I. Introduction
Progress in the structural biology of proteins comes from both experimental and theoretical efforts. Computational methods are capable of delivering fast structural information, ranging from low-resolution protein structural class definition to high-quality information based on homology modeling. Experimental methods that concentrate on obtaining high-resolution information are hampered by inherent time cost, and lack the capacity to provide low-resolution structural information expediently. We present a comprehensive overview of low-resolution structural determinants to correlate NMR-based chemical shift data with protein structural data in order to provide meaningful information expeditiously; i.e., prior to the intensive effort required to perform complete resonance assignments and, from these, derive three-dimensional structural information. With a historical synopsis of developments in the field, we present the underlying concepts, placing emphasis on the nuclear chemical shift, protein secondary structure, and the physical connection between them. Results from this effort have demonstrated that fast, reliable protein structural information can be obtained directly from NMR spectra prior to the complete determination of high-resolution three-dimensional structures. These methods do not provide an alternative to traditional spectroscopy-based techniques, but rather compliment them by providing low-resolution structural information very quickly. We discuss the degree to which chemical shifts of a particular nuclear species in the protein backbone can be used as a low-resolution structural parameter that correlates with a variety of protein structural parameters.
The nuclear chemical shift, first observed in nuclear magnetic resonance (NMR) spectra in 1950 by Proctor and Yu for the 14N nuclei [1] and by Arnold et al in 1957 for the 1H nuclei [2], is among the most reliable known indicators of biomolecular structure. The development of most modern experimental pulse sequences is driven by the goal of increasing the resolution and sensitivity with which chemical shifts can be measured. In addition to structural information [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], chemical shifts provide detailed information about hydrogen bonding interactions, ionization and oxidation states, the ring current influence of aromatic residues, and the nature of hydrogen exchange dynamics[14]. Several excellent review articles describe a variety of experimental and computational methods to correlate chemical shifts with protein three-dimensional structural information [14], [15], [16], [17], [18], [19], [20]. However, these methods rely on the determination of the chemical shift of each atomic resonance in the molecule [21], which remains a challenging and time consuming procedure, despite efforts to automate the process [22], [23], [24], [25], [26], [27], [28], [29]. Moreover, it is not always possible to obtain complete assignments for a particular data set, especially for proteins undergoing conformational changes.
In addition to advances in traditional NMR-based methods, recent efforts have engendered important new approaches to investigating the structural biology of proteins. Two examples are structural proteomics and empirical correlation methods. Structural proteomics seeks to screen large numbers of proteins rapidly to identify new structural folds and to select specific sets of molecules for complete structural investigation [30], [31]. Empirical correlation methods, one example of which is homology modeling [32], [33], seek to define empirical relationships between experimental structural information and other known physio-chemical parameters in order to predict protein structure, function, and dynamics. The development of correlation methods has been fueled over the past decade both by the need for high-throughput strategies and by the advent of readily accessible, well-organized public repositories that make possible the efficient querying and mining of an unprecedented quantity of experimental information. The original, and perhaps best-known, repository of this kind is the Research Collaboratory for Structural Bioinformatics (RCSB) [34], [35], [36], [37], originally developed by Brookhaven National Laboratory, which became available to the public in 1971. Since that time, web-based repositories have become increasingly sophisticated, and are now a cornerstone of many structural biology researches.
Here, we present a comprehensive overview of empirical correlation methods whose aim is to correlate NMR-based chemical shift data with protein structural data in order to provide meaningful information expeditiously; i.e., prior to the intensive effort required to perform complete resonance assignments and, from these, derive three-dimensional structural information. We will begin with a historical synopsis of developments in the field, dating to almost 20 years ago, that includes a general discussion of fundamental, underlying concepts, placing emphasis on the nuclear chemical shift, protein secondary structure, and the physical connection between them. We will then focus on a recent effort by us to develop methods that establish and refine an empirical correlation between averaged chemical shift (ACS) and protein secondary structure content (SSC) by extensively mining chemical shift information from the BioMagResBank (BMRB) [38] and protein structural information from the PDB. We also present an overview of empirical methods for a range of applications, such as prediction of redox-state of the cystines or identification of cis-Prolines. Results from this effort have demonstrated that fast, reliable protein structural information can be obtained directly from NMR spectra prior to the complete determination of high-resolution three-dimensional structures. These methods do not provide an alternative to traditional spectroscopy-based techniques, but rather complement them by providing low-resolution structural information very quickly. This makes possible the high-throughput characterization of protein secondary structural content, and, thereby, the large-scale screening and integration of data required to accelerate efforts in fields such as structural proteomics. Further development of such empirical correlation methods for this purpose will potentially also lead to new experimental and computational protocols.
2. Empirical methods for correlating nuclear chemical shifts with protein structural data: Secondary structure index
NMR chemical shifts provide detailed information on the structure and electronic properties of biological molecules in the solution, noncrystalline and crystalline states. Chemical shifts are perhaps the most information-rich parameter obtainable from NMR; however, the physical basis for particular nuclei assuming specific chemical shift values based on the conformational state of a biomolecule is not fully understood. Although, ab initio or density functional calculations [39], [40], [41] for small peptide units provide some insight into the relevant mechanisms, at present the tools of computational chemistry remain insufficient to determine the high-resolution structure of a protein purely from chemical shifts. Consequently, we must rely upon predictive models that determine whether there exist correlations between three-dimensional structures and chemical parameters obtained from NMR. In the absence of a reliable theory, predictive models must follow semi-empirical or empirical approaches. Empirical methods use databases of previously assigned homologous molecules to predict chemical shifts of new systems. This approach is promising, particularly because the number of assigned spectra available in electronic databases continues to increase; however, it requires a reasonable level of similarity between the target and reference molecules. New combinations of semi-empirical methods have also been developed recently [42] thus paving ways to novel methods of protein structure determination from chemical shifts [43], [44], [45]. Empirical and semi-empirical methods are expected to play a significant role in NMR based structure determination in the near future.
2.1. Secondary chemical shifts in proteins
One intuitive assessment that can be made with some reliability from the chemical shift dispersion of an NMR spectrum (e.g., the 1H spectrum of a protein) is whether the associated structure is folded or disordered. Making this determination continues to be the main goal of research efforts concerned with correlating chemical shifts and protein structure. Researchers soon learned that such straightforward and simple methods tend to predict and contribute to the final high-resolution structures. Obtaining high-quality NMR spectra is a relatively easy task, and a requisite first step toward reducing chemical shift values to a meaningful structural parameter. The empirical-statistical approach capitalizes on the vast and rapidly increasing amount of information contained in repositories of protein chemical shift data, combining these data with three-dimensional structural information to establish empirical correlations. The clear-cut trend between ‘1H chemical shifts and secondary structure in proteins,’ which led to the definition of a ‘secondary structure shift,’ was initially recognized by Dalgarno et al. [6]. This term is also referred to as ‘conformation-dependent shift’ [46] or simply ‘secondary chemical shift.’ The secondary chemical shift of a particular protein nucleus ‘i’ is defined as
| [1] |
Here, is the observed chemical shift and is the corresponding ‘random coil’ value. Though alternate definitions include a correction for ring current shifts ( ), where is the ring current contribution [12]), the general definition expressed by Eq. [1] accounts through for any other variations introduced by the protein.
