Abstract
Sequence-specific binding of a transcription factor to DNA is the central event in any transcriptional regulatory network. However, relatively little is known about the evolutionary plasticity of transcription factors. For example, the exact functional consequence of an amino acid substitution on the DNA-binding specificity of most transcription factors is currently not predictable. Furthermore, although the major structural families of transcription factors have been identified, the detailed DNA-binding repertoires within most families have not been characterized. We studied the sequence recognition code and evolvability of the basic helix–loop–helix transcription factor family by creating all possible 95 single-point mutations of five DNA-contacting residues of Max, a human helix–loop–helix transcription factor and measured the detailed DNA-binding repertoire of each mutant. Our results show that the sequence-specific repertoire of Max accessible through single-point mutations is extremely limited, and we are able to predict 92% of the naturally occurring diversity at these positions. All naturally occurring basic regions were also found to be accessible through functional intermediates. Finally, we observed a set of amino acids that are functional in vitro but are not found to be used naturally, indicating that functionality alone is not sufficient for selection.
Keywords: biophysics, evolution, microfluidics
It has been recognized that phenotypic differences amongst species, especially closely related ones, do not necessarily arise from substantial differences in each species' collection of genes, but often appear, together with other factors such as alternative splicing and posttranslational modifications, through the differential expression of similar gene repertoires (1, 2). The rewiring of transcriptional regulatory networks can be achieved through the evolution of transcription factors (TFs) (trans-acting regulatory elements) and their DNA-binding sites (cis-acting elements). Cis-acting regulatory elements have been studied in detail and are dominant sources for the gradual evolution of transcriptional regulatory networks (3–5). The evolvability of trans-acting elements, or TFs, is less well understood. It is clear that changing a trans-acting element will have a much broader impact on a transcriptional regulatory network, because all target cis-regulatory elements of the trans-element will be affected, whereas a more gradual change can be achieved through evolving individual cis-regulatory elements. Nonetheless, trans-acting elements have evolved over time, giving rise to an ever increasing number of TFs in more complex genomes (6). Although it is relatively simple to identify mutations in both cis- and trans-acting elements, the functional consequences of these mutations are more difficult to glean, especially for trans-elements where it is not yet possible to predict what effects an amino acid substitution has on DNA-binding specificity and affinity. A substantial amount of literature exists on the mutability of Zn finger TFs using selection strategies (7–9), but most other families remain relatively uncharacterized. Here we focused on the evolvability of the helix–loop–helix family of TFs by systematically determining the evolutionary plasticity and DNA-binding repertoire of one TF member of this family.
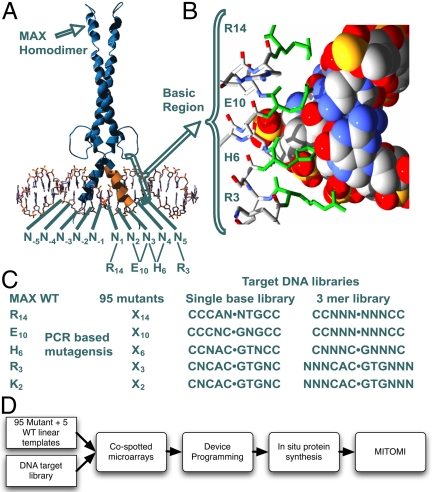
The bHLH TFs comprise the third-largest TF family after the Zn finger and Homeo box families (6, 10). Functionally, bHLH TFs are involved in a wide variety of processes but have been primarily implicated in cellular proliferation, differentiation, and determination (11). E12 and E47 were the first bHLH TFs to be identified and characterized (12, 13), followed by the determination of the protein structure of the Max homodimer (14) as well as the Max/Myc and Max/Mad heterodimers (15). All bHLH crystal structures show a conserved and highly similar metastructure with identical positioning of the basic region into the major groove of DNA. Because of the alpha-helical geometry, roughly every third to fourth amino acid residue of the basic region is positioned proximal to the major groove and is capable of making base-specific contact (Fig. 1 A and B, and Fig. S1). By using our amino acid and DNA-numbering scheme, residue Arg-14 contacts C−1, Glu-10 contacts A−2 and C−3, and His-6 primarily contacts C−3 (Fig. S1). Residues Arg-3 and Lys-2 tend to be unstructured but in certain circumstances have been identified to contact base pairs 4 and 5. This amino acid–base connectivity is observed in all Max structures as well as in the crystal structure of Pho4 (16). Even in bHLH TFs with a different set of 14 and 6 residues, such as E47 (17) and SREP (18), Glu-10 is still positioned to contact A−2 and C−3. Furthermore, positions R14, E10, and H6 are conserved in six of seven bHLH TFs in yeast (19) and are the most commonly observed combination of residues in human bHLH TFs (Fig. S2B).
