Abstract
It is known that the extracellular matrix regulates normal cell proliferation, and it is assumed that anchorage-independent malignant cells escape this regulatory function. Here we demonstrate that human M24met melanoma cells remain responsive to growth regulatory signals that result from contact with type I collagen and that the effect on proliferation depends on the physical structure of the collagen. On polymerized fibrillar collagen, M24met cells are growth arrested at the G1/S checkpoint and maintain high levels of p27KIP1 mRNA and protein. In contrast, on nonfibrillar (denatured) collagen, the cells enter the cell cycle, and p27KIP1 is down-regulated. These growth regulatory effects involve contact between type I collagen and the collagen-binding integrin α2β1, which appears restricted in the presence of fibrillar collagen. Thus melanoma cells remain sensitive to negative growth regulatory signals originating from fibrillar collagen, and the proteolytic degradation of fibrils is a mechanism allowing tumor cells to escape these restrictive signals.
The extracellular matrix (ECM) is a complex structure of collagens, glycoproteins, elastin, and proteoglycans that, in addition to providing a scaffold for tissues, regulates many fundamental cellular processes such as proliferation, survival, migration, and differentiation (1, 2). It is now appreciated that the ECM provides a complex combination of insoluble signals that, in concert with cell–cell contacts and soluble signals provided by growth factors, affect gene expression and influence cell functions (3–5). It is known that normal cells need contact with the ECM to progress into the cell cycle, and loss of this requirement is a hallmark of malignant cells (6). The mechanisms by which the ECM regulates the growth of anchorage-dependent cells have received renewed attention over the past years and have been the subject of investigations that have pointed to cell cycle regulatory proteins that are influenced by contact between cells and the ECM (7–9). On binding to specific cell-surface receptors such as integrins, the ECM exerts its control on cell proliferation by modulating the expression of G1 regulators in a growth factor-dependent and independent manner. In most conditions, there is cooperative action between growth factors and ECM that results in nonredundant signals that transcriptionally up-regulate cyclins D and E and down-regulate the cyclin-dependent kinase (CDK) inhibitors p21CIP1, p27KIP1, and p57KIP2 (10). This cooperative action permits cells to pass through the G1 restriction point and to complete the cell cycle. However, there are other conditions where the ECM has been shown to send growth inhibitory rather than growth stimulatory signals. These conditions depend not only on the composition of the ECM but also on its physical state. Molecules of type I collagen in vivo are organized into striated fibrils. These structures, which are responsible for the resistance of many tissues, are lost on proteolytic degradation of the collagen. These changes in the physical nature of collagen can affect the growth of normal cells. For example, fibrillar type I collagen inhibits cell growth (11, 12), but this effect is lost in the presence of nonfibrillar collagen. Vascular smooth muscle cells remain arrested in G1 phase in the presence of fibrillar collagen but proliferate when grown on nonfibrillar collagen. This negative growth regulatory control involves an increase in the levels of p21CIP1 and p27KIP1 (13).
Much less is known about the growth regulatory effect of the ECM on malignant cells, and it has often been assumed that because malignant cells are anchorage independent, they are not sensitive to growth regulatory signals originating from the ECM. However, several lines of evidence suggest that integrin-mediated contact between malignant cells and the ECM influences their behavior (14–16). For example, reexpression of the α2 integrin in poorly differentiated mammary carcinoma cells that had lost α2β1 expression resulted in a dramatic phenotypic alteration characterized by a reduction in cell spreading, motility, and invasion when these cells were grown on fibrillar collagen matrices (17). Integrin-mediated contact with the ECM can also influence cell survival. For example, reexpression of αvβ3 integrin in melanoma cells that had lost αv allowed them to recognize cryptic binding sites that became available on fibrillar collagen proteolyzed by tumor-derived proteases. Binding of αvβ3 to these cryptic sites protected these cells from undergoing apoptosis (18). These observations suggest that the morphology, motility, and survival of tumor cells can be regulated by the composition and physical nature of the ECM.
The composition and physical nature of the ECM are under the influence of a variety of matrix-degrading proteases including the matrix metalloproteinases (MMPs), whose activity in the extracellular milieu is controlled by specific inhibitors such as tissue inhibitors of MMPs (TIMPs) (19, 20). In cancer, a change in the balance between MMPs and TIMPs in favor of the inhibitors inhibits tumor growth (21, 22). In our laboratory, we have previously demonstrated that overexpression of TIMP-2 in human M24met melanoma cells inhibited the growth of primary tumors implanted in scid mice. Growth inhibition was also observed in M24met cells cultured in the presence of fibrillar collagen and in which the proteolysis of the collagen was prevented in the presence of recombinant TIMP-2 or by TIMP-2 cDNA transfection. In contrast, nonfibrillar collagen had no inhibitory effect (23). These data suggested that preventing fibrillar collagen degradation by MMPs expressed by these cells had a direct inhibitory effect on their growth.
In this study, we now provide evidence that fibrillar collagen inhibits cell cycle progression in malignant melanoma cells and that this inhibition involves adhesion through the α2β1 integrin and up-regulation of the cyclin inhibitor p27KIP1.
Materials and Methods
Reagents.
Unless otherwise noted in the text, chemicals were from Sigma.
Antibodies.