2.2. Early efforts toward low-resolution structural information
Initial attempts to correlate chemical shift information with protein structure were carried out in the 1960s by Jardetzky and co-workers [47]. Subsequently, several groups [6], [48], [49], [50] explored the possibility of correlating protein chemical shift data with elements of regular secondary structure (helices, sheets, and turns). Szilagyi, in his comprehensive historical perspective of chemical shifts in proteins [14], credits Dalgarno et al. [6] for being the first to observe a clear-cut trend relating chemical shifts and secondary structure in proteins. On the basis of early chemical shift data from two proteins (bovine pancreatic trypsin inhibitor (BPTI) and partial assignments from hen-egg lysozyme), these authors noted that secondary chemical shifts of α-carbon protons tend to be shifted up-field for α-helical and downfield for β-sheet regions. However, progress in this area has been slow since 1987, when Jimenez et al., [51] reported secondary structure shifts of − 0.35 ppm, on average, for Hα resonances in helices, and + 0.40 ppm, on average, for Hα resonances in β-sheets based on chemical shift data from the fully assigned (1H) spectra of five proteins. The work of Wishart and co-workers [13] provided the first extensive compilation and statistical analysis of chemical shift data in proteins, facilitating a resurgence of empirical methods for correlating chemical shifts to various structural parameters of proteins, and forming the precursor to assignment-independent NMR techniques to determine the secondary structure content of proteins. This “circular dichroism-like” (CD-like) approach has been applied successfully to a number of proteins.
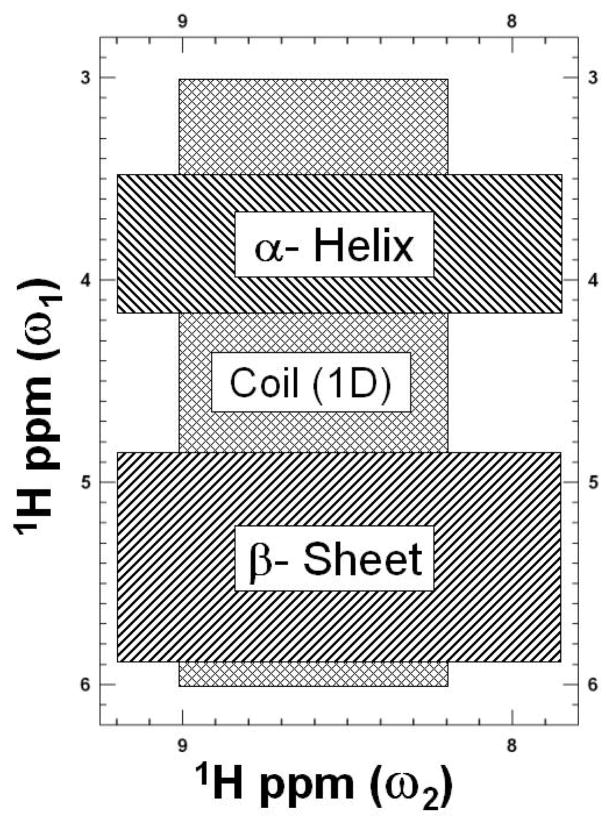
In the method developed by Wishart et al. [13], the normalized integration of amide resonances in the region between 8.20 and 9.00 ppm in a 1D proton NMR spectrum (recorded H2O solution) provides the number of residues in coil regions of the protein (Figure 1). The number of residues in β-sheets is equal to twice the value of the normalized integral of low-field shifted resonances between 4.85 and 5.90 ppm (Figure 1,). The number of residues in a helical conformation is then obtained as the difference: = (number of β-sheet residues) − (number of coil residues). The 2D method relies on counting cross-peaks in the fingerprint (HN/Hα) region of a simple 1H COSY or DQF (double-quantum filtered)-COSY spectrum. The number of cross-peaks (〈C〉) in the map region 8.2–9.00 ppm (ω2) and 3.0–6.00 ppm (ω1) is proportional to the number of residues in the random coil state; Ncoil = 0.9 × 〈C〉. The number of cross peaks (〈B〉) found in the 8.20–9.00 ppm (ω2) and 4.85–5.90 ppm (ω1) is equal to half the number of residues in the β-sheet conformation; Nβ = 2.0 × 〈B〉. The number of residues in a helical conformation can then be determined by counting cross-peaks (〈A〉) in the region 8.20–9.00 ppm (ω2) and 3.4–4.10 ppm (ω1). This number is to be corrected by the number of Gly residues; Nα = 2.0 × [〈A〉 − 2.0 number of Gly]. These regions are indicated in Figure 1 by various shades. It must be noted that the figure corrects for the double counting of peaks lying in overlap regions. Estimates of secondary structure elements based on this method agree surprisingly well with those from X-ray crystallography or from NOESY analysis, and it has been suggested [13] that this simple NMR ‘rule of thumb’ gives significantly better estimates than does CD or FT-IR. It is important to note that when this important work appeared in 1991, the field of structural biology was rapidly evolving. In light of the structural biology tools available today, we have reanalyzed the results of Wishart et al. [13], and present a summary of our findings in Figure 2 and Table 1.
Figure 1.
Schematic description of the peak counting method developed to determine protein secondary structure content. A typical double quantum filter COSY (DQFC) spectrum is shaded to highlight the regions of important structural information. The two hatched blocks marked by β-sheet and α-helix, correspond to the areas used to estimate the β-strand and α-helix, respectively. The hatched region marked Coil (1D) used to estimate the random coil content. Coil information is also obtained from one-dimensional NMR spectra. Some peaks appear in overlapping regions and hence are counted twice when making secondary structure estimates.
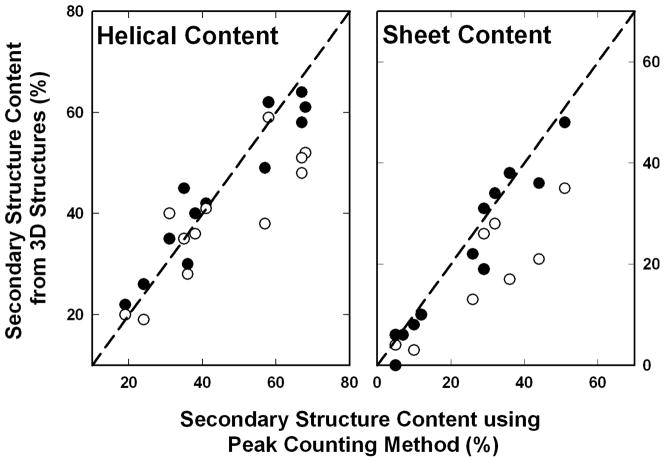
Figure 2.
Comparison of the empirical correlations derived to estimate secondary structure content from proteins by peak counting method. Filled and open circles show the secondary structure content estimated from the three-dimensional structures originally and using recent structural biology tools, respectively. The dashed lines show the ideal linear correlation.
Table 1.
Validation of the early empirical correlation methods
| Protein Name | a PDB | b SSC (method 1) | b SSC (method 2) | b SSC (PROMOTIF) | |||
|---|---|---|---|---|---|---|---|
| α % | β % | α % | β % | α % | β % | ||
| Acyl Carrier Protein | 1ACP | 61 | 0 | 68 | 5 | 52 | 0 |
| α Purothionin | 2PLH | 35 | 22 | 31 | 26 | 40 | 13 |
| BPTI | 1PIT | 26 | 36 | 24 | 44 | 19 | 21 |
| Calmodulin | 1A29 | 62 | 8 | 58 | 10 | 59 | 3 |
| Cytochrome C | 1AKK | 49 | 6 | 57 | 7 | 38 | 0 |
| E. Coli Thioredoxin | 1XOA | 40 | 31 | 38 | 29 | 36 | 26 |
| HEW Lysozyme | 1E8L | 45 | 19 | 35 | 29 | 35 | 0 |
| Human Thioredoxin | 1AIU | 42 | 34 | 41 | 32 | 41 | 28 |
| Insulin | 9INS | 58 | 10 | 67 | 12 | 48 | 0 |
| Parvalbumin | 3PAT | 64 | 6 | 67 | 5 | 51 | 4 |
| Ribonuclease A | 1A2W | 22 | 48 | 19 | 51 | 20 | 35 |
| Staph. Nuclease | 1JOR | 30 | 38 | 36 | 36 | 28 | 17 |
In 1991, it is indeed true that peak counting to determine protein secondary structure content in the absence of resonance assignments was a superior method. Based on the original estimation of secondary structure content from three-dimensional structures, a linear correlation of 0.89 and 0.94 was obtained for determining the helical and sheet content, respectively, from the peak counting method. However, using tools such as PROMOTIF [52], a generally accepted standard for secondary structure estimates, we find the correlation for estimating both helical and sheet structural contents is 0.74. Whilst the peak counting method has demonstrated utility, it has not been widely adopted; primarily it utilizes homonuclear spectra, which tend to get crowded for proteins of modest sizes. At the same time, this procedure has led to one of the most widely used methods, the chemical shift index (CSI), which utilizes both homonuclear and heteronuclear (13C) chemical shift information (vide infra).