Fig. 1.
Structure of a Max homodimer bound to DNA (14). (A) The bases are numbered around the symmetry axis (CAC●GTG = N−3N−2N−1●N1N2N3). The respective amino acids and their contacts are shown below the DNA base numbers. (B) Detailed cross-section of the basic region–DNA interface. Four of the five residues permutated in this study are shown in green. (C) The Max mutant library and the corresponding DNA library that was tested for each mutant set. (D) Schematic overview of the experimental setup. Linear mutant templates and DNA targets are synthesized, sequentially cospotted onto an epoxy glass substrate, which in turn is aligned to a microfluidic device. On-chip mutants are synthesized by using in vitro transcription/translation and measured for binding to the cospotted target DNA sequence with MITOMI.
We characterized all single amino acid substitutions of five DNA-contacting residues of Max to understand the extent to which transcriptional regulatory networks may evolve through direct changes in trans-acting elements. Simultaneously, we were interested in cataloguing the DNA-binding repertoire of a TF family by establishing an empirical table of DNA-binding domains and their corresponding DNA specificities. It has been postulated that it should be possible to generate a TF–DNA recognition code, linking amino acid primary sequence with DNA specificity. In principle, the generation of this code should be possible, but the complexity of the protein–DNA-binding interface has thus far prevented researchers from establishing a comprehensive, unified DNA-recognition code (20–23). Current work with protein-binding microarrays (24) has led to catalogues of putative binding motifs for TF families, but this technique often provides limited information on specific TFs and often fails to resolve binding differences between individual members of the same family (25). On the other hand, with microfluidic affinity analysis methods (26) it is now possible to probe the DNA-binding properties of a TF with exquisite precision and sensitivity, which in turn can serve as a highly informative dataset for the development of computational methods to further automate and unify these various approaches.
Results
Mutant Library Generation and Microfluidic Affinity Analysis.
We systematically generated a comprehensive mutant library of the DNA-binding domain of the bHLH TF Max by using a synthesis-based mutagenesis strategy. To generate our TF library, we mutated each of the four residues known to make DNA base-specific contacts as well as one proximal amino acid (14–18) with all 19 possible amino acid point substitutions (Fig. 1). We used a primer extension, multistep-PCR strategy for the rapid synthesis of these mutants, allowing us to generate TFs with novel DNA-binding domains from a library of long oligomers. We screened each of these 95 mutants and five WT controls against a small single-base library and a larger 3mer library (NNN library) covering 64 DNA sequences for a total of over 6,400 interactions, each measured at least in triplicate (Fig. 1 C and D). To achieve the scale and sensitivity required for such a measurement, we used a highly integrated microfluidic platform (27) coupled to on-chip synthesis of the mutants and mechanically induced trapping of molecular interactions (MITOMI) for relative affinity measurements (26). Combining an on-chip protein synthesis strategy for the generation of TF mutants, use of MITOMI for affinity measurements, and the throughput of the microfluidic platform allowed us to rapidly screen the entire matrix of TF mutants versus a large DNA target library with high fidelity and sensitivity.
Sequence Specificity of Max Mutants.