The following antibodies were used: mAb anti-BrdUrd (clone Bu20a, DAKO), mAb anticyclin E conjugated to Sepharose beads (clone HE-111, Santa Cruz Biotechnology), polyclonal anti-p21CIP1 (C-19, Santa Cruz Biotechnology), polyclonal anti-p27KIP1 (C-19, Santa Cruz Biotechnology), polyclonal anticyclin D1 (M-20, Santa Cruz Biotechnology), mAb anticyclin E (clone HE-12, Santa Cruz Biotechnology), polyclonal anti-CDK2 (M-2, Santa Cruz Biotechnology), mAb anti-Aspergillus niger glucose oxidase (negative control, clone DAK-G01, DAKO), mAb anti-α1 integrin (clones 5E8D9, Upstate Biotechnology, Lake Placid, NY, and clone FB12, Chemicon), mAb anti-α2 integrin (clone P1E6, Chemicon and GIBCO/BRL), mAb anti-α3 integrin (clone P1B5, Chemicon and GIBCO/BRL), mAb anti-α2β1 integrin (clone BHA2.1, Chemicon), monoclonal anti-β1 integrin (clone 6S6, Chemicon and clone P4C10, GIBCO/BRL), nonconjugated goat anti-mouse Ig (DAKO), fluorescein-conjugated goat anti-mouse Ig (DAKO), peroxidase-conjugated goat anti-mouse Ig (Santa Cruz Biotechnology), and peroxidase-conjugated donkey anti-rabbit Ig (Amersham).
Cells.
M24met human melanoma cells (gift from R. Reisfeld, Scripps Research Institute, La Jolla, CA) were maintained in RPMI-1640 medium enriched with 10% heat-inactivated FBS (Omega Scientific, Tarzana, CA), supplemented with 100 units/ml penicillin G, 100 μg/ml streptomycin sulfate (Irvine Scientific) and 2 mM l-glutamine (Gemini Biological Products, Calabasas, CA). One day before each experiment, cells were trypsinized and seeded in new tissue culture dishes at less than 10% confluency to minimize cell–cell contacts. These cells were trypsinized, plated, and cultured on ECM proteins (described below), harvested by incubation in the presence of 2.5 mg/ml Clostridial collagenase in serum-free medium for 15–20 min at 37°C, washed in PBS (137 mM NaCl/2.6 mM KCl/10 mM Na2HPO4/1.7 mM KH2PO4, pH 7.4), and used for the analyses described below.
ECM Proteins.
Nonfibrillar and fibrillar collagen was prepared from pepsin-solubilized acid-dissolved bovine dermal type I collagen (Vitrogen 100, Collagen Corp., Palo Alto, CA). To obtain fibrillar gels, the collagen solution was neutralized in PBS and 0.1 M NaOH to reach a final pH ranging between 7.2 and 7.4. The final collagen concentration was adjusted to 1 mg/ml with PBS, and the preparation was allowed to form a thick gel for at least 1 h at 37°C in tissue culture dishes (Falcon) before cells were plated. Alternatively, cells were embedded into the collagen gel by quickly adding suspended cells in freshly neutralized collagen at 4°C before polymerization at 37°C. Nonfibrillar collagen was prepared by diluting the unneutralized collagen in 0.01 M HCl to reach a final concentration of 1 mg/ml. This acid-treated collagen was then plated in tissue culture dishes (0.1 ml/cm2) and allowed to attach overnight at 37°C. To prepare dishes precoated with other ECM proteins, fibronectin and its 40-kDa and 120-kDa trypsin-generated fragments (GIBCO/BRL), vitronectin or laminin (Promega) was diluted to a final concentration of 10 μg/ml in PBS, placed in tissue culture dishes (0.1 ml/cm2), and allowed to attach at 4°C overnight. After removal of the solution, dishes coated with nonfibrillar collagen, fibronectin, fibronectin fragments, vitronectin, or laminin were washed twice with PBS before the addition of the cells.
Cell Cycle Analysis.
Cells in suspension were permeabilized in the presence of 0.5% Nonidet P-40 and their nuclei stained in the presence of 10 μg/ml of propidium iodide (PI) containing 0.2 mg/ml RNase A (Calbiochem) and 0.5 mM EDTA. Cell cycle parameters of 10,000 nuclei from these cells were acquired with a FACScan flow cytometer (Becton Dickinson) by using gates to exclude multiple cells. The data generated were analyzed with cellquest (Becton Dickinson) and modfit lt (Verity, Topsham, ME) softwares. When indicated, cells were pulse labeled in culture with 10 μM BrdUrd for 2 h at 37°C. At indicated time points, cells were harvested, permeabilized, and stained with PI, and their DNA was acid denatured by addition of 1 M HCl. The presence of BrdUrd incorporated into the DNA was then examined by indirect immunofluorescence and flow cytometry in the presence of anti-BrdUrd mAb followed by FITC-conjugated secondary antibody (24). The threshold for positive staining was based on background staining with an isotype control antibody. Green (FITC) and red (PI) fluorescence was quantified by dual parameter flow cytometry.
Quantification of Apoptotic Cells.
Collagenase-treated M24met cells (106) in suspension were washed once in PBS at 4°C and analyzed by using the ApoAlert kit (CLONTECH). After addition of FITC-conjugated Annexin-V (0.5 μg/ml) and PI (1.2 μg/ml), cells were incubated for 15 min in the dark and analyzed by dual parameter flow cytometry. As positive control, we used DAOY medulloblastoma cells undergoing apoptosis (data not shown).
Immunoprecipitation, Western Blotting, and in Vitro Kinase Assays.