Recently, Moreau et al., [53] have presented a method similar to the above mentioned approach using heteronuclear (13C and 15N) chemical shifts in addition to the 1H chemical shifts. This method is called PASSNMR (Prediction of the Amount of Secondary Structure by Nuclear Magnetic Resonance). The goal of this approach and many of the other methods discussed in this article is to predict the amount of secondary structure in proteins for structural genomics applications. Overall reliability of the PASSNMR approach for helical, sheet and coiled structures are 72%, 74% and 42%, respectively and the using only the 15N-1H data, these values drop to 49% for helix and 50% for sheets [53].
2.3. The chemical shift index method
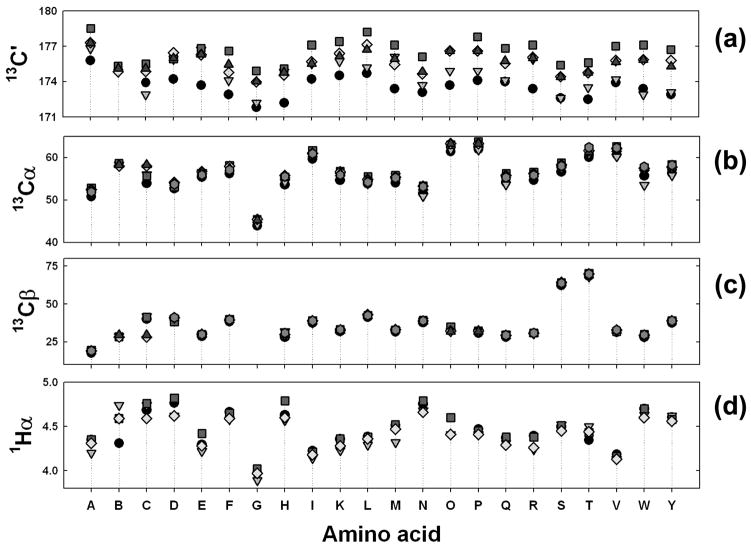
The chemical shift index (CSI) is the first user-friendly tool for converting secondary chemical shifts to useful protein structural information [54], [55]. This method was introduced to identify the secondary structural element of each residue in a sequence-dependent manner. Prior to CSI, it was necessary to obtain the complete set of sequence-specific assignments to even get an initial glimpse of the secondary structure. The original CSI method, based on the experimental chemical shift values of a set of proteins, some empirical adjustment, and intuition, was used to develop a reference table of chemical shifts. Table 2 reproduces the values reported in the original references [55], [56]. The observed chemical shifts of the particular nuclei are then compared with the respective reference values using a set of rules. The method assigns three indices, −1, 0, or 1, depending on whether the observed chemical shift is near the average value, or at one of the extremes. Consecutive occurrences of like indices are used to identify the presence of secondary structure. To further increase accuracy, a jury system averages assignments from multiple chemical shifts (1Hα, 13Cα, 13Cβ and 13C′) to arrive at a consensus assignment.
Table 2.
Chemical shift reference values used for the CSI methoda
| Residue | 1Hα±0.1 ppm | 13Cα±0.7 ppm | 13Cβ±0.7 ppm | 13C′±0.5 ppm |
|---|---|---|---|---|
| Ala | 4.4 | 52.5 | 19.0 | 177.1 |
| Cys(red) | 4.7 | 58.8 | 28.6 | 174.8 |
| CYS(ox) | 58.0 | 41.8 | 175.1 | |
| Asp | 4.8 | 54.1 | 40.8 | 177.2 |
| Glu | 4.3 | 56.7 | 29.7 | 176.1 |
| Phe | 4.7 | 57.9 | 39.3 | 175.8 |
| Gly | 4.0 | 45.0 | 173.6 | |
| His | 4.6 | 55.8 | 32.0 | 175.1 |
| Ile | 4.0 | 62.6 | 37.5 | 176.8 |
| Lys | 4.4 | 56.7 | 32.3 | 176.5 |
| Leu | 4.2 | 55.7 | 41.9 | 177.1 |
| Met | 4.5 | 56.6 | 32.8 | 175.5 |
| Asn | 4.8 | 53.6 | 39.0 | 175.5 |
| Pro | 4.4 | 62.9 | 31.7 | 176.0 |
| Gln | 4.4 | 56.2 | 30.1 | 176.3 |
| Arg | 4.4 | 56.3 | 30.3 | 176.5 |
| Ser | 4.5 | 58.3 | 62.7 | 173.7 |
| Thr | 4.4 | 63.1 | 68.1 | 175.2 |
| Val | 4.0 | 63.0 | 31.7 | 177.1 |
| Trp | 4.7 | 57.8 | 28.3 | 175.8 |
| Tyr | 4.6 | 58.6 | 38.7 | 175.7 |
For example, a 1Hα chemical shift that is greater or less than the CSI reference value ±0.1 ppm (Table 2) is assigned an index value of + 1 or − 1, respectively. Chemical shifts within the range ±0.1 ppm are assigned an index value of zero. Any group of four or more − 1 (not necessarily consecutive) indices uninterrupted by a + 1 identifies a helix and, likewise, any group of three or more consecutive + 1 indices identifies a β-strand. All other combinations are designated as coil. In other words, the selection criterion for secondary structure identification is set to exceed 70% of the ‘local density’ of CSIs over a window of five (for helices, 4/5 = 80%) or four (for sheets, 3/4 = 75%) residues. Termination points of helices or β– strands are defined as being either at the first appearance of a CSI value of opposite sign to an adjacent, high ‘local density’ set of values, or at the first appearance of two consecutive zero-valued CSIs. The procedure was claimed to be accurate to 90–95% after testing it on about 50 proteins [54]. Since its original description, the CSI method has been refined to account for joint probability by defining ‘consensus’ CSI values according to a simple majority rule (two out of three or three out of four) when more than two chemical shift indices are available [56]. This improvement has substantially increased the predictive power of the method.
The reliability of predictions based on the CSI method critically depends on the threshold values provided by the reference chemical shifts (Table 2). Though a quick comparison of these values with their respective random coil values might suggest approximate agreement (vide infra), some of the empirical adjustments made to provide a best fit to observed secondary structure can highly skew the distribution of chemical shifts for some residues in some structures.
2.4. Alternate methods to chemical shift index
CSI-based determination of residue-specific secondary structure is straightforward, and has become routine. NMR data processing software, such as NMRView [57], [58], allows easy implementation of these procedures following the chemical shift assignment process. In the last few years, several alternative methods have been developed that use a range of novel computational tools ranging from probability-based index identification to neural network programming. These methods include, chronologically: (1) probability-based secondary structure identification (PSSI) [59]; (2) secondary structure from chemical shift and sequence (PsiCSI) [60], (3) prediction algorithm for amino acid types with their ;secondary structure in proteins (PLATON) [61]; (4) protein energetic conformational analysis from NMR chemical shifts (PEACAN) [62], and (5) a two-dimensional cluster analysis method referred to as 2DCSi [63]. Here, we briefly describe these methods in turn, and discuss some of the advantages they offer relative to conventional CSI-based approaches.