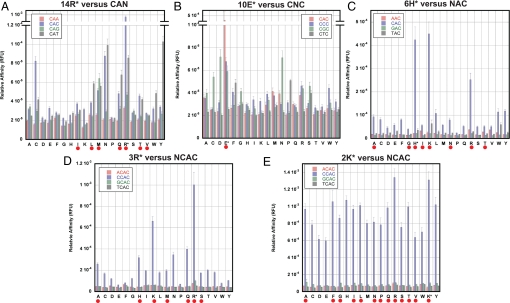
In the initial round of screens, we tested each set of mutants against base substitutions in the location where the WT residue contacts DNA, as determined from the Max and Pho4 crystal structures (Fig. 2, Dataset S1). This small-scale screen revealed a broad diversity in the function of each position. Substitutions in the Arg-14 position resulted in several changes in sequence specificity (Fig. 2A). Substituting hydrophilic-neutral amino acids such as tyrosine, glutamine, and asparagine, as well as the hydrophobic amino acids leucine and tryptophan, for Arg-14 changed the sequence specificity from CAC to CAT. Only methionine caused a specificity change from CAC to CAG. CAA was not accessible by any substitution. This situation drastically changed for the Glu-10 WT position (Fig. 2B), where essentially any substitution caused a drop in affinity to near-baseline levels, indicating that Glu-10 is absolutely required for sequence-specific binding. In positions H6, R3, and K2, no change in sequence specificity was observed with any substitution, but the mutants were able to modulate affinity (Fig. 2 C–E). Together, these results show that the three main positions (6, 10, 14) each exhibit a unique function, with position 14 defining specificity, position 10 being absolutely necessary, and positions 6 and 3 being able to modulate overall binding affinity (position 6 and 3 are also found to modulate specificity based on the NNN screen described below). Position 2 has no major effect on either specificity or affinity and thus is probably not forming any sequence-specific contacts and only limited nonspecific interactions. The basic region is therefore surprisingly rigid in respect to the possible DNA sequence specificities that are accessible through single-site mutations. The only three sequences that could be recognized are CA[C,T,G], albeit with a wide range of affinities.
Fig. 2.
Dataset of mutants of positions 14 (A), 10 (B), 6 (C), 3 (D), and 2 (E). Each graph shows the 20 amino acids per position and their binding specificity for the indicated DNA sequence. The WT MAX amino acid residue is denoted by an asterisk, and naturally occurring amino acids in other bHLH TFs are indicated by a red dot.
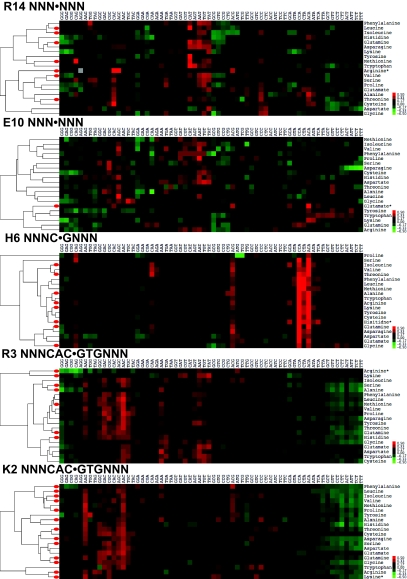
To ascertain that we were not missing any higher-order sequence-specificity changes, we expanded the DNA sequence space to a 3mer centered on the base being contacted by the corresponding amino acid residue. To represent the large amount of affinity data, we plotted the data in the form of heat maps and clustered the amino acids according to their DNA-specificity profile (Fig. 3, Figs S3–S7, and Dataset S2). The results for Arg-14 of the larger screen corroborate the results obtained from the small single-base screen, as no additional sequence specificities beyond CA[C,T,G] were accessible. The situation was similar for Glu-10, which we previously determined to be essential. When screening H6, we found some additional sequences that were not observed in the single-base screen. We observed a cluster of amino acids that recognize CAAC as well as the WT CCAC. A second, more generic, sequence motif we observed is a split binding motif with the two e-box half-sites being separated by four bases: CAC GCGC GTG. We found a similar motif in positions Arg-14 and Glu-10, where CAC GC GTG and CAA GC TTG were recognized by WT, which we observed previously (26). We also found another split binding-site motif of the sequence CAG CG CTG in positions 14, 10, and 6. This observation suggests that the loop region of bHLH TFs allows these factors to recognize motifs split by GC, CG, or GCGC, albeit with low affinity. It is interesting that the spacer is also sequence specific. Position 3 can be subdivided into two major functional groups, one that prefers N[G/A]C CACGTG and a second smaller group that prefers N[G/A]T CACGTG. In