Resuspended M24met cells (106–107 cells/ml) were lysed at 4°C in RIPA buffer (50 mM Hepes, pH 7.5/150 mM NaCl/1 mM EDTA/2.5 mM EGTA/10 μg/ml aprotinin/10 μg/ml leupeptin/0.5% Nonidet P-40/0.1% Triton-X-100), and the lysates were clarified by centrifugation at 14,000 × g and 4°C (25). The protein concentration in the supernates was determined with the DC Protein Assay system from Bio-Rad. These lysates were either directly examined by PAGE and Western blot analysis or were used for immunoprecipitation. Immunoprecipitation in these lysates was performed by incubating equal amounts of proteins in the presence of an anticyclin E mAb bound to Sepharose beads (10 μl/sample). After the addition of 50 μl of a 1:1 slurry of protein A-Sepharose to increase the volume of the pellets, the beads were collected by centrifugation at 14,000 × g at 4°C. Immunoprecipitates were washed three times in RIPA buffer, suspended in ×2 Laemmli buffer (125 mM Tris, pH 6.8/20% glycerol/4% SDS/10% β-mercaptoethanol/0.05% bromophenol blue), and eluted by boiling. The proteins in these eluates or in total cell lysates were resolved by SDS/PAGE on 10% polyacrylamide gels and transferred to nitrocellulose membranes (26). Membranes were probed with anticyclin D1, anticyclin E, anti-CDK2, anti-p21CIP1, or anti-p27KIP1 antibodies (each 0.2 μg/ml) for 1–2 h at room temperature, and the immunoreactive complexes were detected by enhanced chemiluminescence (ECL, Amersham) by using peroxidase-conjugated goat anti-mouse immunoglobulins (0.2 μg/ml) or donkey anti-rabbit immunoglobulins (dilution 1/2,500–1/5,000) secondary antibodies. For the cyclin E-associated kinase assay, the immunoprecipitates were washed twice with RIPA buffer and once with H1 kinase buffer (50 mM Tris⋅HCl, pH 7.4/10 mM MgCl2/1 mM DTT). The kinase activity in these immunoprecipitates was performed with 10 μg of histone H1 as substrate and 10 μCi of [γ32-P] ATP per reaction, as described (27).
RNA Extraction and Northern Blotting.
Total RNA was isolated by using RNeasyR Mini Kit (Qiagen, Chatsworth, CA). Five micrograms of RNA for each lane was electrophoresed on a 1% formaldehyde agarose gel and blotted onto Zeta-probe membrane (Bio-Rad). Blots were sequentially hybridized with [32P]-labeled cDNA probes (p27KIP1 and 18S rRNA) for 16 h at 42°C in ultrasensitive hybridization buffer (Ambion, Austin, TX). Human p27KIP1 cDNA was purchased from American Type Culture Collection and the cDNA probe for 18S rRNA from Ambion. After each hybridization, the blot was washed in ×2 SSC (300 mM NaCl/30 mM sodium citrate, pH 7.0/16.7 mM NaH2PO4/0.067% SDS) and in ×0.1 SSC at 42°C before autoradiography at −80°C.
Integrin Experiments.
To determine the expression of integrins, M24met cells (3 × 105) were collected with cell dissociation buffer (GIBCO/BRL), washed with medium containing 10% FBS, and incubated for 30 min at 4°C in the presence of 1 μg of antiintegrin or negative control antibody in 100 μl PBS containing 0.1% BSA. After two washes in PBS containing 0.1% BSA, cells were resuspended in 50 μl of the same buffer containing 3 μg of FITC-conjugated goat anti-mouse IgG and incubated for 20 min at 4°C. After three washes in PBS–BSA, green fluorescence was detected and quantified by flow cytometry.
To determine the integrins involved in adhesion of M24met cells to collagen, we used nontissue culture-treated 48-well cluster plates coated with nonfibrillar or fibrillar collagen as described above. Nonspecific binding sites were blocked by addition of 1% heat-denatured BSA in PBS for 1 h at 4°C, and negative control wells were coated with 1% BSA only. Cells were trypsinized, and trypsin was neutralized by addition of medium containing 10% FBS. Cells were washed and resuspended in adhesion medium (culture medium enriched with 1 mM MgCl2 and 0.2 mM MnCl2), preincubated for 10 min in the presence of specific antiintegrin or isotype control mAbs (10 μg/ml), and plated at 20,000 cells per plate. After 20 min, nonattached cells were removed by gentle washing (three times) with adhesion medium by using a hand pipette. To avoid artifacts because of nonspecific absorption of Thiazolyl blue crystals in the matrix, cells attached on fibrillar collagen were released by collagenase digestion and further treated in suspension. Attached cells or collagenase-treated cells were stained in the presence of 0.25 mg/ml Thiazolyl blue for 2 h at 37°C. The crystallized reaction product was dissolved with DMSO and quantified by dual densitometry at 550–650 nm.
