Abstract
Linkage analysis has been widely used to identify from family data genetic variants influencing quantitative traits. Common approaches have both strengths and limitations. Likelihood ratio tests typically computed in variance component analysis can accommodate large families but are highly sensitive to departure from normality assumptions. Regression-based approaches are more robust but their use has primarily been restricted to nuclear families. In this paper, we develop methods for mapping quantitative traits in moderately large pedigrees. Our methods are based on the score statistic which in contrast to the likelihood ratio statistic, can use nonparametric estimators of variability to achieve robustness of the false positive rate against departures from the hypothesized phenotypic model. Because the score statistic is easier to calculate than the likelihood ratio statistic, our basic mapping methods utilize relatively simple computer code that performs statistical analysis on output from any program that computes estimates of identity-by-descent. This simplicity also permits development and evaluation of methods to deal with multivariate and ordinal phenotypes, and with gene-gene and gene-environment interaction. We demonstrate our methods on simulated data and on fasting insulin, a quantitative trait measured in the Framingham Heart Study.
Keywords: Genetic linkage, Quantitative trait locus, Score test, Extended pedigrees, Variance component analysis
1 Introduction
After a hiatus following the seminal paper of [Haseman and Elston 1972], linkage analysis of quantitative traits has been the subject of considerable research since the middle 1990s. Notable contributions have come from Blangero and colleagues (e.g., [Blangero and Almasy, 1997, Almasy and Blangero, 1998], etc.). Their research and the related software SOLAR uses a likelihood analysis of a components of variance model that depends on the critical assumption that phenotypes are conditionally multivariate normal, given identity by descent (IBD) counts.
A strength of this method is its ability to adapt, subject to some computational problems mentioned below, to general pedigree structures. Its weakness is the associated computational burden and the lack of robustness of its false positive error rate when the underlying assumption of multivariate normality is not satisfied.
The original Haseman-Elston method is simpler computationally; and because its test statistic is based on a regression, not a likelihood, its false positive error rate is asymptotically robust against the failure of the normality assumption. However, it is not fully efficient under the working assumption of multivariate normality and as originally conceived was applicable only to sibling pairs. Although it can be made fully efficient and extended from sibling pairs to more general pedigrees [Sham et al. 2002], it loses some of it conceptual simplicity and no longer seems to enjoy its once favored position.
Recently several authors [e.g., Tang and Siegmund, 2001, Wang and Huang, 2002, Chen, Broman and Liang, 2004, Bhattacharjee et al., 2008] have pointed out that a components of variance approach, using what amounts to a score statistic, combines some of the attractive features of both methods. Its false positive rate can be made asymptotically robust by using a nonparametric estimate of variability, and because nuisance parameters need be estimated only once, under a null model, the computational burden is reduced. Hence more complex models involving, say gene-gene or gene-environment interaction, can be considered. Various “regression based” or Haseman-Elston statistics are special cases.
The purpose of this paper is to delineate the similarities and differences between the likelihood and score statistic/regression approaches, and to give several examples of these differences. In some cases the two methods yield very similar results, as they should if the normality assumption is reasonable. In other cases the lack of robustness of the likelihood ratio method can suggest quite different conclusions. We have not tried to achieve systematic understanding of these similarities and differences, which would require the analysis of many more examples, both simulated and real, and most likely can only accumulate with incremental experience. To illustrate the relative simplicity of our methods, our analysis uses MERLIN [Abecassis et al., 2002] or LOKI [Heath 1997] as a front end to estimate IBD distributions, and a small suite of R functions [R Development Core Team 2006], which can be easily expanded to deal with more complex cases. These R functions are available on request, and we welcome feedback regarding their use and suggestions for future development, should that appear to be desirable.
As a simple illustration, we present simulated data based on a sample of size N = 200 from an idealized three generation pedigree. To confirm the flexibility of our methods to accommodate varying pedigree structures and missing data, we discuss simulated data from a set of N = 330 two generation pedigrees that closely resemble the largest two generation pedigrees of the Framingham Heart Study (FHS). We also present one analysis of actual FHS data based on our model for gene-environment interaction and discuss briefly our experience with some other analyses of actual FHS data, which includes use of the substantially larger three generations pedigrees that have recently become available. A short description of the FHS study follows.
The Original FHS cohort comprised 5,209 adults between 28 and 62 years of age in about two-thirds of the households in the town of Framingham, Massachusetts in 1948 [Dawber et al. 1951]. These subjects have been examined every two years since the inception of the study. In 1971, a second cohort was initiated, the Framingham Offspring cohort (n=5,124), consisting mainly of children of the Original cohort and their spouses, who have been examined approximately every four years since 1971 [Kannel et al. 1979]. To date, 29 and 8 exams have been completed for the Original and Offspring cohort, respectively. At each examination, extensive data are gathered, including routine medical history, medication usage, physical examination, and laboratory assessment of cardiovascular disease (CVD) risk factors. A genome scan has been performed on the largest 330 families from the Original and Offspring cohorts (1,702 genotyped individuals), using both short tandem repearts (STR) and a dense set of approximately 100,000 single nucleotide polymorphisms (SNP) [Cupples et al. 2007]. Recently a third generation has been added to the FHS study [Splansky et al. 2007].
In addition to a simulation study based on the two generation Framingham pedigree structures comparing and contrasting likelihood ratio and robust score statistic, we illustrate the various approaches with data based on fasting insulin measured in the Offspring cohort of the FHS.
Remark
Although our primary interest is in population-based data, as in the FHS, the robustness to false positive errors achieved by our score statistic is based on an analysis conditional on phenotypes. Hence one still has robustness if pedigrees are ascertained, although power loss is expected in the absense of a suitable adjustment for ascertainment in the estimation of nuisance parameters, such as the phenotypic variance (e.g., Peng and Siegmund, 2004).
2 The basic variance component model
Let Yi be the vector of phenotypes for all members of a pedigree i, and Xi be the covariate measurements (matrix) for all pedigree members. The standard variance component model [Lange et al. 1976, Amos 1994] assumes that the vector Y has mean value (conditional on the observed covariates) E[Y|X]= m + aX and covariance matrix
Here m is the overall mean; a is a vector of covariate effects; is the phenotypic variance, which is assumed to be the sum of genotypic and environmental variances, and , respectively; and are additive and dominance variance components; φij is the kinship coefficient; is the probability that pair i and j share k alleles IBD; and the ellipsis denotes a linear combination of interaction variance components, which we have not written explicitly. For example, if there are pairwise additive-additive interactions, and no other pairwise nor higher order interactions, then , and the omitted terms equal . See Tang and Siegmund [2002] for the general case of pairwise interactions and no higher order interactions.