2.4.1. Probability-based secondary structure identification (PSSI) [59]
PSSI assigns the secondary structure type (β-strand, coil, or α-helix) to each amino acid on the basis of the joint probability, derived from the observed 1HN, 15N, 1Hα, 13Cα, 13Cβ and 13C′ chemical shift data corresponding to each structure type. Based on their observed chemical shifts, nuclei are associated with particular secondary structures in a two-step process. In the first step, a joint probability is defined. Given δn, the chemical shift value of a particular nucleus ‘n’ in amino acid ‘i’, the secondary structure type of the amino acid is ascribed by evaluating the joint probability of the three secondary structure types, Ps,i, given by
| [2] |
Where Fs,j represents the probability for amino acid i at the secondary structure type s (s= α-helix (α), β-strand (β) or coil (C)). Gs,j is given by a Gaussian distribution
| [3] |
Where, δ̄n,s, j and σn,s,i are the center and width of the Gaussian distribution. A secondary structure type is initially assigned based on the joint probability of each type (e.g., s=α-helix, if Pα,i > PC,i and Pα,i > Pβ,i). The total probability is also set to 1 so that the residues will be in one of the three secondary structures. In the second step, which is optional, the resulting probability values are smoothed or filtered. For example, if a local density of either a β-strand or a coil exceeds one-half for a five-residue window, its secondary structure type is adjusted; ββCβC will be adjusted to βββββ (if the P value of the last residue > 0.35) or ββββC (if P < 0.35). Other rules for the end residues of a β-strand or α-helix, and short separated segments, can also be employed [59]. Global assessment of the PSSI method has suggested a significant improvement in both accuracy (~88%) and confidence over a set of 36 proteins. A JAVA interface developed by one of the authors (Y.J. Wang) is also available http://pronmr.com/yunjunwang_files/yjw_pssi.html).
2.4.2. Secondary structure from chemical shift and sequence (PsiCSI) [60]
PsiCSI combines both chemical shift-based and sequence-based methods to further increase the accuracy of secondary structure assignments [60]. In addition, it is designed to best utilize all the available data. PsiCSI begins by refining the CSI methodology; it assigns three separate potentials (scaling) ranging from 0 to 1 to reflect the relative likelihood of a given chemical shift value being associated with a given state of secondary structure (CSI assigns three indices). This approach is similar to the PSSI approach, though the actual method of calculating the potential differs. Like CSI and PSSI, PsiCSI reduces noise by polling nearby shifts. PsiCSI examines a small window of shifts (three residues) centered on the residue in question. Potentials derived from these shifts, along with the estimated residue-dependent reliabilities (i.e., probability of the assignment being correct) of these potentials, are fed into a first layer of neural networks to derive a second set of refined potentials. Multiple shifts are used to further increase accuracy. Additional information from 15N shifts and from Psipred (secondary structure prediction from protein sequence) [64], [65] predictions is also used. Rather than utilizing a simple jury system, PsiCSI trains a second layer of neural networks. Every possible combination of the available data for the residue (i.e., refined potentials from the first layer of networks and Psipred potentials) is fed into separate neural nets. Reliabilities for each combination are estimated, and the best-performing combination (for that residue type) is used to provide potentials for the next layer of neural networks. Finally, as with Psipred, a last neural net is used to take into account local interactions in a manner similar to that in which the first layer of neural nets is used to average out chemical shift noise. However, because the accuracy of the inputs at this stage is much higher, it is possible to utilize a much larger window (17 vs. 3 residues) to take into account more subtle interactions between distant residues. The most reliable outputs from the second layer, along with the estimated reliabilities, are fed into this final neural net to obtain the PsiCSI prediction.
PsiCSI achieves an accuracy of 89% (per residue), which is a significant improvement over the 82.8% (z > 12) accuracy observed for CSI. A server to use PsiCSI with sequence and chemical shift data is available from Samudrala’s group (http://protinfo.compbio.washington.edu/psicsi/).
2.4.3. Prediction algorithm for amino acid types with their secondary structure in proteins (PLATON) [61]
Prediction algorithm for amino acid types with their secondary structure, or PLATON, uses a database query approach to predict the secondary structure of a particular residue from its chemical shift values [61]. The method bases its prediction on a database consisting of reference chemical shift patterns (CSP) from the assigned chemical shifts of 51 proteins of known 3D structure. This reference CSP database is used for extracting distributions of amino acid types, along with their most likely secondary structures, for comparing single amino acid with query CSPs. The chemical shift pattern is a vector of Booleans describing relative positions of chemical shifts, and is defined by an optional combination of chemical shifts. The starting point for the definition of the CSP is the creation of an N-dimensional chemical shift space. N is determined by the kind of nuclei for which chemical shifts are available in the databases; for example, 13Cα, 13Cβ, 13C′, and 1Hα, or subgroups of those. The CSP is assigned “+” or “−” elements, depending on the position of the investigated chemical shift with respect to a reference value, for all nuclei considered. The positions of the elements are defined by the axes of the chemical shift space; for example, CSP (13Cα, 13Cβ, 13C′) = + − +. The chemical shift value of an amino acid is compared to the value at this point. If the observed value is larger, a “+” is assigned, and if it is smaller, a “−” is assigned. Hence, for all dots in this selection, the CSP (13Cα, 13Cβ, 13C′) = + + + is obtained. The chemical shift space can be further subdivided by introducing reference points into the two halves of each dimension to allow for a distinction of otherwise identical CSPs. The new reference value defines another coordinate system in the upper right quadrant. Practically, the second and higher-order reference points are chosen according to a statistical analysis of all amino acid species having the same three-digit CSPs in the original coordinate.
Results obtained for the 10 investigated proteins indicate that the percentages of correct amino acid species in the first three positions in the ranking list range from 71.4% to 93.2% for the more favorable penalty function. According to the authors, the advantage of this method over those that rely on averaged chemical shift values lies in its ability to increase database content by incorporating newly derived CSPs, and therefore to improve PLATON’s performance over time. The source code for PLATON is available from one of the authors (D. Laudde, http://www.bioforscher.de/).
2.4.4. Protein energetic conformational analysis from NMR chemical shifts (PECAN) [62]
Protein energetic conformational analysis from NMR chemical shifts (PECAN) is an energy model that predicts elements of secondary structure by optimizing a combination of sequence information and residue-specific statistical energy functions to yield energetic descriptions. The energetic model presents a framework for combining the interdependent information from sequence and chemical shifts in a manner that optimizes their joint predictive potential. PECAN uses a database containing ~37,000 residues from 310 protein sequences to construct a statistical potential that is used to predict the secondary structure. An additional, non-overlapping database containing ~12,000 residues from 97 protein sequences is used to determine the model that is independent of the dataset. Equivalent unbiased criteria were used in selecting the members of each dataset, which consists of proteins with known structure and assigned chemical shifts. According to the authors, there is a marked increase in accuracy in the predicated secondary structure. The reader is referred to the original paper-describing PECAN (and supporting information) for details of the mathematical model. A web server is available at: http://bija.nmrfam.wisc.edu.