To study the role of integrins on cell cycle, cells in suspension were preincubated with antiintegrin mAbs (10 μg/ml), plated on collagen-coated culture dishes, and incubated for 20 h at 37°C. Adherent cells were collected by collagenase treatment and analyzed for distribution in the phases of the cell cycle, as described above. Alternatively, experiments were performed on immobilized antiintegrin mAbs. Culture wells were coated for 2 h at 4°C with 25 μg/ml of unconjugated goat anti-mouse IgG in PBS and washed three times with PBS. Nonspecific adhesion sites were blocked by addition of 1% heat-denatured BSA in PBS for 1 h at 4°C. After one wash with PBS, wells were coated with 10 μg/ml of a specific antiintegrin antibody for 16 h at 4°C. Wells were washed three times in PBS, and cells were plated and incubated overnight at 37°C (28). After washing with PBS, cells were lysed in the presence of RIPA buffer (for Western blot analysis) or 0.5% Nonidet P-40 in DNA staining solution (for cell cycle analysis).
Results
Fibrillar Collagen Blocks Cell Cycle Progression in the G1 Phase Without Affecting Apoptosis.
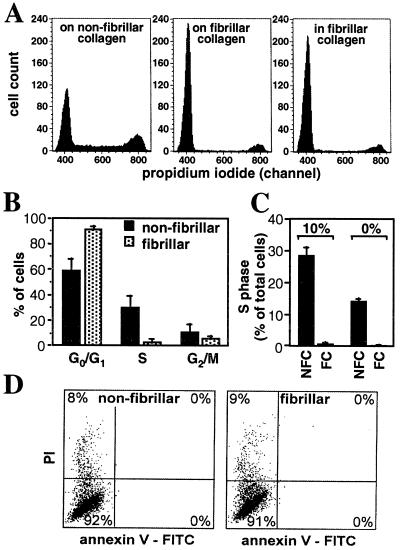
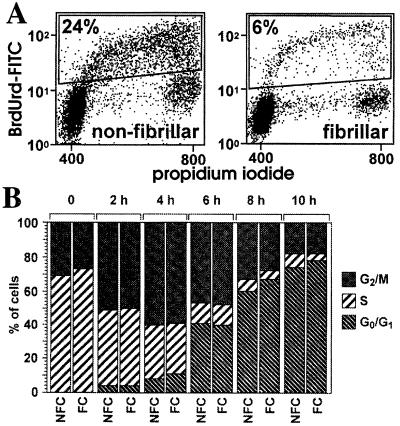
To determine whether the growth inhibitory effect we had previously observed when M24met melanoma cells were grown in the presence of fibrillar collagen was the result of a specific effect on cell cycle and/or on programmed cell death, we performed a series of experiments in which M24met cells plated overnight in the presence of nonfibrillar or fibrillar collagen were examined by flow cytometry for distribution of the DNA content (cell cycle) and for modifications of the plasma membrane (apoptosis) (Fig. 1). We also compared cells that were plated on top of a fibrillar collagen gel with cells that were embedded in the fibrillar collagen gel. An analysis of DNA content distribution indicated a significant difference between cells plated in the presence of fibrillar collagen and cells plated on nonfibrillar collagen. In the experiment shown (Fig. 1A), 51% of the cells were in G0/G1 phase, 29% in S phase, and 20% in G2/M phase on dishes coated with nonfibrillar collagen. In contrast, in the presence of fibrillar collagen, 90% of the cells were in G0/G1 phase, 2% in S phase, and 8% in G2/M phase, and no significant difference was observed between cells plated on top of the collagen gel or inside the collagen gel. Over a total of 10 independent experiments, the average percentage of cells in S phase was 30 ± 9% and 3 ± 2% when cells were cultured in the presence of nonfibrillar and fibrillar collagen, respectively (Fig. 1B). The inhibitory effect of fibrillar collagen on cell cycle progression did not involve serum-derived growth factors, because similar levels of inhibition on the percentage of cells in S phase were observed in the presence or absence of FBS when cells were plated on fibrillar collagen (Fig. 1C). No changes in apoptosis were detected between cells cultured on fibrillar or nonfibrillar collagen, as determined by analysis of the membrane polarity (Annexin-V) and permeability (PI) (Fig. 1D). We performed flow cytometric analyses on M24met cells labeled with the thymidine homolog BrdUrd to confirm that the changes in DNA distribution observed in the presence of fibrillar collagen reflected an inhibition of entry into S phase. In the experiment shown (Fig. 2A), 24% of the cells cultured on nonfibrillar collagen had taken BrdUrd as they were synthesizing new DNA, whereas only 6% of the cells plated on fibrillar collagen had taken BrdUrd, suggesting that they were blocked in G1. To examine whether the progression through other phases of the cell cycle was affected by the presence of fibrillar collagen, we performed a pulse–chase experiment in the presence of BrdUrd. The data (Fig. 2B) indicated that the physical nature of the collagen did not affect the kinetics of the progression of BrdUrd-labeled cells (cells in S phase) through the S phase (2 h), the G2 and M phases (4 h), and the return to the G1 phase (6–10 h). From these data, we concluded that the presence of fibrillar collagen specifically arrested M24met cells in G1 phase.
Figure 1.
The presence of fibrillar collagen blocks the cell cycle of M24met cells in the G1 phase without stimulating apoptosis. (A) M24met human melanoma cells were cultured for 20 h on top of a fibrillar collagen, embedded into the collagen gel, or plated in tissue culture dishes precoated with nonfibrillar collagen. Cells collected by collagenase treatment were permeabilized and stained with PI for nuclear DNA content analysis by flow cytometry. The mean fluorescence intensity of cells with a relative DNA content of 1 (G0/G1 phases) was 400 and that of cells with a relative DNA content of 2 (G2/M phases) was 790. (B) Distribution of cells in different phases of the cell cycle (average + SD), based on 10 independent experiments. (C) In some experiments, we compared cells cultured in the presence or absence of serum. The histograms present the percentage of cells in S phase, as the average + SD in one representative experiment performed in triplicate. NFC, nonfibrillar collagen; FC, fibrillar collagen. (D) Unpermeabilized cells were incubated in the presence of PI and FITC-conjugated-Annexin V and analyzed by bivariate flow cytometry for apoptosis. Note in both conditions a small (8–9%) percentage of PI-positive cells but an absence of cells binding Annexin-V at their surface.