position 2, N[G/A]C is the main sequence cluster being recognized with a strengthening of affinity for AAG as well as the N[C/T][G/C] sequence cluster, rather than N[G/A]T. Position 2 therefore has no major effect on flanking-base specificity, which is determined by position 3. As mentioned above, two major specificity clusters are observed: N[G/A]C and N[G/A]T. Even though 5 aa cause a shift to the N[G/A]T cluster: E,D,W,C, and K, only lysine is used naturally to change specificity (see also Fig. S2). We have previously shown that Max and Pho4 have significantly different flanking-base specificities than Cbf1, recognizing NCC and [A/G]GT, respectively (26). This flanking-base preference is explained in our dataset (Fig. 3) with the observed sequence specificities in position 3, particularly in base position N−4. Finally, a complex picture emerges when comparing the physical characteristics of the amino acids with their sequence-specificity profile (see Figs. S3–S8). The overall specificity profile varies drastically in each position, although small clusters of amino acids with similar physical properties and sequence specificities are observed. By using principal component analysis, we determined that the observed variances in sequence specificity and affinity as a function of amino acid properties in positions 14, 6, 3, and 2 are significantly correlated with the pI of the substituted amino acids (Table S1). Position 10, on the other hand, is dominated by residue size and volume; surface size and volume were also found to play a significant role in position 6.
Fig. 3.
Large-scale dataset of all 20 amino acid mutants in the five positions measured against a full 3mer (64 DNA sequences). Affinity to a specific sequence is shown by a color gradient from green to red (low to high affinity, respectively). Each row has been normalized to better visualize the DNA-binding preference of each amino acid. The amino acid binding specificities are clustered, and the distances between amino acid specificities are shown on the left side of each graph.
Comparison of the in Vitro Data with the Naturally Occurring Basic Region Diversity.
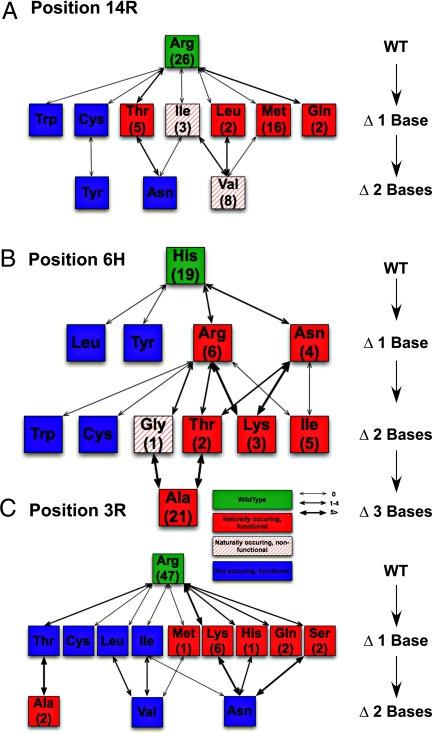
After having experimentally characterized the possible amino acid repertoire of the bHLH basic region, we were interested in determining the naturally occurring basic region diversity. To this end, we extracted the annotated basic regions from all ∼240 bHLH TFs listed in the InterPro database and performed a sequence alignment. From this alignment, we extracted 85 nonredundant basic regions, which aligned into nine major branches (Fig. S2A). Two of these branches consisted of basic regions known not to bind DNA, as seen in the ID TF subfamily. The sequence alignment shows that Glu-10 is conserved in all 64 remaining basic regions, in concordance with our experimental measurements. When only considering the diversity in the functionally dominant positions, the diversity further collapses into 15 families (Fig. S2B), for which we predicted the binding specificity by using the experimentally determined values (Fig. S2C). For the functionally dominant positions 14, 6, and 3, we plotted the mutational distance of all naturally observed amino acids and those amino acids we find to be functional in vitro (Fig.4). We defined amino acid substitutions to be “functional” if they showed at least twice the affinity as that observed for proline in our single-base screen. Proline serves as a convenient baseline in our dataset as it strongly inhibited binding when substituted in positions 14, 10, 6, and 3. We find that all naturally occurring amino acids are functional except for isoleucine and valine in position 14 and glycine in position 6. Valine in position 14 is mainly observed in branch number 8 of the basic region alignment, with one instance occurring in branch number 7. Branch number 8 consists of the Ascl bHLH TFs and