2.4.5. Two-dimensional cluster analysis method 2DCSi [63]
2D CSI (“two-dimensional cluster analyses of chemical shifts to identify protein secondary structure”) analyzes paired, two-dimensional scattering diagrams of six chemical shift data sets; i.e., six different chemical shifts (1Hα, 1HN, 13Cα, 13Cβ, 13C′, and 15NH) are used to identify the secondary structure of amino-acid residues in proteins. In a three-step approach, first the data sets of chemical shifts and protein secondary structures are collected and cross-referenced. Second, 15 cluster scattering diagrams are plotted for paired chemical shifts of the six data sets, and the clusters as a function of the secondary structure are examined. Third, score matrices created for each of 20 amino acids are used to determine the secondary structure of the residues. The probability score is estimated based on two parameters: P r(ξ| χ1, χ2), the probability of a ξ state for observed chemical shifts χ1 and χ2, and τ(χ), the sum of all 14-probability scores. The pair, (χ1,χ2), can take values: χ2=cβ, χ1=cα, c′, nh, hα, or hn; χ2=cα, χ1= c′, nh, hα, or hn; χ2=c′, χ1= nh, hα, or hn; χ2=nh, χ1= hα, or hn; and χ2= hα, χ1= hn. Here, cα, cβ, c′, nh, hα and hn are the chemical shift values of 13Cα, 13Cβ, 13C′, 1HN, 1Hα, and 15NH, respectively. In addition, ξ can be helix (H), extended In addition, ξ can be helix (H), extended structure (E), or random coil (C), defined as neither helix nor extended structure. From the two-dimensional cluster analysis, three situations of Pr (ξ| χ1,χ2) can arise: (i) (χ1,χ2) falls outside all clustered elliptical areas; (ii) (χ1, χ2) falls onto one and only one elliptical area; (iii) (χ1, χ2) falls onto an intersection area of two ellipses. A set of rules is then used to make the prediction. These rules are: (a) add up probability scores of each column in the scoring matrix to obtain the total score τ(χ) for secondary structure states, and (b) identify any secondary structural state ζ if and only if τ(χ) ≥ 0.8×λ, where 0.8 represents the decision threshold (decided based on the target data of 601 entries), λ is the total number of resonances used.
2DCSi uses a dataset containing ~40,706 residues from 336 non-redundant proteins. The performance of 2DCSi is compared [63] with that of CSI and psiCSI using a set of 45 reference-corrected proteins for the prediction accuracy of three secondary structure states. Though the authors mention web-server (http://www.ncku.2dsci.idv.tw/) is available for using 2DCSi, it is not possible to access the program.
2.4.6. Comparison of the methods
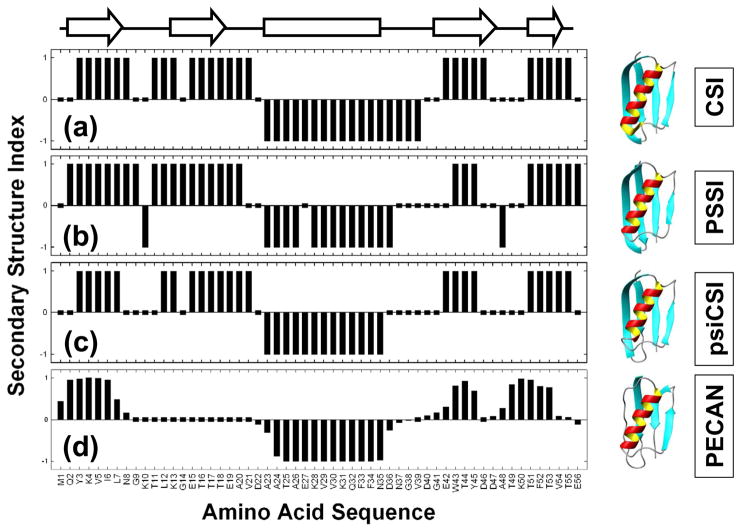
In order to compare the performance of the different methods, we calculated the secondary structure index (+1, 0 and −1, to represent β-strand, coil and α-helix, respectively) of a small protein (Protein G). Figure 3 shows the secondary structure index calculated using: (a) CSI, (b) PSSI, (c) psiCSI and (d) PECAN. We are unable to make a similar calculations using 2DCSi and PLATON as these codes were not available at the time of this study at the URLs mentioned in the respective manuscripts. The chemical shift values of protein G were obtained from the biological magnetic resonance bank (BMRB) (access number bmr5654.str). Figure 3 also shows the secondary structure estimated from an NMR-determined three-dimensional structure (the average structure corresponding to PDB ID 1GB1) using the Kabsch-Sanders algorithm in MOLMOL [66]. The secondary structures estimated by these respective methods are also superimposed on the three-dimensional structure in Figure 3. The codes for the above-mentioned programs were used with no further modification. Protein G is one of the most extensively studied proteins by either NMR or X-ray crystallography [67], [68], [69], [70]. It has 56 residues and is classified as an alpha and beta (α+β) protein [71], [72]. One important feature of the comparison is that no two methods give the same results, likely because their criteria for secondary structure identification based on the chemical shift data are inherently different. Therefore, it is important to be aware of how each method determines the secondary structure, and to exercise caution when using this information as a structural constraint upon 3D structural models. In general, the methods exhibit a broad consensus as to the location of most helix and strand core segments in protein structures; however, the termini of the segments are inconsistently defined.
Figure 3.
Plot of the secondary structure indices calculated for Protein G using four different methods, (a) CSI, (b) PSSI, (c) psiCSI and (d) PECAN. Secondary structure indices, +1, 0 and −1 correspond to α-helix, coil and β-strand, respectively. The chemical shift information is obtained from BMRB (bmr5664.str) and the programs CSI, PSSI, psiCSI and PECAN are used with their default setup. The arrows and the bar at the top of the figure are the secondary structure determined from the ensemble averaged NMR structures (RCSB file 1GB1) and the respective secondary structures are also superimposed on the 3D structure, using the molecular rendering program MOLMOL [66].
In our experience, in addition to the choice of algorithm, the choice of reference chemical shift (often referred to as the random coil chemical shift) used to determine the secondary chemical shift itself can introduce significant variation in secondary structure estimations. This issue is addressed in the following section.
2.5. Effect of reference chemical shifts on protein secondary structure estimation
Reference (random coil) chemical shifts are integral to defining the secondary chemical shifts in proteins that translate into protein secondary structure information. As discussed previously, though various techniques for estimating protein secondary structure from chemical shift data are widely employed and seem fairly reliable, at least for folded proteins, the choice of reference chemical shift values can significantly alter the outcome of secondary structure estimation. Random coil chemical shifts are the characteristic chemical shifts of the nuclei constituting the amino acid residues of disordered proteins. The effect of a particular secondary structure on the observed chemical shift known as the secondary chemical shifts are predominantly influenced by non-covalent interactions, such as secondary structural changes, hydrogen bonds, and aromatic stackings.
The primary goal of the work presented by Mielke and Krishnan [73] was to evaluate the effect on secondary structure prediction of using differing random coil chemical shift reference tables in conjunction with the CSI algorithm or, in principle, any of the alternative methods. The secondary structure content (the total percentage of helical and sheet content) of a set of 396 folded proteins was calculated using the consensus CSI method. Corresponding structural information was calculated from the three-dimensional structural coordinates of the proteins. A comparison of the results obtained using five different reference tables for CSI calculations to those obtained using a structure-based method allows a critical evaluation of the reliability of the standard protocol for evaluating secondary structure from chemical shift information using CSI.
Here we highlight some of these findings based on five different reference random coil chemical shift value sets and their respective use in estimating protein secondary structure. In general, the results show that none of the reference random coil data sets chosen for evaluation fully reproduces the actual secondary structures. Among the reference values generally available to date, most tend to be good estimators only of helices. On the basis of this, we recommend the experimental values measured by Schwarzinger et al. [74], and the statistical values obtained by Lukin et al. [75], as good estimators of both helical and sheet content.