Figure 2.
The presence of fibrillar collagen blocks the G1/S transition without affecting other phases. (A) Cells were incubated for 2 h in the presence of BrdUrd, washed, and further cultured. At indicated times, cells were collected by collagenase treatment, permeabilized, and dual labeled for DNA content with PI and for BrdUrd incorporation by indirect immunofluorescence. The nuclei were analyzed by bivariate flow cytometry. The dot-plot analysis of BrdUrd incorporation (y axis) vs. PI staining (x axis) was performed at the end of the incubation with BrdUrd. The percentage of BrdUrd-positive cells is indicated (Top Left). (B) Histograms of pulse–chase analysis showing the PI-based distribution of the BrdUrd positive cells over time. NFC, nonfibrillar collagen; FC, fibrillar collagen.
p27KIP1 Is Up-Regulated by Fibrillar Collagen.
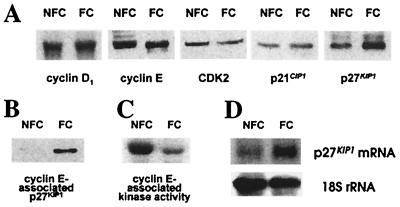
We investigated whether the expression or activity of key regulators of the G1 phase and more specifically, the G1 restriction point, was affected by the presence of fibrillar collagen. In a first set of experiments, the expression of cyclin D1, cyclin E, CDK2, p21CIP1, and p27KIP1 was examined by Western blot analysis on total cell lysates obtained from M24met cells cultured overnight on nonfibrillar or fibrillar collagen. The data (Fig. 3A) demonstrated no significant differences in the amounts of cyclin D1, cyclin E, CDK2, and p21CIP1, but a marked increase in levels of p27KIP1 when cells were cultured on fibrillar collagen. Because p27KIP1 can block the G1/S transition by forming an inhibitory complex with CDK2 associated with cyclin E, we examined the amount of p27KIP1 associated with cyclin E by performing an immunoprecipitation on cell lysates with an anticyclin E antibody. The data indicated an increased amount of p27KIP1 associated with cyclin E when M24met cells were cultured in the presence of fibrillar collagen (Fig. 3B). In vitro phosphorylation of histone H1 was then used to demonstrate that the increased association of p27KIP1 with cyclin E resulted in a substantial decrease in cyclin E-associated kinase activity (Fig. 3C). By Northern blotting analysis, a 3.8-fold increase in p27KIP1 mRNA was detected in M24met cells cultured in the presence of fibrillar collagen in comparison with nonfibrillar collagen (Fig. 3D). Thus, the presence of fibrillar collagen blocked M24met cells at the G1 checkpoint through up-regulation of p27KIP1, at least in part because of a pretranslational mechanism and a subsequent inhibition of the cyclin E-associated kinase activity.
Figure 3.
p27KIP1 is up-regulated and cyclin E-associated kinase activity is down-regulated in the presence of fibrillar collagen. M24met cells were cultured for 20 h on nonfibrillar collagen-coated plates (NFC) or on a fibrillar collagen gel (FC) and collected by collagenase digestion. (A) Aliquots of cell lysates containing 25 μg of proteins were analyzed by Western blotting by using antibodies against cyclin D1, cyclin E, CDK2, p21CIP1, and p27KIP1. (B) Aliquots of cell lysates were immunoprecipitated in the presence of an anticyclin E mAb, and the immunoprecipitates were analyzed by Western blotting with an anti-p27KIP1 antibody. (C) Similar immunoprecipitates were analyzed for kinase activity in the presence of histone H1 and [γ32P]ATP. (D) Total mRNA was extracted from cultured cells, and aliquots of 5 μg were analyzed by Northern blotting for p27KIP1 mRNA and 18S rRNA.
Growth Inhibition Is Specific to Fibrillar Collagen.
To determine whether other ECM proteins could affect the G1/S transition of M24met cells, we performed similar cell cycle analysis on M24met cells plated overnight on various ECM proteins. In comparison with nonfibrillar collagen, we observed an increase rather than a decrease of the percentage of cells in S phase, of 1.8-fold on laminin, 1.65-fold on fibronectin, 1.5-fold on the 40-kDa α-chymotrypsin-generated cell/heparin binding fragment of fibronectin, 1.4-fold on the 120-kDa α-chymotrypsin-generated cell-binding fragment of fibronectin, and 1.3-fold on vitronectin. Addition of up to 500 μg/ml fibronectin on fibrillar collagen failed to allow the cells to enter the cell cycle, whereas adding recombinant human interstitial collagenase (2.5 μg/ml) to cells plated in fibrillar collagen for 24 h resulted in a 2-fold increase in the percentage of cells in S phase. Thus the data suggested that the fibrillar structure of the collagen was a key element that controlled cell cycle progression.
α2β1 Integrin Is Involved in Adhesion to Collagen and Cell Cycle Progression.