If there is a quantitative trait locus (QTL) located at t, we are interested in the covariance conditional on the proportion of alleles shared IBD at location t. For future developments, as discussed below, it is convenient to reparameterize the variance components. To this end, let and . Here and are respectively the additive and dominance variance components due to the QTL at t, and again the ellipses stand for omitted interaction variance components [Tang and Siegmund, 2002]. Then
Here ρij is the unconditional phenotypic correlation of the pair i and j, πtij is the identity by descent proportion for pair i and j at t, and is the indicator variable that pair i and j share k alleles IBD at t. Note that a test of α = 0 is actually a test of no linkage, because α is made up of a positive linear combination of all variance components related to the putative QTL. We shall call α and δ the generalized additive effect and generalized dominance deviation, respectively.
Under the “working” assumption that Y, conditional on πtij, is multivariate normal, the marginal log likelihood for a QTL located at t has the following form:
where mX = E(Y|X), ρ is the vector of unconditional correlations for different types of relative pairs, and the sum is over all pedigrees.
In the following section we shall make the additional simplifying assumption that and interaction variance components are all 0 and that there are no environmental correlations between different individuals. The importance of this assumption is to allow us to write , thus reducing the very large number of relative-pair-specific nuisance parameters to only one, . However, the general case is important to keep in mind. The hypothesis α = 0 is the hypothesis of interest, that t is unlinked to the phenotype, and the robust score test given below of α = 0 is a legitimate test of that hypothesis regardless of whether the simplified model is correct or not. This stands in contrast to the case for association, where under the usual assumptions the efficient score for an additive effect has a noncentrality depending only on the putative additive variance, so a locus acting only through dominance or interaction will show no indication of association unless these other effects are included explicitly in the model [Dupuis et al. 2007].
2.1 Efficient score test
Likelihood ratio statistics (LRT) have long been used for testing H0:α = 0; see for e.g. [Blangero and Almasy, 1997, Almasy and Blangero, 1998]. As an alternative to the LRT, Tang and Siegmund [2001] and others [Putter et al. 2002; Wang and Huang 2002] suggested using the efficient score statistic. The latter has some advantages over the LRT because it does not require maximum likelihood estimation at each genomic location tested for linkage. In addition, the type-I error of the score test can be made robust to departures from normality, a problem which can greatly inflate the type-I error of the LRT [Allison et al. 1999].
To obtain the efficient score to test for a QTL at location t, one takes the first derivative of the likelihood function with respect to α evaluated at α = 0 [Tang and Siegmund 2001]:
| (1) |
where
| (2) |
has elements wij and Aπ is a matrix whose elements are the pairwise centered IBD proportions.
The efficient score is standardized by an estimate of its variance to form a test statistic to determine the presence or absence of linkage at location t. If the multivariate normality assumption holds, one can estimate the variance using the expectation of the second derivative of the likelhood function evaluated at α = 0:
| (3) |
where the entries of Σ−1 are represented by σij.
Tang and Siegmund [2001] suggested estimating the variance conditional on phenotypes in order to create a statistic robust to departure from the normality assumption under the hypothesis of no linkage:
| (4) |
The robust score statistic, obtained by dividing the efficient score by the square root of the variance estimate, is maximized over all genome locations t: maxt Zt, where . These statistics depend on the parameters mX and Σ, which must be estimated, but they need be estimated only once—under the null hypothesis of no linkage, where α = 0.
The asymptotic expectation of Zt for a small value of α at a marker completely linked to a QTL is the ratio of the expectation of (1) to the square root of the expectation of (4), where the latter is calculated under the hypothesis of no linkage. Since the matrix W in (1) is a quadratic function of the phenotypes, its conditional expectation given the IBD proportions, Aπ, depends only on the basic covariance model for Σπ and not on the working hypothesis of multivariate normality. Hence without invoking the normality hypothesis, one can show that the expectation of (1) is
(This argument is substantially more complicated when the unknown parameters in W are replaced by estimators.) The expectation of (4) is approximately
Because W is quadratic in the phenotypes, the latter expectation involves fourth moments of phenotypes. If the working hypothesis of multivariate normality is approximately satisfied, the last expression simplifies to Σn E[tr(Σ−1Aπ)2], which in turn leads to simplification of the asymptotic noncentrality parameter, but to achieve robustness of the false positive error, we must estimate the denominator consistently even if the working hypothesis of normality fails to hold.
A second component of the efficient score is easily derived by differentiating with respect to δ. For the following reasons, however, we assume for most of what follows that δ = 0. (a) This assumption reduces the number of nuisance parameters to be estimated, hence simplifying subsequent calculations. (b) A two-dimensional efficient score leads to a two-dimensional statistic, which requires a larger rejection threshold; in addition the fact that δ ≤ α means that the possible increase in the noncentrality parameter only rarely leads to a useful increase in the power of the statistic. We return to this second point below, which has related consequences for trying to model gene × gene interactions.
The preceding calculations assume that markers are fully informative. It is easy to show that for partially informative markers, the efficient score is computed by replacing πtij with π̂tij, the expected proportion of alleles IBD given marker genotypes.
In order to standardize the efficient score in extended pedigrees, it follows from (3) and (4) that one must estimate the variances and covariances in IBD proportion between pairs of relative pairs. Simple theoretical calculation may be used in the case of fully informative markers, and in the simulation section we refer to statistics computed with the theoretical covariance values as “Theoretical.” When markers are only partially informative, there are several different possibilities, which we discuss in more detail below.
In principle the variances and covariances of π̂tij can be estimated by simulation, as suggested by Lebrec [2007]. This is computationally demanding even with fairly simple pedigree structures and seems not to be feasible with moderately large pedigrees. The computationally simplest solution is to replace Cov(πtij, πtkl) by the empirical values (π̂tij − φtij)(π̂tkl − φtkl) in equations (3) and (4). We call this the “Empirical” estimate of covariances in what follows.
If one were repeatedly sampling pedigrees of a given structure, it would be natural to regard these variances and covariances as parameters to be estimated from all the data. For example the variance of π̂tij could be estimated by the sample variance of all relative pairs of the same relationship as i and j. It is easy to compute the correlations (covariances) between pairs of relative pairs when markers are fully informative. In many cases, e.g., for pairs of sibling pairs, this correlation is 0. There is no obvious bias in using these theoretical values, unlike the case of variances, which are always over-estimated when theoretical values for fully informative markers are used. We have combined the use of empirical variances with theoretical correlations, where the estimator of Cov(π̂tij, π̂tkl) becomes Cor(πtij, πtkl){Σ(π̂tij − φtij)2/np1 Σ(π̂tkl − φtkl)2/np2}1/2, and the sum is taken over all np1 and np2 relative pairs of the same type. We refer to this estimator as “Estimated Variances” in the simulations section, where these different estimators are compared. See Bhattacharjee et al. [2008] for a more systematic comparison of different estimates, albeit restricted to nuclear families.