E12/E47. Here, arginine 9 and 11 are highly conserved and known to make nonspecific and specific DNA contacts (17) and thus are likely needed to reconstitute binding. Interestingly, we consistently find cysteine, tryptophan, and tyrosine to be functional, even though those amino acids are never naturally observed, suggesting that these 3 aa are possibly under negative selection pressure. Even in position 2, where the selection pressure is reduced, cysteine, tryptophan, and tyrosine are still omitted. The ability of cysteine to form disulfide bonds may be one reason for its negative selection, whereas the observed specificity of tryptophan may not be sufficient for in vivo usage. The reason for the omission of tyrosine remains unclear. We find in each position functional amino acids that are only one mutational step removed from the WT but are not naturally observed, indicating that the ability to bind DNA specifically by itself is not sufficient to cause selection and in vivo usage. The notion that a negative pressure must exist that prevents certain amino acids from being used in the basic region is further strengthened by the fact that a mutation from arginine to cysteine in position 14 is only one mutational step away and does not lead to changes in sequence specificity, but nonetheless is never observed. It is of course also possible that affinity plays a major role in TF evolution, which could explain some, but not all, of the observed amino acid usage patterns.
Fig. 4.
A graphical representation of the mutational distance of the amino acids, which naturally occur and/or have been determined to be functional in our in vitro assay. The WT amino acid is shown with a green background, with all naturally occurring amino acids shown in red. Functional amino acids are solidly shaded, whereas nonfunctional amino acids are indicated by a hatched shading. Numbers in parentheses below the amino acid abbreviation indicate the number of times the amino acid is observed in the basic region alignment tree (Fig. S2B). The edge thickness corresponds to the observed amino acid substitution rates (41, 42). (A) In position 14, all functional amino acids, with the exception of valine, are one mutational step away from the WT, including the second-most commonly observed amino acid, methionine. Tryptophan and cysteine are both functional and only one mutational step away from the WT, but they are not naturally observed. Tyrosine and asparagine are also not observed and are two mutational steps away. (B) Position 6 shows naturally occurring and functional amino acids up to three mutational moves away from the WT. Particularly interesting is the polar distribution of histidine and alanine, the two most common naturally occurring amino acids. Again, functional amino acids are observed one and two mutational steps away from the WT, which nonetheless are not observed naturally. (C) Position 3 resembles the picture seen in position 14. Here, the two most functional amino acids, arginine and lysine, are also the most common with 47 and six counts, respectively.
Finally, we were interested in determining whether our systematic single amino acid substitutions and in vitro sequence-specificity measurements were sufficient to predict the observed sequence specificities of naturally occurring bHLH TFs. We extracted known experimental data from literature for members of the functional branches of the basic region alignment tree (branches 1–3 and 6–9 of Fig. S2A) and summarized this information in Table S2. Most of this sequence-specificity information was derived from noncomprehensive EMSAs, or SAABs (selected and amplified binding), which can distort the actual sequence specificity. Nonetheless, in most cases we correctly predict the observed specificity. Only branches 3, 8, and 9 show disagreements, and the latter two branches show internal ambiguity due to the limitations of the experimental methods used.