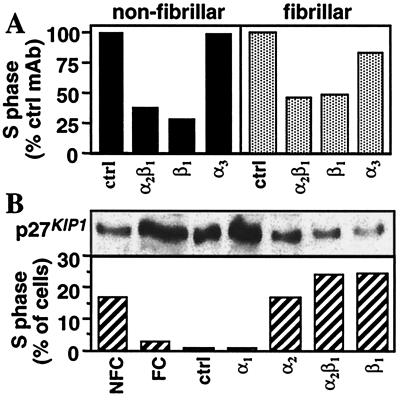
Cell attachment on the ECM is mediated by adhesion receptors and in particular by integrins. Integrins that mediate cell contact with collagen are α1β1, α2β1 and α3β1 (29). We first determined by flow cytometry that M24met cells expressed large amounts of the α2β1 and α3β1 collagen-binding integrin receptors in the absence of α1β1 (not shown). Using blocking mAbs, we then demonstrated that attachment of M24met cells to both forms of collagen was predominantly mediated by the α2β1 integrin. A 95% inhibition of adhesion to nonfibrillar collagen and a 66% inhibition to fibrillar collagen were achieved with an anti-α2β1 mAb. In contrast, the inhibition achieved with an anti-α3β1 mAb was only 43% and 4% on nonfibrillar and fibrillar collagen, respectively. To determine whether the α2β1 integrin played a positive or negative role in cell cycle regulation, we performed a series of experiments in which M24met cells were preincubated and cultured for 20 h with anti-α2β1, anti-β1 or anti-α3 function blocking mAbs. Some inhibition of adhesion was still observed with the anti-α2β1 and anti-β1 mAbs, and only cells that had attached were then examined for cell cycle distribution. The data (Fig. 4A) indicated that cells plated on nonfibrillar collagen in the presence of an anti-α2β1 or an anti-β1 antibody were inhibited at the G1/S transition (62% and 73% inhibition of S phase, respectively). On fibrillar collagen, the antibodies further inhibited cell cycle progression at the G1/S transition (53% and 50% inhibition of S phase, respectively). There was only little inhibition of cell cycle progression in the presence of an anti-α3. Thus the data pointed to α2β1 playing a positive role in M24met cell proliferation. Confirmation of such a positive role was obtained by performing a series of experiments in which antibodies against the α2β1 integrin were used as agonists and stimulators of integrin clustering rather than antagonists. This was accomplished by culturing M24met cells for 20 h on anti-α2, anti-α2β1, or anti-β1 mAbs immobilized on antimouse IgG-coated plates. It has been previously demonstrated that under these conditions, blocking antiintegrin mAbs act as agonists rather than antagonists (28). These experiments (Fig. 4B) indicated that interaction between α2β1 integrins and these antibodies stimulated cell entry into S phase and simultaneously down-regulated the levels of p27KIP1. This effect was specific because it was not observed with an anti-α1 antibody or an isotype control antibody.
Figure 4.
Integrin α2β1 is involved in M24met cell cycle progression. (A) Resuspended M24met cells were preincubated in the presence of specific antiintegrin mAbs and cultured for 20 h on nonfibrillar or fibrillar collagen. Attached cells were then collected by collagenase treatment, and the proportion of cells in S phase was estimated by PI staining. Data in histograms represent the proportion of cells in S phase as a percentage of the S phase observed in the presence of a negative control antibody (23% on nonfibrillar collagen and 4% in the presence of fibrillar collagen). (B) Cells were cultured for 20 h on immobilized anti-α1, anti-α2, anti-α2β1, anti-β1, negative control antibodies, or nonfibrillar (NFC) and fibrillar collagen (FC). The proportion of cells in S phase was quantified by PI staining of their nuclei, and the amount of p27KIP1 in total cell lysates (15 μg proteins/lane) was determined by Western blot.
Discussion
In this work, we used an in vitro model that reconstitutes the fibrillar structure of type I collagen to examine the effect of the physical nature of the ECM on tumor cell proliferation. We provide evidence that contact with fibrillar collagen inhibits the proliferation of malignant and highly metastatic M24met cells and that this inhibition depends on the organization of the collagen into fibrils. Inhibition of proliferation is the result of a specific growth arrest at the G1/S transition that involves an increase in p27KIP1 mRNA and protein and a subsequent inhibition of cyclin E-associated kinase activity. We demonstrate that the collagen-binding α2β1 integrin is directly involved in p27KIP1 regulation and cell cycle progression in those cells. Its effect, however, depends of the physical state of the collagen and is restricted in the presence of fibrils.
