10.1002/rcm.3213. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Bottari P, Turecek F, Aebersold R, Gelb MH. Anal. Chem. 2004;76:4104–4111. doi: 10.1021/ac049905b. [DOI] [PubMed] [Google Scholar]
- 15.Lee MR, Shin I. Org. Lett. 2005;7(19):4269–4272. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 16.Hong CY, Chen YC. J. Chromatogr., A. 2007;1159:250–255. doi: 10.1016/j.chroma.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Yan F, Chen L, Tang Q, Wang R. Bioconjugate Chem. 2004;15:1030–1036. doi: 10.1021/bc049901d. [DOI] [PubMed] [Google Scholar]
- 18.Murov SL, Carmichael I, Hug GL. Handbook of Photochemistry. 2nd ed. New York: Marcel Dekker Inc.; 1993. pp. 298–305. [Google Scholar]
- 19.Ottl J, Gabriel D, Marriott G. Bioconjugate Chem. 1998;9(2):143–151. doi: 10.1021/bc970147o. [DOI] [PubMed] [Google Scholar]
- 20.Smet M, Liao LX, Dehaen W, Dominic VM. Org. Lett. 2000;2(4):511–513. doi: 10.1021/ol991373b. [DOI] [PubMed] [Google Scholar]
- 21.Whiteaker JR, Zhao L, Zhang H, Feng LC, Piening BD, Anderson L, Paulovich AG. Anal. Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed JK, Smith JR, Li P, Greene AS. Proteomics. 2007;7:2371–2374. doi: 10.1002/pmic.200700219. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, Yang F, Chen L, Meehan EJ, Huang M. J. Struct. Biol. 2007;162:40–49. doi: 10.1016/j.jsb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis PJ. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Curry S, Mandelkow H, Brick P, Franks N. Nat. Struct. Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu T, Oguro Y, Teramura Y, Takeoka S, Okai J, Anraku M, Otagiri M, Tsuchida E. Biochim. Biophys. Acta. 2004;1675(1–3):21–31. doi: 10.1016/j.bbagen.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Schilling B, Row RH, Gibson BW, Guo X, Young MM. J. Am. Soc. Mass Spectrom. 2003;14:834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- 28.Rinner O, Seebacher J, Walzthoeni T, Mueller L, Beck M, Schmidt A, Mueller M, Aebersold R. Nat. Methods. 2008;5(4):315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadeau OW, Wyckoff GJ, Paschall JE, Artigues A, Sage J, Villar MT, Carlson GM. Mol. Cell. Proteomics. 2008;7(4):739–749. doi: 10.1074/mcp.M800020-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]