2.2 Type-I error and power approximations
For fully informative markers, one can derive a theoretical type-I error approximation to evaluate the significance of the maximum score statistic over the genome. For fully informative markers equally spaced at distance Δ, an approximation is
Here Φ and ϕ are the standard normal cumulative distribution function and probability density function, respectively, C and L are respectively the number of chromosomes and the genetic length of the region under consideration, and ν = ν[b(2rΔ)1/2] where ν(2x) ≈ x−1[Φ(x) − 1/2]/[xΦ(x) +ϕ(x)].
The parameter r is related to the recombination rate and depends on the constellation of relative pairs. If there were only sibling pairs, it would be 0.04/cM. Usually the number of sibling pairs will dominate more distant relatives to the extent that the value 0.04 can be used more generally. A proper accounting for other relatives is to use a weighted average of pair-specific values, where some of the pair-specific values are as follows: Grandparent-grandchild, 0.02; half siblings, 0.04, first cousins and double first cousins, 0.053, avuncular pairs, 0.05.
If there is a marker at 0 recombination distance from the QTL τ and E(Zτ) = ξ, power to detect the QTL can be approximated by
where ν = ν[b(2rΔ)1/2], as above.
The approximation is slightly more complicated when τ lies between markers (cf. Siegmund and Yakir, 2007, Chapter 6).
For a human genome (22 autosomes averaging 150 cM in length), markers spaced 5 cM apart and a recombination parameter β = 0.04/cM, the threshold for a genome wide 0.05 significance level is b = 3.73, corresponding to a logarithm of odds (LOD) score of 3.02. For an intermarker spacing of 1 cM, the corresponding threshold is b = 3.91 (LOD=3.32). Both of these intermarker spacings require a noncentrality parameter of about ξ = 5 to achieve 90% power and about ξ = 3.75 to achieve 50% power.
For cases where there is a preponderance of moderately large pedigrees, containing large sibships/nuclear families and more distant relatives, the distribution of ZiΔ is right skewed, and the approximation given above can be anti-conservative. A modified approximation, which involves more computation but can be substantially more accurate, is discussed in an appendix. In many cases this modification has only a modest effect, which may not justify the added complications in its calculation. For an example where the effect is substantial and leads to quite different conclusions, see Shi et al. [2007].
3 Extensions to the basic variance component model
An advantage of the fact that numerical computations for the robust score statistic are relatively simple (compared to the likelihood ratio statistic) is that one can accommodate more complex models without encountering severe computational impediments. Examples considered below are gene × covariate interactions, multivariate phenotypes, and ordinal phenotypes.
A possibility that we have not yet considered in detail is a model for gene × gene interaction. As noted above, the noncentrality parameter of a statistic designed to test for an additive effect at a linked marker also involves fractions of the dominance variance and interaction variance components. This limits the noncentrality that can be obtained from efficient scores orthogonal to the efficient score for additivity [Tang and Siegmund, 2002], and thus the increase in power to detect linkage that might in principle be obtained from a more complex model involving interaction variance components. This possibility will be considered in the future, but our understanding of the relevant theory suggests that models for gene × gene interaction be given lower priority than those discussed below. This contrasts with the case in experimental genetics based on a backcross or intercross and in association analysis under standard assumptions, where interactions not included in the model do not contribute to the noncentrality.
3.1 Gene × covariate interactions
The likelihood ratio test has been extended to incorporate gene × covariate interactions for a binary covariate by Towne et al. [1997]. Later Diego et al. [2003]. introduced a quite different model for continuous covariates. Peng, Tang and Siegmund [2005] suggested a model for gene × covariate effects for arbitrary covariates (which appears to reduce to the model of Towne et al. for binary covariates) and derived the appropriate robust score tests. In this section we discuss the score test for general pedigrees.
Let x be the vector of a covariate of interest, i.e., one of the columns of X, the matrix of all covariates. Under the working assumption that all genetic effects are additive and there are no gene × gene interactions, we replace the additive genetic effect, say Ai, by Ai + xiCi. Here Ai represents a “pure” genetic effect, while Ci is that part of the genetic effect that interacts with the covariate xi. Under the assumption of convenience that both Ai and Ci are additive and without gene × gene interactions, the revised covariance matrices incorporating a gene × covariate interaction are [Peng, Tang and Siegmund 2005]:
Adding a constant to all the xi or multiplying by a constant changes the relative value of the variance components, so it is often convenient to standardize the covariates to have mean value 0 and variance 1. In the case of a two-valued covariate (without standardization) the model is “nonparametric.” It simply assigns one of three variance components to each relative pair, according to both having covariate value equal to one, both having it equal to 0, or one having covariate one while the other has covariate 0.
Conditional on IBD at a locus t, we have
Under the working (additive) model, , β = σac, ; but as in the preceding section, these parameters may involve other variance components as well. The matrix Aπ is defined in the previous section as the matrix of centered IBD proportions, Bx, π is the matrix with entries (xi + xj)(πtij − φij) and Γx, π is a matrix with entries xixj(πtij − φij). When the QTL is unlinked to location t, α = β = γ = 0; if there is no gene × environment interaction, β = γ = 0.
The efficient score under the working assumption of multivariate normality is
where has elements uij.
The conditional covariance matrix of the efficient score is Vη, with entries , E[ℓαℓβ|x, Y], etc., where, for example, .
The global test of linkage is obtained by maximizing over all genome locations t: . In practice, when the IBD proportions are not observed, one can substitute π̂tij, an estimate of the IBD proportions. Possible estimators of the covariance between IBD proportions are as discussed in the previous section.
Because α and γ are both (sums of) variance components, hence non-negative, there are constraints on the score statistic, as discussed by Peng, Tang and Siegmund [2005]. There is also a constraint arising from the Cauchy-Schwarz inequality on the parameter β, but by ignoring this last constraint we can implement the non-negativity constraints via a quadratic optimization. This approach has the advantage of generalizing immediately to higher dimensional problems. Observe that for multivariate normal data, the constrained score statistic can be obtained by a constrained maximization of the log likelihood, which is quadratic, so the maximization can be obtained from a quadratic programming algorithm. While the efficient score ℓη is only approximately trivariate normal, for these purposes we regard it as exactly multivariate normal.
In principle we could include the constraint on β by means of a more general nonlinear optimization. Since we have ignored this constraint, we must be careful if a large value of the statistic arises from a large value of ℓβ in the presence of small values of ℓα and ℓγ.