Discussion
We systematically determined the sequence specificity of a large set of bHLH mutants, including amino acid substitutions that are not naturally observed. With our dataset, it is possible to determine the evolutionary trajectory a bHLH TF can take via single amino acid substitutions, rather than solely cataloguing the diversity of an existing family. We found that the basic region of bHLH TFs is extremely restricted by the number of residues that retain function. We also observed that all naturally observed basic regions are accessible without intermediate loss of function. This suggests that, even after a gene duplication event, a bHLH TF retains viability, which may be followed by a slow gradual drift through the functional amino acid space. Furthermore, the sequence-specificity space that bHLH TFs can explore through single amino acid substitutions is extremely restricted. Nature has explored the entire available sequence-specificity repertoire of the basic region, indicating that the bHLH family evolved from a new structural motif efficient at recognizing a particular sequence but consequently was not able to significantly diversify. One obvious solution to a limited sequence repertoire is dimerization (28), which allows the multiplexing of a small number of sequence specificities into a larger repertoire. A similar sequence repertoire restriction exists in the Zn finger family, but there nature was able to evolve a more modular scaffold, which allows for the multiplexing of several Zn finger domains, explaining the numerical dominance of that specific family (29, 30). Overall, our experiments show that evolving new and considerably different sequence specificities is not easily achievable within the bHLH family, but rather must involve larger topological changes or repositioning of the DNA-binding interface, seeding a new family of TFs (31). These findings can likely be extended to the bZip family, because of the considerable structural similarities the bZip family of TFs shares with the bHLH family. Additionally, as the four largest TF families frequently sense DNA sequence by inserting an alpha–helix into the major groove of DNA, the findings here may apply to all TFs using this mode of sequence-specific binding. To evolve new transcriptional regulatory network connectivity by slow, gradual drift of TF specificity through single-site mutations therefore is possible, but it also seems to be suboptimal, suggesting that network evolution is probably mainly driven by reshuffling of existing DNA binding domains, evolving new DNA binding sites (32, 2, 33), gene duplications (31), or other more plastic regulatory methods, such as posttranscriptional regulation of gene expression (34).
Our experiment provides a bHLH-specific “lookup table” that can be used to predict the sequence specificity of most bHLH TFs (Table S2) and demonstrates the possibility of generating similar tables for the remaining major TF families. Moreover, because of the dominant role bHLH TFs play in development, we were able to link our dataset to a number of cases where a mutation in the basic region of a bHLH TF gives rise to a disorder (35–37). For example, a R14W mutation in HES7 causes spondylocostal dystosis (35), a R14H mutation in MYCN causes Feingold syndrome (37), and a E10K mutation in NeuroD1 causes maturity-onset diabetes of the young (36). All of these mutations can be understood on a molecular level by using our dataset.
Even though we generated a systematic library of single-site amino acid substitutions, the experimental approach is extremely flexible and can be applied to higher-complexity mutants containing multiple substitutions as well as other protein libraries (38). Additionally, the approach presented here is not only applicable to measuring protein–DNA interactions but can also be used to measure protein–protein interactions or screens of enzymatic function. Coupled with current advances in gene synthesis, our approach will provide a rapid path to characterizing the relationship between protein sequence and function, broadly impacting fields such as molecular evolution, directed evolution, and protein design. Furthermore, datasets obtained with such experimental approaches should prove instructive for the development of accurate molecular simulations.
Materials and Methods
Method Overview.
Max mutant linear-expression templates and DNA targets were synthesized essentially as described previously (26). The linear-expression templates and DNA targets were sequentially cospotted onto an epoxy-coated glass substrate (CEL Associates), followed by alignment of the array to a microfluidic device, essentially as described (26). The fluidic devices contained either 640 or 2,400 unit cells, depending on the size of the experimental dataset obtained. Mutants were synthesized on-chip by introducing a wheat germ in vitro transcription/translation mixture (Promega) spiked with a tRNAlys−bodipy−fl conjugate (Promega) for the residue-specific labeling of proteins with a fluorophore. The device was scanned once after 60-min incubation at 30° C on a hot plate. For detection, the MITOMI membrane was closed, the channels washed for 5–10 mins with PBS, followed by a second scan. The array scans were quantified by using Genepix Pro 6.0 (Axon Laboratories). The binding profiles were clustered by using Gene Cluster 3.0 (Michiel de Hoon, University of Tokyo) and displayed by using Java Treeview (39). Basic regions were extracted and modified by using custom-written python code and aligned by using Clustal X (40). Sequence logos were generated by using SEQLOGO 0.4 (EMBL-EBI). Experimental details are reported in the SI Text.
Supplementary Material
Acknowledgments.
We thank M. Kreitman and J. Widom for critical reading of the manuscript. Financial support was provided in part by the National Institute of Health Director's Pioneer Award and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907688106/DCSupplemental.