In normal cells, p27KIP1 and CDK2 are important targets for cell cycle regulation in response to extracellular signals generated by growth factors, anchorage to the matrix, and cell–cell contacts. Because most tumor cells are known to be anchorage independent, it has been thought that their proliferation is no longer affected by signals originating from the ECM. Our data clearly demonstrate that this is not the case, as we provide evidence that contact with fibrillar collagen results in a p27KIP1-dependent inhibition of cell cycle progression in malignant melanoma cells. This work, in conjunction with two recent publications demonstrating that p27KIP1 can be up-regulated by growth factors (30) and cell–cell contacts (31), emphasizes the key role played by p27KIP1 in the regulation of the proliferation of malignant cells in response to signals originating from the extracellular milieu. The intracellular concentration of p27KIP1 is mainly controlled at the posttranscriptional level. In growth-stimulated normal cells, although p27KIP1 mRNA levels are constant during the cell cycle, the p27KIP1 protein undergoes rapid degradation through a process initiated by phosphorylation by its target enzyme, CDK2, and completed by the ubiquitin/proteasome pathway (32–35) or by proteolytic processing (36). However, transcriptional up-regulation of the p27KIP1 gene was recently observed after interleukin-6 treatment in interleukin-6 sensitive melanoma cell lines (37). The role of p27KIP1 in tumor cell proliferation has been the subject of recent studies. Significantly decreased amounts of p27KIP1, compared with that of adjacent normal tissue, were reported in several human cancers, and it has been suggested that low levels of p27KIP1 are a prognostic marker of poor outcome (reviewed in refs. 38 and 39). The mechanisms leading to decreased levels of p27KIP1 in cancer cells are still poorly understood but because mutations of the gene encoding p27KIP1 are rare (40, 41), alterations in mechanisms regulating its degradation are more likely to be involved. Whether increased p27KIP1 levels in M24met cells cultured on fibrillar collagen result only from increased transcriptional/translational activity or also from dysregulation of its degradation is currently investigated.
Our data demonstrate that the integrin α2β1 is involved in attachment of M24met cells on fibrillar and nonfibrillar collagen and plays a key role in growth control. In this regard, two nonmutually exclusive mechanisms can be involved in the inhibition of proliferation of M24met cells in the presence of fibrillar collagen. As a first possibility, clustering of α2β1 is required to generate a positive growth stimulatory signal, and lack of binding results in absence of such signal. Therefore, clustering could be inhibited on fibrillar collagen as a result of a restriction in the number and/or distribution of α2β1 binding sites. On breakdown of the fibrils, the sites would be redistributed in a manner that facilitates integrin clustering and down-regulation of p27KIP1. This hypothesis is supported by our experiments, in which we demonstrate that agonist mAbs against α2β1 promote cell spreading, down-regulate p27KIP1, and stimulate cell cycle progression. It is also consistent with the observation that α2β1 expression in melanoma increases as tumors progress from a radial growth phase to a vertical growth phase (42). However, a second possibility is that the type of signal generated by contact between α2β1 and collagen depends on the physical nature of the collagen. In the presence of fibrils, a dominant signal that actively up-regulates p27KIP1 would be generated. This possibility was suggested by Koyama et al. (13) and was further supported by Zutter et al. (17), who showed that the morphological changes of carcinoma cells are inhibited in the presence of fibrillar collagen only in cells overexpressing α2β1 but not in the parental cells that do not express α2β1.
In summary, our data illustrate the importance of the integrity of the ECM in controlling the growth of malignant cells. As the ECM around tumor cells becomes proteolytically modified, growth stimulatory signals mediated by integrins that affect tumor cell behavior are generated. Thus, proteolytic degradation of the ECM in tumors has a double effect. It allows cancer cells to locally invade tissues but also to escape negative growth regulatory signals. In this respect, inhibition of ECM degradation by protease inhibitors will have not only an antiinvasive and metastatic effect but also a cytostatic effect on tumor cell proliferation.
Acknowledgments
This work was supported by grant RO1 CA42919 from the National Institutes of Health (Y.D.). P.H. was the recipient of a Postdoctoral Fellowship Supplementation Support from the University of Southern California/Norris Cancer Center, of a Career Development Fellowship from the Childrens Hospital Los Angeles Research Institute, and of a Postdoctoral Fellowship from the Fonds de la Recherche Scientifique of the Université Catholique de Louvain. We thank Prof. Y. Eeckhout for his support and critical review of the manuscript and Mrs. J. Rosenberg for assistance in preparing this manuscript. P.H. is grateful to Lora W. Barsky and Dr. E. Monika Smogorzewska for training in flow cytometry.
Abbreviations
- ECM
extracellular matrix
- CDK
cyclin-dependent kinase
- PI
propidium iodide
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170290997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170290997