Basic approximations for the significance level and power must be modified, as discussed in Peng, Tang and Siegmund [2005] to account for the constrained three-dimensional statistic. For example, for 1 cM intermarker spacing a threshold of about 4.59 (LOD=4.57) is required for a genome wide false positive rate of 0.05. Noncentrality parameters of approximately 5.49 (4.15) produce power of approximately 90% (50%). By comparing these results with those discussed above for a one-dimensional genome scan, one finds a loss of efficiency equivalent to about 20% of the sample size if one uses the model for gene × covariate interaction when no interaction exists.
It is worth noting that if gene × covariate interaction of the form of our model does exist, the parameter α that enters into the noncentrality parameter of the simple one-dimensional statistic based on (1) would become α+Corr(xi, xj)γ. Hence if correlation between relatives of the covariate is large, perhaps due to similar environments, then the one-dimensional statistic already captures a substantial fraction of the noncentrality that is potentially available for detecting linkage. (Note, however, that this correlation need not be large: for the covariate of sex, one would expect it to be 0.)
This framework can accommodate both binary and continuous covariates and is easily extended to multiple covariates [Peng, Tang and Siegmund 2005]. However, the number of degrees of freedom increases with the number of covariates and may reduce power to detect linkage. We have computed the above linkage statistic incorporating gene × body mass index (BMI) interaction to evaluate linkage to fasting plasma insulin in FHS and present the results in the “Examples” section.
3.2 Multivariate models
Extensions to the basic variance component model to accommodate multivariate traits have been proposed by several authors [e.g., Hooper et al. 1982; Almasy et al. 1997; Williams et al. 1999; Amos et al. 2001]. Extensions have focused on the likelihood ratio statistic for testing linkage to multivariate phenotypes. More recently, Wang and Elston [2007] proposed a multivariate regression-based linkage approach, arguing that the variance component likelihood ratio statistics are sensitive to violation of the normality assumption but that regression based methods are more robust; see also Wang [2003]. Below we derive a robust score test for bivariate phenotypes, which can be generalized directly to an arbitrary number of phenotypes and will alleviate the problems of inflated type-I error probability when the data deviate from the multivariate normality assumption.
Let Y be a vector of length 2n, where Y1 … Yn are the values of the first phenotype for individuals 1 to n and Yn+1 … Y2n are the equivalent values for the second phentoype.
One can write the unconditional covariance of Y as
where ⊗ represents the Kronecker product of two matrices and
We denote the inverse of the (unconditional) covariance matrix by
where and are matrices of size n.
Under the working model of pure additivity, the variance conditional on the IBD proportion at t is:
where
say. The likelihood function is
Under the null hypothesis of no linkage, α = β = γ = 0. To derive the score statistic, we take the first derivative with respect to each parameter and evaluate at Σa = 0:
where
The robust covariance Vθ has elements , E[ℓαℓγ∣Y], etc.
These are all weighted sums of Cov(πtij, πtkl), the covariance between IBD proportions, which can be estimated by the approaches described in Section 2.1. The global bivariate linkage statistic maximized over all genome location t is written as .
With regard to constraints implied by non-negativity of variance components and approximations to false positive error rates and power, the discussion for gene × covariate interaction applies essentially without change.
3.3 Ordinal Phenotypes
In some cases phenotypes are measured on an ordered categorial scale rather than as a continuous variable. This may be a matter of covenience, or the categories may have medical meaning. The model developed above can be used for ordinal data, with only a minor conceptual adjustment. If an actual, observed phenotype is denoted by Y, the modeled phenotype, which is unobserved, is the penetrance Ỹ = E(Y|Genotype). For relatives, we assume that actual phenotypes are conditionally independent given genotypes, and proceed as above. Although the model of multivariate normality cannot be defended, the defense of that model is weak under the best of conditions, and it is used only to suggest the form of a test statistic, which then must stand on its own. Because we evaluate the statistic itself and the false positive rate conditional on the phenotypes, the statistic can be calculated and a suitable significance threshold determined regardless of the fact that the phenotypes do not do not satisfy the multivariate normal model.
4 Simulation study
We designed a simulation study with several goals in mind. The first objective is to test our software written in R that builds on existing software for IBD estimation, to achieve efficient, accurate analysis of moderately large pedigrees. Secondary objectives include verification of the accuracy of theoretical approximations, development of simple rules to evaluate different pedigrees, and comparison of the recovery of IBD information for pedigrees vs sibling pairs and SNPs vs STRs. Finally, we want to compare different estimators of covariance and to study the effect of non-normality of phentoypes.
To compare efficiency of relative pairs and pedigree structures, we selected two study designs: a sample of 200 three-generation pedigrees with 10 members (Figure 1) and a set of appproximately 330 multiplex pedigrees, chosen to mimic the largest families selected from the first two FHS generations. To evaluate the effect of marker spacing, we generated simulated genotypes for five scenarios. The first set of three scenarios consisted of a single chromosome of length 150 cM, with markers spaced every 1, 5 and 10 centiMorgan (cM), where the markers at 1 cM were di-allelic markers with allele frequencies of 0.3 and 0.7, while markers spaced at greater than 1 cM interval had four equally frequent alleles. In addition we selected two other scenarios that closely resemble typical scans of a single chromosome, with SNPs or STR markers. We used chromosome 20 with an approximate length of 98 cM (60 Mb), and simulated STR markers with frequencies and genetic locations taken from Marshfield clinic panel 8A (http://research.marshfieldclinic.org/genetics/), which was used in genotyping the first and second FHS generations. Di-allelic markers were generated to mimic SNPs that were selected in a previous linkage report using a subset of SNPs from a 100,000 genome-wide SNP scan [Cupples et al. 2007].
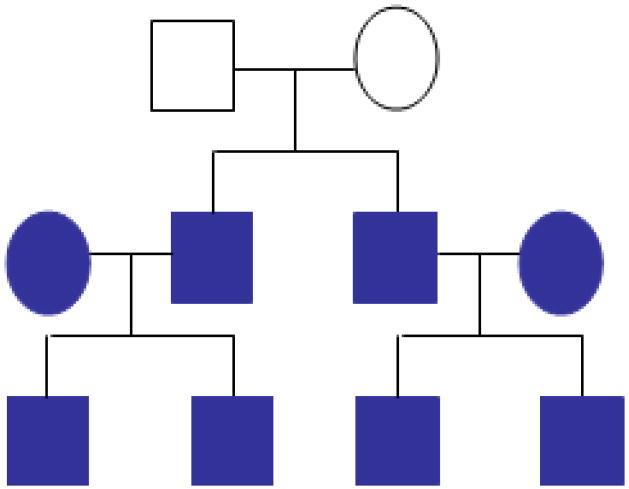
Figure 1.
Simulated pedigree. Phenotypes and genotypes from the grandparents (open symbols) are assumed missing.