List of reference chemical shift values
There are several reference random coil chemical shift tables in the literature, and these can be classified into two types: those measured experimentally, and those derived statistically. The complete details of these tables, including a description of the experimental conditions under which they were obtained, and their respective references, are given in Table 3. Of the various references listed in Table 3, only six different random coil chemical shift values that follow the recommendations of Wishart and Nip [20] are used for further analysis. In what follows, these five sets are identified by the initials of the first and last authors of the references as KW, WS, SD, LH, WJ and WM; Wüthrich et al. [21], [76], Wishart and Sykes [19], [77], [78], Schwarzinger et al. [74], Lukin et al. [75], Wang and Jardetzky [79], and Wang et al. [80], respectively (shown as block letters in Table 3). Of these six datasets, our study uses the first five for the analysis. Of the five chosen data sets, three were experimentally derived, while two were obtained using statistics-based approaches. We have re-referenced KW and WS, originally referenced to TMS/Dioxane, to DSS. Since reference table LH does not derive 1Hα values, the 1Hα reference values of Wang and Jardetzky [79] were used for structure estimation using LH. Though the experimental values of Plaxco et al. [81] are relevant for the comparison, these were not considered for the analysis due to lack of heteronuclear chemical shift values.
Table 3.
List of various reference random coil chemical shift data
| Experiment- based random coil shifts | |||||||
|---|---|---|---|---|---|---|---|
| Sample | Nuclei | Solvents | Reference | T (° C) | pH | Correction | Reference |
| H-GG-X-A-OH | 1H, 13C | D2O | TMS | 35 | Varied | None | [192], [193] |
| Apamin, BPTI | 1H, 15N | 90% H2O/10% D2O | TSP | 50, 65 | 2.2–4.6 | None | [194], [195] |
| GG-X-GG | 13C | D2O/10, 20, or 30% acetonitrile or TFE | TSP | 25 | 2.0–3.5 | None | [196] |
| H-GG-X-A-OH (KW) | 15N | 90% H2O/10% D2O | TSP | 35 | 2.0 and 5.0 | Sequence- corrected | [21], [76] |
| H-GG-X-GG-OH | 1H | 90% H2O/10% D2O and TFE% varied | DSP | 278–318 | 5.0 | None | [197] |
| GG-X-Y-GG, Y=A, P (WS) | 1H, 13C, 15N | 95% H2O/5% D2O | DSS | 25 | 5.0 | Nearest neighbor | [77], [78] |
| Ac-GG-X-GG-NH2 | 1H | 90% H2O/10% D2O (50mM Sodium Phosphate) 2, 3, 6, 8 M GuHCl |
DSS | 20 | 5.0 | None | [81] |
| Ac-GG-X-GG-NH2 (X=phosphorylated amino acid) | 1H, 13C, 15N | 90% H2O/10% D2O | DSS | 25 | 2.0–9.0 | None | [198] |
| Ac-GG-X-GG-NH2(SD) | 1H, 13C, 15N | 90% H2O/10% D2O and 8M Urea | DSS | 20 | 2.3 | None | [74] |
| Ac-GG-X-GG-NH2 | 1H, 13C, 15N | 90% H2O/10% D2O | DSS | 20 | 2.3 | Sequence- corrected | [74], [90] |
| Statistically derived random coil shifts | |||||||
| Method | Nuclei | Solvent condition | Referenced to | T (° C) | pH | Correction | Reference |
| Manual | 1H, 13C, 15N | Aqueous solution | DSS | - | - | None | [77], [78] |
| Probability-based (LH) | 13C, 15N | Aqueous solution | DSS | - | - | None | [75] |
| Probability-based (BMRB) | 1H, 13C, 15N | Aqueous solution | DSS | - | - | None | [79] |
| Probability-based (WJ) | 1H, 13C, 15N | Aqueous solution | DSS | - | - | Neighboring residue effect | |
| Data base derived (WM) | 13C, | Aqueous solution | DSS | - | - | Re—references using RefDB | [80] |
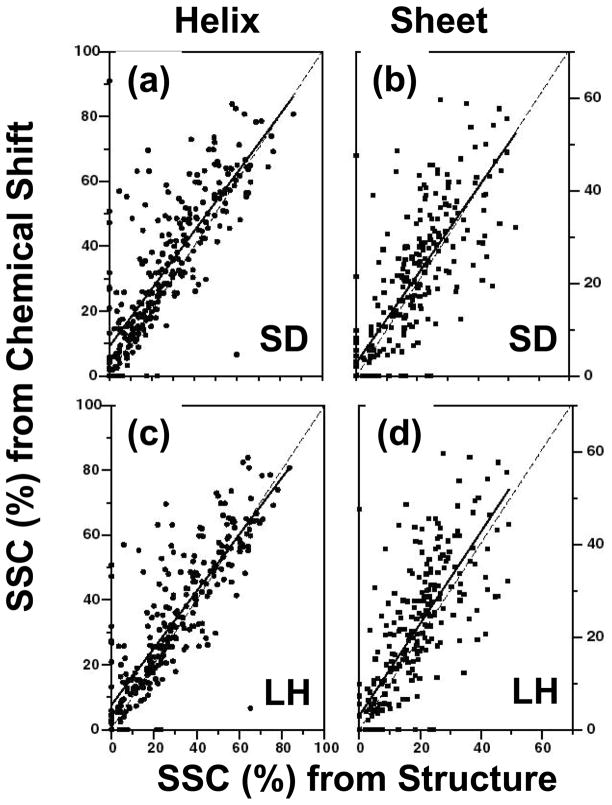
Figure 4 shows the results of estimating the percentage of helical (left panels) and sheet (right panels) content determined from the random coil chemical shift tables, SD and LH, respectively, using CSI, versus the same content calculated from relevant three-dimensional structures. The dashed lines in the figures correspond to an ideal correlation, and the solid lines to an unbiased linear regression analysis of the data. Table 4 lists the coefficients (slope and intercept) of the fit, and the correlation coefficients of the regression analysis. Chemical shift values corresponding to protein atoms were obtained from BMRB NMR-STAR files [38]. Only proteins with 50 or more amino acid residues were considered, since these are expected to contain a significant amount of secondary structure. Further, only proteins with at least 70% of their residues assigned chemical shifts were considered. As nearly all recently submitted BMRB chemical shifts are referenced using the widely accepted standard procedure recommended by Wishart [77], no re-referencing was performed. The consensus chemical shift index (CSI) of the proteins was calculated using the procedure outlined by Wishart and Sykes [55], using nuclei that are known to be highly sensitive to secondary structural changes (1Hα, 13Cα, 13Cβ and 13C′). Structure files were obtained from RCSB( http://www.rcsb.org/pdb/) [35], [82]. Since most BMRB NMR- STAR files identify several corresponding PDB structures, it was necessary to examine each entry and choose by inspection the most appropriate PDB ID number. When possible, the PDB ID corresponding to the “best” NMR structure was chosen, though in some cases it was necessary to choose the best X-ray structure (resolution < 2.5 Å). A total of 396 proteins was found to be suitable, and downloaded from the Protein Data Bank. The total percentage of sheet and helix (α and 310) was determined using the program PROMOTIF (http://www.biochem.ucl.ac.uk/~gail/promotif/promotif.html) [52], which uses the atomic coordinate files obtained from the RCSB. Uncertainties in the former were obtained by a linear model bootstrapping procedure using the R statistical package (www.cran.us.r-project.org) with 512 bootstrap replicates. Based on this analysis, several distinct features are observed.
Figure 4.
Plots of secondary structure content (SSC) in percentage determined from chemical shifts and three-dimensional coordinates. Panels (a) helical and (b) sheet content for the original SD [74], [90] random coil reference values correspond to (Table 3), while (c) and (d) show the corresponding correlations for using the random coil chemical shifts of LH [75]. The dashed line corresponds to an ideal correlation, while the solid line represents the linear regression analysis results (Table 5).