References
- 1.Weaver V M, Roskelley C D. Trends Cell Biol. 1997;7:40–42. doi: 10.1016/S0962-8924(97)30078-6. [DOI] [PubMed] [Google Scholar]
- 2.Lukashev M E, Werb Z. Trends Cell Biol. 1998;8:437–441. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
- 3.Lelievre S, Weaver V M, Bissell M J. Recent Prog Horm Res. 1996;51:417–432. [PMC free article] [PubMed] [Google Scholar]
- 4.Huhtala P, Humphries M J, McCarthy J B, Tremble P M, Werb Z, Damsky C H. J Cell Biol. 1995;129:867–879. doi: 10.1083/jcb.129.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werb Z, Tremble P M, Behrendtsen O, Crowley E, Damsky C H. J Cell Biol. 1989;109:877–889. doi: 10.1083/jcb.109.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzac F, Retta S F, Albini A, Melchiorri A, Koteliansky V E, Geuna M, Silengo L, Tarone G. J Cell Biol. 1994;127:557–565. doi: 10.1083/jcb.127.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottazzi M E, Assoian R K. Trends Cell Biol. 1997;7:348–353. doi: 10.1016/S0962-8924(97)01114-8. [DOI] [PubMed] [Google Scholar]
- 8.Giancotti F G, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 9.Aplin A E, Howe A K, Juliano R L. Curr Opin Cell Biol. 1999;11:737–744. doi: 10.1016/s0955-0674(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 10.Sherr C J, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 11.Schor S L. J Cell Physiol. 1980;41:159–175. doi: 10.1242/jcs.41.1.159. [DOI] [PubMed] [Google Scholar]
- 12.Martin G R, Sank A C. In: Peptide Growth Factors and Their Receptors. Sporn M B, Roberts A B, editors. Berlin: Springer; 1990. pp. 463–477. [Google Scholar]
- 13.Koyama H, Raines E W, Bornfeldt K E, Roberts J M, Ross R. Cell. 1996;87:1069–1078. doi: 10.1016/s0092-8674(00)81801-2. [DOI] [PubMed] [Google Scholar]
- 14.Varner J A, Cheresh D A. Curr Opin Cell Biol. 1996;8:724–730. doi: 10.1016/s0955-0674(96)80115-3. [DOI] [PubMed] [Google Scholar]
- 15.Ben Zeev A. Curr Opin Cell Biol. 1997;9:99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- 16.Keely P, Parise L, Juliano R. Trends Cell Biol. 1998;8:101–106. doi: 10.1016/s0962-8924(97)01219-1. [DOI] [PubMed] [Google Scholar]
- 17.Zutter M M, Santoro S A, Staatz W D, Tsung Y L. Proc Natl Acad Sci USA. 1995;92:7411–7415. doi: 10.1073/pnas.92.16.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petitclerc E, Stromblad S, von Schalscha T L, Mitjans F, Piulats J, Montgomery A M, Cheresh D A, Brooks P C. Cancer Res. 1999;59:2724–2730. [PubMed] [Google Scholar]
- 19.Birkedal Hansen H. Curr Opin Cell Biol. 1995;7:728–735. doi: 10.1016/0955-0674(95)80116-2. [DOI] [PubMed] [Google Scholar]
- 20.Murphy G. Matrix Biol. 1997;15:509–518. [Google Scholar]
- 21.Koop S, Khokha R, Schmidt E E, MacDonald I C, Morris V L, Chambers A F, Groom A C. Cancer Res. 1994;54:4791–4797. [PubMed] [Google Scholar]
- 22.DeClerck Y A, Perez N, Shimada H, Boone T C, Langley K E, Taylor S M. Cancer Res. 1992;52:701–708. [PubMed] [Google Scholar]
- 23.Montgomery A M, Mueller B M, Reisfeld R A, Taylor S M, DeClerck Y A. Cancer Res. 1994;54:5467–5473. [PubMed] [Google Scholar]
- 24.Larsen J K. In: Flow Cytometry. Darzynkiewicz Z, Robinson J P, Crissman H A, editors. San Diego: Academic; 1994. pp. 377–388. [Google Scholar]
- 25.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnette W N. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 27.Gorospe M, Liu Y, Xu Q, Chrest F J, Holbrook N J. Mol Cell Biol. 1996;16:762–770. doi: 10.1128/mcb.16.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stromblad S, Becker J C, Yebra M, Brooks P C, Cheresh D A. J Clin Invest. 1996;98:426–433. doi: 10.1172/JCI118808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santoro S A, Zutter M M, Wu J E, Staatz W D, Saelman E U, Keely P J. Enzymology. 1994;245:147–183. doi: 10.1016/0076-6879(94)45010-2. [DOI] [PubMed] [Google Scholar]
- 30.Kim J W, Kim H S, Kim I K, Kim M R, Cho E Y, Kim H K, Lee J M, Namkoong S E. Gynecol Oncol. 1998;69:230–236. doi: 10.1006/gyno.1998.5003. [DOI] [PubMed] [Google Scholar]
- 31.St. Croix B, Sheehan C, Rak J W, Florenes V A, Slingerland J M, Kerbel R S. J Cell Biol. 1998;142:557–571. doi: 10.1083/jcb.142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen H, Gitig D M, Koff A. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montagnoli A, Fiore F, Carrano A C, Draetta G F, Hershko A, Pagano M. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirane M, Harumiya Y, Ishida N, Hirai A, Miyamoto C, Hatakeyama S, Nakayama K, Kitagawa M. J Biol Chem. 1999;274:13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- 37.Kortylewski M, Heinrich P C, Mackiewicz A, Schnierthauer U, Klingmuller U, Nakajima K, Hirano T, Horn F, Behrmann I. Oncogene. 1999;18:3742–3753. doi: 10.1038/sj.onc.1202708. [DOI] [PubMed] [Google Scholar]
- 38.Sgambato A, Cittadini A, Faraglia B, Weinstein I B. J Cell Physiol. 2000;183:18–27. doi: 10.1002/(SICI)1097-4652(200004)183:1<18::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 39.Slingerland J, Pagano M. J Cell Physiol. 2000;183:10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi S, Koeffler H P, Hinton D R, Miyoshi I, Melmed S, Shimon I. J Endocrinol. 1998;157:337–341. doi: 10.1677/joe.0.1570337. [DOI] [PubMed] [Google Scholar]
- 41.Dahia P L, Aguiar R C, Honegger J, Fahlbush R, Jordan S, Lowe D G, Lu X, Clayton R N, Besser G M, Grossman A B. Oncogene. 1998;16:69–76. doi: 10.1038/sj.onc.1201516. [DOI] [PubMed] [Google Scholar]
- 42.Duncan L M, Bouffard D, Howard C, Mihm M C, Jr, Byers H R. Mod Pathol. 1996;9:938–943. [PubMed] [Google Scholar]