To evaluate the robustness of the score statistics, we generated trait values that were uncorrelated with the genotypes. To provide suitable background genetic correlation, the simulated traits have a total heritability of 50%, which is similar to some of the values observed for BMI in the FHS [Atwood et al. 2002]. We first generated normally distributed traits, and in order to assess the effect on the type-I error of departures from the normality assumptions, we quantile transformed the trait distributions to χ2 and t-distributions with 7 degrees of freedom. The χ2 distribution is skewed, and the two distributions have kurtosis of 2.0 and 1.7, respectively. To evaluate power, we simulated a single QTL explaining 25% of the variance of the phenotype. The QTL was located mid-way between markers, which represents the worst case scenario in terms of power to detect a QTL.
For each scenario, we generated 1000 simulation replicates and evaluated the type-I error and power of four statistics: the robust score statistic with 3 different ways of estimating the covariance in IBD proportion: 1) theoretical (Theoretical); 2) theoretical correlation but empirical variance (Estimated Var); 3) empirical covariance estimates (Empirical); and the score statistic with non-robust estimate of the variance using the empirical estimate of IBD proportion covariances. Theoretical power results are included under the heading of “Complete IBD information.” Thresholds corresponding to a chromosome-wide error rate of 5% were determined theoretically and used to evaluate power and type-I error rates.
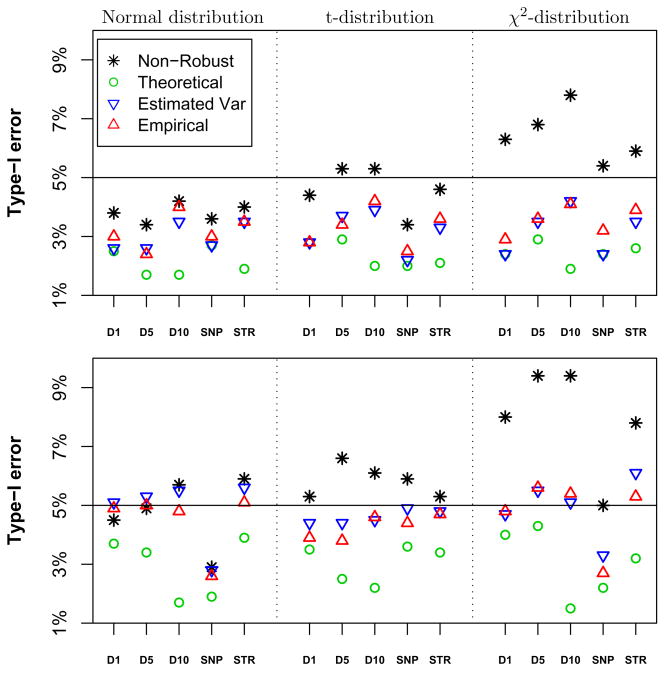
As expected, for normally distributed traits, all statistics have type-I error rate close to or below 5% (Figure 2). However, for χ2 distributed traits, the non-robust score statistics have inflated type-I error, while all three robust score statistics have type-I error rate close to or below 5%, with the statistic computed using theoretical variances being the most conservative. These observations hold for both study designs: 200 three-generation pedigrees and 330 Framingham pedigrees. Note that the score statistics using empirical covariances (Empirical) or estimated variances with theoretical correlation (Estimated Var) yield very similar type-I error rates.
Figure 2.
Type-I error for (top) 200 three-generation pedigrees; and (bottom) 330 families similar in size to FHS. The leftmost panel are normally distributed trait values, center are t-distributed and rightmost are χ2 distributed trait values. D1: 151 SNPs every 1 cM; D5: 31 STRs every 5 cM; D10: 16 STRs every 10 cM; SNP: 190 SNPs ~ 0.6cM; and STR: 18 STRs ~ 5.8cM.
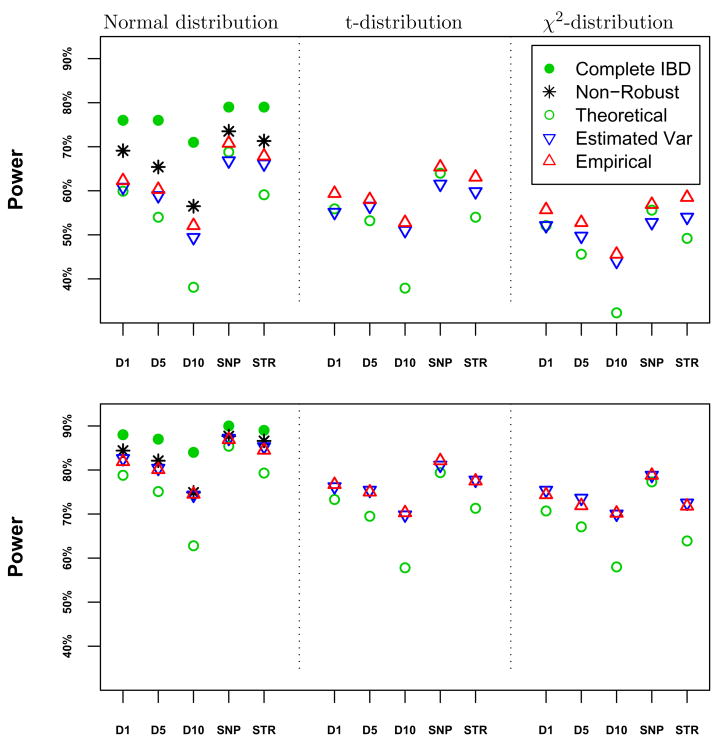
The power of all statistics is comparable, with the exception of the robust score using theoretical variances, which shows lower power due to overestimates of the variances in IBD proportions (Figure 3). The non-robust and robust statistics with empirical variance estimate have equivalent power for the extended pedigree study designs, while for the three-generation pedigrees, the non-robust statistics tended to have higher power for normally distributed traits. Overall, two versions of the robust statistics, “Empirical” and “Estimated Var,” which differ only in how the covariance in IBD counts is estimated, have very similar power. Because the “Empirical” version is much easier to implement and little is gained by using the more complex denominator, in the next section we only use the “Empirical” robust statistic to evaluate linkage in the FHS.
Figure 3.
Power for (top) 200 three-generation pedigrees; and (bottom) 330 families similar in size to FHS. The leftmost panel are normally distributed trait values, center are t-distributed and rightmost are χ2 distributed trait values. D1: 151 SNPs every 1 cM; D5: 31 STRs every 5 cM; D10: 16 STRs every 10 cM; SNP: 190 SNPs ~ 0.6cM; and STR: 18 STRs ~ 5.8cM.