Table 4.
Linear regression analysis of CSI vs. structure-based helical and sheet content estimates
| Random coil reference | Helical content (%) | Sheet content (%) | ||||
|---|---|---|---|---|---|---|
| Intercept | Slope | CCa | Intercept | Slope | CCa | |
| KW [21], [76] | 5.33±0.88 − 3.45±1.02 |
0.91±0.02 0.82±0.03 |
0.82 0.77 |
4.56±0.59 8.11±0.58 |
0.95±0.02 1.03±0.03 |
0.77 0.67 |
| WS [77], [78] | 1.21±1.06 | 0.80±0.03 | 0.86 | 5.79±0.69 | 0.84±0.03 | 0.73 |
| SD [74] | 6.6±0.85 | 0.93±0.02 | 0.88 | 4.27±0.61 | 0.91±0.03 | 0.77 |
| LH [75] | 5.97±0.89 | 0.89±0.02 | 0.90 | 4.12±0.58 | 0.98±0.02 | 0.79 |
| WJ [79] | 6.92±0.77 | 0.94±0.02 | 0.88 | 6.6±0.61 | 0.96±0.03 | 0.73 |
Correlation coefficient.
Figure 4 contains several significant outliers along both the abscissa and ordinate. Points along the abscissa are primarily representative of poor quality chemical shift data (inappropriate references and assignments), while points along the ordinate might represent large discrepancies between the chemical shift data and corresponding three-dimensional structures. Though removing these outliers might have affected the correlations (Table 4), they were left in the data set in order that our results reflect as accurately as possible the quality of available experimental information.
The correlations (Figure 4 and Table 4) suggest that chemical shift-based methods for predicting secondary structure content are better indicators of helical regions than sheet regions in proteins. This could be due to insufficient sensitivity of secondary chemical shifts for identifying sheets. Ambiguity in the definition of a β-sheet, by contrast with that of an α-helix, may also contribute to this error [83], [84]. In calculating the secondary structure content from three-dimensional coordinates, we have used the program PROMOTIF, which uses the DSSP (database of secondary structure assignments) algorithm of Kabsch and Sander [85]. Definitions of secondary structure by PROMOTIF [52] closely follow IUPAC convention rule 6.3, and have been widely accepted amongst crystallographers. Other commonly used programs for secondary structure determination include STRIDE (secondary structure assignment from atomic coordinates) and DEFINE (determine the secondary and first level supersecondary structure) [85]. Cuff and Barton [86] have performed a comprehensive comparison of these three methods (DSSP, STRIDE and DEFINE), and shown that DSSP and STRIDE have an overall and segment-wise agreement of 95%. As the secondary structure definitions are based on the coordinates of a model derived by X-ray crystallography or NMR, any algorithm will be affected by the quality of the underlying data. The best estimation rate varies widely depending on the choice of algorithm [86], [87], [88]. However, of the many different methods of defining secondary structure proposed, DSSP has most successfully stood the test of time, and is widely used in the field of structural biology. Consequently, using PROMOTIF to perform NMR-based secondary structure calculations seems well justified. Moreover, any variation in the secondary structure content determined from three-dimensional coordinates, though it might alter correlations with secondary structure predicted from CSI using a given reference set of random coil values, will not influence systematic variations arising from the use of different reference sets.
2.6. Note on random coil chemical shifts
Variations in the random coil values
Reference (random coil) chemical shifts used in many of the methods for secondary structure estimation vary widely (Figure 5 and Table 5). The degree of variation in the estimated secondary structure contents using the various reference random coil chemical shift sets suggests the importance of investigating the origin of differences between the values they contain. Figure 5 shows a plot of the reference random coil shifts of 13C′, 13Cα, 13Cβ and 1Hα in panels a, b, c and d, respectively, for all the amino acids. Residue types are identified by their single-letter amino acid codes, with B and O corresponding to reduced cystine and cis-proline, respectively. The largest differences in the random coil values are seen for the 13C′ nuclei, more modest differences for the 13Cα and 1Hα spins, and the least variability for the 13Cβ nuclei. Visual inspection of the 13C′ chemical shifts (Figure 5a and Table 5) shows that in general the experimental reference values obtained in for an aqueous solution at 35 °C (reference shifts KW, marked as black circles) are lowest, and the experimental values obtained in aqueous solution with 8M Urea (reference shifts SD, marked as black squares) are the highest (see also Table 5). Values in the third experimental reference shift set (WS, filled squares) tend to be close to those in SD, while those in both statistical sets (LH and WJ, shown as stars and plus signs, respectively) fall mostly between the limits of WH and SD. Figure 5 suggests that contributions from random coil reference shifts corresponding to carbonyl nuclei, which are perhaps the most sensitive to protein structural changes, introduce the largest variability.
Figure 5.
Plots of the variation in the reference random coil values as a function of amino acid type. Panels (a), (b), (c) and (d) correspond to random coil values of 13C′ (carbonyl carbon), 13Cα, 13Cβ and 1Hα, respectively. The six different reference value sets are represented by symbols: black circles (KW) [21], [76], grey triangles (WS) [77], [199], black squares (SD) [74], [90], grey diamonds (LH) [75], black triangles (WJ) and grey circles (WM) [59], [80]. Plots (b) and (c) have all the six sets and plots (a) and (b) have only 5 and 4 sets, respectively (Table 5). Amino acids along the X-axis are given in single letter codes, with ‘B’ and ‘O’ representing oxidized cystine and cis-proline, respectively.
Table 5.