References
- 1.King MC, Wilson AC. Evolution at 2 levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 3.Isalan M, et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mustonen V, Kinney J, Callan CG, Lassig M. Energy-dependent fitness: A quantitative model for the evolution of yeast transcription factor binding sites. Proc Natl Acad Sci USA. 2008;105:12376–12381. doi: 10.1073/pnas.0805909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moses AM, Chiang DY, Kellis M, Lander ES, Eisen MB. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol Biol. 2003;3:19. doi: 10.1186/1471-2148-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 7.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse dna target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 8.Rebar EJ, Pabo CO. Zinc-finger phage-affinity selection of fingers with new dna-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 9.Segal DJ, Dreier B, Beerli RR, Barbas CF. Toward controlling gene expression at will: Selection and design of zinc finger domains recognizing each of the 5′-gnn-3′ dna target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:RESEARCH0030. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massari ME, Murre C. Helix–loop–helix proteins: Regulators of transcription in eukaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murre C, McCaw PS, Baltimore D. A new dna binding and dimerization motif in immunoglobulin enhancer binding, daughterless, myod, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 13.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 14.Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by max of its cognate dna through a dimeric b/hlh/z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 15.Nair SK, Burley SK. X-ray structures of myc-max and mad-max recognizing dna. molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu T, et al. Crystal structure of pho4 bhlh domain-dna complex: Flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal-structure of transcription factor e47—e-box recognition by a basic region helix–loop–helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 18.Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 angstrom resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 19.Robinson KA, Lopes JM. Survey and summary: Saccharomyces cerevisiae basic helix–loop–helix proteins regulate diverse biological processes. Nucleic Acids Res. 2000;28:1499–1505. doi: 10.1093/nar/28.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews BW. Protein-dna interaction—no code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 21.Luscombe NM, Thornton JM. Protein-dna interactions: Amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 22.Pabo CO, Nekludova L. Geometric analysis and comparison of protein-dna interfaces: Why is there no simple code for recognition? J Mol Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 23.Havranek JJ, Duarte CM, Baker D. A simple physical model for the prediction and design of protein-dna interactions. J Mol Biol. 2004;344:59–70. doi: 10.1016/j.jmb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the dna-binding specificities of zinc fingers with dna microarrays. Proc Natl Acad Sci USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger MF, et al. Variation in homeodomain dna binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 27.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 28.Amoutzias GD, Bornberg-Bauer E, Oliver SG, Robertson DL. Reduction/oxidation-phosphorylation control of dna binding in the bzip dimerization network. BMC Genomics. 2006;7:107. doi: 10.1186/1471-2164-7-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavletich NP, Pabo CO. Zinc finger dna recognition—crystal-structure of a zif268-dna complex at 2.1-a. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe SA, Nekludova L, Pabo CO. Dna recognition by cys(2)his(2) zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 31.Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. Embo Reports. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 33.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 34.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and micrornas. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 35.Sparrow D, Guillen-Navarro E, Fatkin D, Dunwoodie S. Mutation of hairy-and-enhancer-of-split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet. 2008;17:3761. doi: 10.1093/hmg/ddn272. [DOI] [PubMed] [Google Scholar]
- 36.Kristinsson SY, et al. Mody in iceland is associated with mutations in hnf-1alpha and a novel mutation in neurod1. Diabetologia. 2001;44:2098–2103. doi: 10.1007/s001250100016. [DOI] [PubMed] [Google Scholar]
- 37.van Bokhoven H, et al. Mycn haploinsufficiency is associated with reduced brain size and intestinal atresias in feingold syndrome. Nat Genet. 2005;37:465–467. doi: 10.1038/ng1546. [DOI] [PubMed] [Google Scholar]
- 38.Gerber D, Maerkl SJ, Quake SR. An in vitro microfluidic approach to generating protein-interaction networks. Nat Methods. 2009;6:71–74. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saldanha AJ. Java treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 40.Larkin MA, et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Collins DW, Jukes TH. Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics. 1994;20:386–396. doi: 10.1006/geno.1994.1192. [DOI] [PubMed] [Google Scholar]
- 42.Bordo D, Argos P. Evolution of protein cores—constraints in point mutations as observed in globin tertiary structures. J Mol Biol. 1990;211:975–988. doi: 10.1016/0022-2836(90)90087-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.