The approximate power for fully informative markers reported in the left hand panels of Figure 3, which provides an upper bound on the power achievable with partially informative markers, is based on the approximation given in Section 2.2, using the notion of an “equivalent number of sibling pairs” to obtain a rough approximation to the noncentrality parameter ξ = E(Zτ). The expression for ξ for sibships given by Tang and Siegmund [2001], reinforced by numerical calculations in some extended pedigrees by Shi [2006], indicates that the ratio of noncentrality parameters of different pedigrees varies relatively slowly as a function of the phenotypic correlation provided that correlation is not too large. This suggests that one can approximate the noncentrality parameter for an additive QTL by determining the noncentrality parameter for an equivalent number of sibling pairs, defined to be the weighted sum of the number of different relative pairs, with weights equal to twice the variance of the number of alleles inherited IBD by the relative pair. Our rough approximation to the noncentrality parameter is the noncentrality parameter for the equivalent number of sibling pairs. Since each pedigree in Figure 1 contains three sibling pairs, four first cousin pairs and 4 avuncular pairs, we evaluate the pedigree as 3 × 1 + 4 × 3/8 + 4 × 1/2 = 6.5 equivalent sib pairs. For our simulations there were 200 pedigrees, so the equivalent number of sibling pairs is 1300. For the FHS pedigrees the number of equivalent sibling pairs was 1646, which is reflected in the greater power obtained for these pedigrees.
The power given in Figure 3 for partially informative markers is obtained directly from the simulated process. An alternative, which would require slightly less calculation, is to simulate only the noncentrality parameter and then use the approximation suggested in Section 2.2. The noncentrality parameter at a (fully informative) marker at distance Δ from a QTL is ξ exp(−βΔ). Thus one might approximate the value of ξ at a QTL τ located in the center of an interval of length Δ by simulating the average noncentrality parameter at the two flanking markers, then dividing this average by exp(−βΔ;/2).
For a numerical example, consider 200 three generation pedigrees with markers every 5 cM on a 150 cM chromosome. The threshold for chromosome wide significance of 5% we used is 2.81. The noncentrality parameter for 1300 sibling pairs and fully informative markers can be calculated from a formula of Tang and Siegmund [2001], which gives the value ξ = 3.5 for power of approximately 0.76. In the case of partially informative markers spaced at 5 cM the simulated values labeled “Empirical” gave an average noncentrality at the two flanking markers of 2.63, which converts to a noncentrality at the QTL of 2.95. Using this value for the noncentrality parameter and the approximation in Section 2.2 gives the value of 0.59 for power, where simulations gave 0.58. Hence the loss of power due to the loss of information from partially informative markers is about 0.76-0.58 = 0.18. In other cases this loss can be greater or less depending on marker informativeness and intermarker spacing, as is readily seen from Figure 3.
Other numerical examples suggest that the power approximation in Section 2.2 is quite accurate (data omitted).
5 Example
To demonstrate the computational feasibility of our approach on real data from moderately large pedigrees, we applied the score statistic to continuous traits measured on the Offspring cohort in FHS. IBD probabilities were computed using the software MERLIN [Abecassis et al., 2002] for STR markers and for SNPs selected to have low level of linkage disequilibrium [Cupples et al., 2007] to avoid biasing IBD probability estimates [Schaid et al., 2002]. The IBD probabilities were the input to our implementation in R of the methods described in the previous sections.
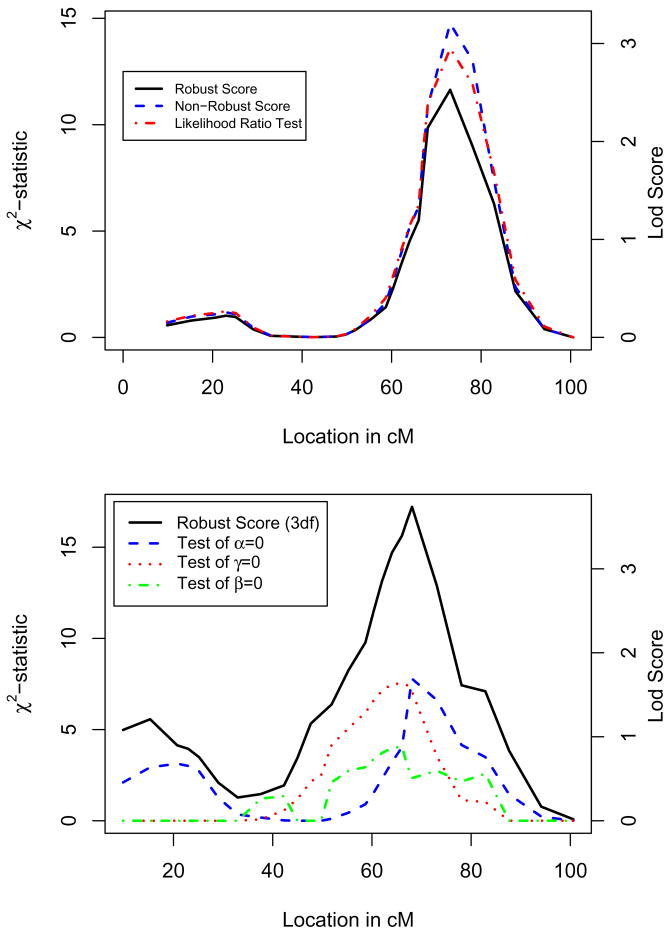
For fasting plasma insulin, linkage evidence has been reported on chromosome 19 [Panhuysen et al. 2003], in the vicinity of the Apolipoprotein E (APOE) gene. The evidence for linkage increased when restricting the analysis to families with higher average Body Mass Index (BMI) [Meigs et al. 2008], which may indicate the presence of gene×BMI interaction. Motivated by this finding, we computed the linkage statistics on chromosome 19 for 15 STR markers with and without incorporating gene × BMI interaction in the linkage scan (Figure 4). Fasting insulin has a skewed distribution, with some individuals showing extremely high values. Not surprisingly, the LRT and non-robust score statistics yielded similar evidence for linkage, while the robust score statistic showed reduced linkage evidence (Figure 4 to panel). A similar, more extreme, discrepancy was obtained for fasting glucose, which has a skewed distribution even after a logarithmic transformation (data not shown).
Figure 4.
Linkage statistics for fasting plasma insulin on chromosome 19 using 15 STR markers with (top panel) and without (bottom panel) BMI × gene interaction.
When allowing for gene×BMI interaction, the linkage statistic went from 11.6 (1 degree of freedom (df)) to 17.2 (3 df). However, because of the increase in the number of degrees of freedom for the score statistic allowing for interaction, this increase in the robust score statistic translates into only a small change in the (chromosome wide) p-value, from 0.005 to 0.004. Similar results were obtained using 170 SNPs covering chromosome 19 (results not shown).
Our implementation of the score statistic may be applied to the full three generation Framingham families, provided that the IBD probabilities are estimated with software capable of handling large pedigrees when they are encountered (e.g. MERLIN supplemented by LOKI [Heath 1997]). An application was recently reported by Schnabel et al. [2009] in the search for genes influencing lipoprotein-associated phospholipase A2 (LpPla2), a biomarkers of inflammation. A small linkage peak was found on chromosome 6 (LOD=2.4; Figure 1 of [Schnabel et al. 2009]) for LpPla2 activity, near the phospholipase A2 (PLA2G7) gene, which regulates LpPla2 activity. However, no other linkage peaks were identified.
6 Discussion
In this paper we have shown that for mapping QTL in extended pedigrees the score statistic can be made robust against false positive errors by estimating the variance of the efficient score conditional on phenotypes. Because nuisance parameters need be estimated only once, under the hypothesis of no linkage, some of the computational impediments to using moderately large pedigrees and somewhat more complex models of inheritance have been mitigated. Software written in the programming language R is currently available for implementing models for univariate and bivariate phenotypes, gene × covariate interaction, and ordinal phenotypes.
Likelihood ratio tests for linkage incorporating gene by covariate interactions were introduced in 1997 [Towne et al. 1997] and can improve the likelihood to detect linkage when such interaction effects are present [Towne et al. 1997; Diego et al. 2003]. However, examples of their use are few and have generally resulted in modest improvement in the linkage scan [Franceschini et al. 2006; North et al. 2007; Diego et al. 2007]. Bivariate analyses have been more extensively used, probably due to the availability of software for computing the likelihood ratio statistic, and there are a few interesting examples where the bivariate evidence for linkage is much stronger than the univariate [Martin et al. 2004; Wang et al. 2007; among others]. However, the likelihood computations for bivariate analyses remain highly computer intensive and have often been restricted to chromosomes showing evidence of linkage in univariate analyses. The score tests proposed in this paper greatly alleviate the computational burden, allowing more complex models, such as bivariate analyses and models incorporating interactions, to be applied to the whole genome.
The problem of robustness can be addressed by transforming the phenotypes to something closer to a multivariate normal distribution. One truly multivariate transformation, available in SOLAR, is based on assuming the phenotypes have a multivariate t distribution, and the methods of this paper are easily adapted to deal with this modeling assumption as well. This helps to reduce the effect of outliers, but does not deal with asymmetry. Another apparently widely used technique is to use the empirical distribution of the phenotypes to force the marginal distribution to be approximately normal. In our (limited) experience, use of this transformation together with the likelihood ratio statistic often produces results similar to the robust score statistic; but it is not easy to interpret the effect of this nonlinear transformation nor to compare the results of different studies.
In the future we plan to extend the methods described in this paper to deal with other problems. Reasonably straightforward examples are gene × gene interaction and longitudinal traits, which can be approached by methods similar to our treatment of gene×covariate interaction. A more complicated example is age of onset data along the lines suggested in Dupuis, Siegmund and Yakir [2007].
Acknowledgments
Supported by the National Institute of Health R01 HG000848 (D.S; J.D.), RO1 HL076784 (E.J.B.), 1R01 AG028321 (E.J.B.), RO1 HL064753 (E.J.B), R21 DK65732, K24 DK080140 (J.B.M), an American Diabetes Association Career Development Award (J.B.M.), the National Heart, Lung, and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195), and the Boston University Linux Cluster for Genetic Analysis (LinGA) funded by the NIH NCRR Shared Instrumentation grant (1S10RR163736-01A1).
9 Appendix: Type I Error Probabilities
In Section 2.2 we provided approximations for the false positive error rate based on the assumption that the number of pedigrees is sufficiently large for a normal approximation to be appropriate. We also stressed that this approximation is conditional on phenotypes, so the asymptotic normality is a consequence of the central limit theorem, not an assumption about the distribution of the phenotypes. When the data involve a small number of relatively large pedigrees, the statistic Zt can have a positively skewed distribution by virtue of within pedigree dependencies of IBD counts and distant relatives, even if the phenotypes are exactly multivariate normal. The approximation given below corrects for this skewness (conditionally on phenotypes if desired).
To state the approximation let γ = E(Zt)3, computed conditionally on the phenotypes or unconditionally and let θ = [−1+(1+2bγ)1/2]/γ. Then for markers equally spaced at an intermarker distance Δ, a chromosomal region containing C chromosomes of total genetic length L, under the hypothesis of no linkage
where ν = ν[b(2rΔ)1/2].
One should note that this is quite a different correction for non-normality from that discussed by Blangero et al. [2000], which is concerned with the lack of robustness of the likelihood ratio statistic when the phenotypes fail to be approximately multivariate normal. In that case the failure of the standard asymptotic distribution is not just a question of sample size, and an appropriate correction involves the kurtosis of the of the efficient score.
For an admittedly extreme example where the skewness correction has a substantial effect, see Shi, Siegmund and Levinson [2007]. Software to apply the skewness correction in moderately large three generation pedigrees is available on request from Jianxin Shi.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J. Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci-mapping procedure. Am J Hum Genet. 1999;65:531–544. doi: 10.1086/302487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Amos CI, de Andrade M, Zhu DK. Comparison of multivariate tests for genetic linkage. Hum Hered. 2001;51:133–144. doi: 10.1159/000053334. [DOI] [PubMed] [Google Scholar]
- Atwood LD, Heard-Costa NL, Cupples LA, Jaquish CE, Wilson PW, D’Agostino RB. Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am J Hum Genet. 2002;71:1044–50. doi: 10.1086/343822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Kuo C-L, Mukhopadhyay N, Brock GN, Weeks DE, Feingold E. Robust score statistics for QTL linkage analysis. Am J Hum Genet. 2008;82:567–582. doi: 10.1016/j.ajhg.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Almasy L. Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol. 1997;14:959–964. doi: 10.1002/(SICI)1098-2272(1997)14:6<959::AID-GEPI66>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol. 2000;19:S8–S14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Chen W-M, Broman K, Liang KY. Quantitative trait linkage analysis by generalized estimating equations: unification of variance components and Haseman-Elston regression. Genet Epidemiol. 2004;26:265–272. doi: 10.1002/gepi.10315. [DOI] [PubMed] [Google Scholar]
- Cupples LA, Arruda HT, Benjamin EJ, D’Agostino RB, Sr, Demissie S, DeStefano AL, Dupuis J, Falls KM, Fox CS, Gottlieb DJ, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Kathiresan S, Kiel DP, Laramie JM, Larson MG, Levy D, Liu CY, Lunetta KL, Mailman MD, Manning AK, Meigs JB, Murabito JM, Newton-Cheh C, O’Connor GT, O’Donnell CJ, Pandey M, Seshadri S, Vasan RS, Wang ZY, Wilk JB, Wolf PA, Yang Q, Atwood LD. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE. Epidemiologic approaches to heart disease: the Framingham study. Am J Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diego VP, Almasy L, Dyer TD, Soler JM, Blangero J. Strategy and model building in the fourth dimension: a null model for genotype × age interaction as a Gaussian stationary stochastic process. BMC Genet. 2003;4(Suppl 1):S34. doi: 10.1186/1471-2156-4-S1-S34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diego VP, Rainwater DL, Wang XL, Cole SA, Curran JE, Johnson MP, Jowett JBM, Dyer TD, Williams JT, Moses EK, Comuzzie AG, MacCluer JW, Mahaney MC, Blangero J. Genotype× Adiposity Interaction Linkage Analyses Reveal a Locus on Chromosome 1 for Lipoprotein-Associated Phospholipase A2, a Marker of Inflammation and Oxidative Stress. Am J Hum Genet. 2007;80:168–177. doi: 10.1086/510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Siegmund DO, Yakir B. A unified framework for linkage and association analysis of quantitative traits. Proc Natl Acad Sci U S A. 2007;104:20210–5. doi: 10.1073/pnas.0707138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini N, MacCluer JW, Goring HHH, Cole SA, Rose KM, Almasy L, Diego V, Laston S, Lee ET, Howard BV, Best LG, Rabsitz RR, Roman MJ, North KE. A Quantitative Trait Loci-Specific Gene-by-Sex Interaction on Systolic Blood Pressure Among American Indians: The Strong Heart Family Study. Hypertension. 2006;48:266–270. doi: 10.1161/01.HYP.0000231651.91523.7e. [DOI] [PubMed] [Google Scholar]
- Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behavior Genetics. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Heath S. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet. 1982;46:373–83. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–90. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- Lange K, Westlake J, Spence MA. Extensions to pedigree analysis. III. Variance components by the scoring method. Ann Hum Genet. 1976;39:485–91. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- Lebrec J. Ph D Thesis. University of Leiden; 2007. Linkage mapping for complex traits: a regression-based approach. [Google Scholar]
- Martin LJ, Cianflone K, Zakarian R, Nagrani G, Almasy L, Rainwater DL, Cole S, Hixson JE, Mac-Cluer JW, Blangero J, Comuzzie AG. Bivariate Linkage between Acylation-Stimulating Protein and BMI and High-Density Lipoproteins. Obesity Research. 2004;12:669–678. doi: 10.1038/oby.2004.77. [DOI] [PubMed] [Google Scholar]
- Meigs JB, Manning AK, Dupuis J, Liu C, Florez JC, Cupples LA. Ordered Stratification to Reduce Heterogeneity in Linkage to Diabetes-related Quantitative Traits. Obesity. 2008;16:2314–22. doi: 10.1038/oby.2008.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North KE, Franceschini N, Borecki IB, Gu CC, Heiss G, Province MA, Arnett DK, Lewis CE, Miller MB, Myers RH, Hunt SC, Freedman BI. Genotype-by-Sex Interaction on Fasting Insulin Concentration. Diabetes. 2007;56:137–142. doi: 10.2337/db06-0624. [DOI] [PubMed] [Google Scholar]
- Panhuysen CI, Cupples LA, Wilson PW, Herbert AG, Myers RH, Meigs JB. A genome scan for loci linked to quantitative insulin traits in persons without diabetes: the Framingham Offspring Study. Diabetologia. 2003;46:579–87. doi: 10.1007/s00125-003-1066-z. [DOI] [PubMed] [Google Scholar]
- Peng J, Tang HK, Siegmund D. Genome scans with gene-covariate interaction. Genet Epidemiol. 2005;29:173–84. doi: 10.1002/gepi.20100. [DOI] [PubMed] [Google Scholar]
- Peng J, Siegmund D. Mapping quantitative traits with random and with ascertained sibships. Proc Natl Acad Sci U S A. 2004;101:7845–50. doi: 10.1073/pnas.0401713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putter H, Sandkuijl LA, van Houwelingen JC. Score test for detecting linkage to quantitative traits. Genet Epidemiol. 2002;22:345–55. doi: 10.1002/gepi.01104. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2006. R Foundation for Statistical Computing; Vienna, Austria: R: A language and environment for statistical computing. URL http://www.R-project.org. [Google Scholar]
- Schaid DJ, McDonnell SK, Wang L, Cunningham JM, Thibodeau SN. Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet. 2002;71:992–995. doi: 10.1086/342666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Dupuis J, Larson MG, Lunetta KL, Robinsc SJ, Zhu Y, Rong J, Ying X, Stirnadel HA, Nelson JJ, Wilson PWF, Keaney JF, Vasan RS, Benjamin EJ. Clinical and genetic factors associated with lipoprotein-associated phospholipase A2 in the Framingham Heart Study. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2008.10.030. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–53. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Ph D thesis. Stanford University; 2006. Two problems in quantitative trait mapping. [Google Scholar]
- Shi J, Siegmund DO, Levinson DF. Statistical corrections of linkage data suggest predominantly cis regulations of gene expression. BMC Proc. 2007;1(Suppl 1):S145. doi: 10.1186/1753-6561-1-s1-s145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund D, Yakir B. The Statistics of Gene Mapping. Springer; Berlin: 2007. [Google Scholar]
- Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB, Sr, Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–35. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- Tang HK, Siegmund D. Mapping quantitative trait loci in oligogenic models. Biostatistics. 2001;2:147–162. doi: 10.1093/biostatistics/2.2.147. [DOI] [PubMed] [Google Scholar]
- Tang HK, Siegmund D. Mapping multiple genes for quantitative and complex traits. Genet Epidemiol. 2002;22:313–327. doi: 10.1002/gepi.01108. [DOI] [PubMed] [Google Scholar]
- Towne B, Siervogel RM, Blangero J. Effects of genotype-by-sex interaction on quantitative trait linkage analysis. Genet Epidemiol. 1997;14:1053–1058. doi: 10.1002/(SICI)1098-2272(1997)14:6<1053::AID-GEPI82>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wang T, Elston RC. Regression-based multivariate linkage analysis with an application to blood pressure and body mass index. Ann Hum Genet. 2007;71:96–106. doi: 10.1111/j.1469-1809.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- Wang K, Huang J. A score-statistic approach for the mapping of quantitative-trait loci with sibships of arbitrary size. Am J Hum Genet. 2002;70:412–424. doi: 10.1086/338659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. Mapping quantitative trait loci using multiple phenotypes in general pedigrees. Hum Hered. 2003;55:1–15. doi: 10.1159/000071805. [DOI] [PubMed] [Google Scholar]
- Wang XL, Deng FY, Tan LJ, Deng HY, Liu YZ, Papasian CJ, Recker RR, Deng HW. Bivariate Whole Genome Linkage Analyses for Total Body Lean Mass and Bone Mineral Density. J Bone Miner Res. 2007;23:447–452. doi: 10.1359/JBMR.071033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]