List of random coil chemical shift values
| AA | 13C′ (ppm) | 13Cα (ppm) | 13Cβ (ppm) | 1Hα (ppm) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KW* | WS* | SD | LH | WJ | KW* | WS* | SD | LH | WJ | WM | KW | WS* | SD | LH | WJ | WM | KW | WS | SD | LH/OJ | |
| A | 175.8 | 176.8 | 178.5 | 177.3 | 177.3 | 50.8 | 51.9 | 52.8 | 52.4 | 52.5 | 51.9 | 17.7 | 19.0 | 19.3 | 19.0 | 19.0 | 19.1 | 4.3 | 4.2 | 4.4 | 4.3 |
| B | -- | 174.8 | 175.3 | 174.8 | 175.1 | - | 58.3 | 58.6 | 58.0 | 58.2 | - | 28.6 | 28.3 | 28.2 | 29.5 | 4.3 | 4.7 | 4.6 | 4.6 | ||
| C | 173.9 | 172.9 | 175.5 | 174.8 | 175.1 | 53.9 | 56.0 | 55.6 | 58.0 | 58.2 | 40.0 | 41.8 | 41.2 | 28.2 | 29.5 | 4.7 | 4.7 | 4.8 | 4.6 | ||
| D | 174.2 | 175.9 | 175.9 | 176.5 | 176.0 | 52.7 | 53.2 | 53.0 | 54.1 | 54.0 | 53.7 | 39.8 | 40.8 | 38.3 | 40.8 | 40.8 | 41.1 | 4.8 | 4.6 | 4.8 | 4.6 |
| E | 173.7 | 176.7 | 176.8 | 176.3 | 176.3 | 55.4 | 56.5 | 56.1 | 56.7 | 56.7 | 55.9 | 28.7 | 29.7 | 29.9 | 30.1 | 29.9 | 29.9 | 4.3 | 4.2 | 4.4 | 4.3 |
| F | 172.9 | 174.1 | 176.6 | 174.8 | 175.5 | 56.2 | 57.5 | 58.1 | 57.9 | 57.5 | 57.1 | 38.4 | 39.3 | 39.8 | 39.3 | 39.4 | 39.6 | 4.7 | 4.6 | 4.7 | 4.6 |
| G | 171.8 | 172.2 | 174.9 | 174.0 | 174.0 | 43.9 | 44.2 | 45.4 | 45.4 | 45.3 | - | - | - | - | - | 4.0 | 3.9 | 4.0 | 4.0 | ||
| H | 172.2 | 174.8 | 175.1 | 174.5 | 174.8 | 53.6 | 54.2 | 55.4 | 55.8 | 55.7 | 55.5 | 28.1 | 32.0 | 29.1 | 29.8 | 29.5 | 30.9 | 4.6 | 4.6 | 4.8 | 4.6 |
| I | 174.2 | 175.5 | 177.1 | 175.7 | 175.5 | 59.6 | 60.6 | 61.6 | 61.0 | 60.8 | 60.9 | 37.5 | 37.5 | 38.9 | 38.9 | 38.4 | 38.8 | 4.2 | 4.1 | 4.2 | 4.2 |
| K | 174.5 | 175.7 | 177.4 | 176.4 | 176.2 | 54.6 | 55.7 | 56.7 | 56.6 | 56.3 | 55.9 | 31.9 | 32.3 | 33.2 | 32.6 | 32.5 | 33.1 | 4.4 | 4.2 | 4.4 | 4.3 |
| L | 174.7 | 175.2 | 178.2 | 177.2 | 176.7 | 53.8 | 53.9 | 55.5 | 54.8 | 54.8 | 54.2 | 41.2 | 41.9 | 42.5 | 42.8 | 42.1 | 42.4 | 4.4 | 4.3 | 4.4 | 4.4 |
| M | 173.4 | 176.1 | 177.1 | 175.5 | 175.9 | 54.0 | 55.7 | 55.8 | 55.3 | 55.4 | 55.2 | 31.7 | 32.8 | 32.9 | 33.0 | 32.9 | 32.9 | 4.5 | 4.3 | 4.5 | 4.5 |
| N | 173.1 | 173.7 | 176.1 | 174.7 | 174.8 | 51.5 | 50.9 | 53.3 | 53.2 | 53.0 | 53.1 | 37.7 | 39.0 | 39.1 | 38.7 | 38.4 | 38.9 | 4.8 | 4.7 | 4.8 | 4.7 |
| O | 173.7 | 174.9 | -- | 176.6 | 176.6 | 61.4 | 61.8 | 63.0 | 63.3 | 63.2 | 33.0 | 31.7 | 34.8 | 32.1 | 31.8 | - | 4.4 | 4.6 | 4.4 | ||
| P | 174.1 | 174.9 | 177.8 | 176.6 | 176.6 | 61.9 | 61.8 | 63.7 | 63.3 | 63.2 | 30.6 | 31.7 | 32.2 | 32.1 | 31.8 | 4.5 | 4.4 | 4.5 | 4.4 | ||
| Q | 174.0 | 174.1 | 176.8 | 175.5 | 175.8 | 54.1 | 53.6 | 56.2 | 55.8 | 55.9 | 55.3 | 28.1 | 30.1 | 29.5 | 29.3 | 29.0 | 29.4 | 4.4 | 4.3 | 4.4 | 4.3 |
| R | 173.4 | 175.9 | 177.1 | 176.1 | 176.0 | 54.6 | 55.7 | 56.5 | 56.3 | 56.2 | 55.8 | - | 30.3 | 30.9 | 30.6 | 30.4 | 30.9 | 4.4 | 4.2 | 4.4 | 4.3 |
| S | 172.6 | 172.6 | 175.4 | 174.4 | 174.4 | 56.6 | 57.4 | 58.7 | 58.3 | 58.2 | 58.0 | 62.3 | 62.7 | 64.1 | 64.1 | 63.8 | 63.8 | 4.5 | 4.5 | 4.5 | 4.5 |
| T | 172.5 | 173.5 | 175.6 | 174.8 | 174.8 | 60.1 | 60.8 | 62.0 | 61.6 | 61.3 | 62.4 | 68.4 | 68.1 | 70.0 | 69.8 | 68.9 | 69.8 | 4.3 | 4.5 | 4.4 | 4.4 |
| V | 173.9 | 174.2 | 177.0 | 175.8 | 175.7 | 60.7 | 60.2 | 62.6 | 62.1 | 62.0 | 62.3 | 31.4 | 31.7 | 31.8 | 32.7 | 32.4 | 32.9 | 4.2 | 4.1 | 4.2 | 4.1 |
| W | 173.4 | 172.9 | 177.1 | 175.9 | 175.9 | 55.7 | 53.5 | 57.6 | 57.5 | 57.5 | 57.8 | 28.1 | 28.3 | 29.8 | 29.1 | 29.6 | 29.6 | 4.7 | 4.6 | 4.7 | 4.6 |
| Y | 172.9 | 173.1 | 176.7 | 175.8 | 175.3 | 56.3 | 55.8 | 58.3 | 57.8 | 57.6 | 58.2 | 37.5 | 38.7 | 38.9 | 38.9 | 38.8 | 38.8 | 4.6 | 4.6 | 4.6 | 4.6 |
AA: Single letter amino acid codes correspond to: A: Ala, R: Arg, N: Asn, D: Asp, B: Cys (reduced), C: Cys (oxidized), Q: Gln, E: Glu, G: Gly, H: His, I: Ile, L: Leu, K: Lys, M: Met, F: Phe, O: Pro (cis), P: Pro (trans), S: Ser, T: Thr, W: Trp, Y: Tyr, and V: Val.
Random coil chemical shifts obtained given by KW [21], [76], WS [77], [78], SD [74], LH [75] and WJ [79]
KW* and WS*: Carbon chemical shifts were originally referenced to TMS/Dioxane. These have been re-referenced to DSS in the calculations. All other chemical shifts are referenced to DSS or TSP, and are obtained from the references listed in Table 3 of the main text.
Sequence-dependent effects
According to Flory [89], a “random coil” is independent of influences from neighboring residues. However, sequence-dependent corrections of random coil chemical shifts have recently been noted using experimental [77], [78], [90] and statistical [79] methods. Schwarzinger et al. [90] have experimentally studied a subset of penta-peptides to investigate the effect of neighboring residues on the observed chemical shift, and elegantly utilized the results to determine the residual secondary structures in partially unfolded proteins. Wang and Jardetzky [79] have recently determined a statistical distribution of nearest neighbor effects from chemical shift data obtained from the BMRB. Though the nearest-neighbor effects determined by the statistical method bear a trend similar to that of the experimental results in a solution of 8M urea for random coil chemical shifts, the former approach inherently assumes that residues that are neither helical nor sheet must be “random coil.” In practice, however, it would be necessary to collect experimental data on at least 8000 different tri-peptide samples to determine nearest-neighbor effects completely. Since this would require a monumental effort, and none of the available databases provide a complete set of experimental random coil chemical shifts, we have not accounted for nearest neighbor effects.
One must be able to define what is a ‘random coil’ of a polypeptide, before addressing the question of what is a ‘random coil chemical shift’? This issue seems to have attracted considerable attention in recent literature, particularly with respect to proteins that are ‘intrinsically unstructured’ [91]. The original definition of Flory [89], corroborated by Tanford [92], defines the random coil state of a peptide as one in which the backbone dihedral angles, φ and ψ, of each amino acid residue are independent of the conformations of neighboring residues. Alternatively, a random coil is sometimes defined as a reference state in which sidechain-sidechain interactions are absent [93], which neglects the intrinsic folding propensities of amino acids. In developing a probabilistic model to estimate the random coil chemical shifts of carbon-13 chemical shifts from protein chemical shift databases (such as BMRB), Wang et al. [80] adopt the following definition: a state in which the geometry of the polypeptide ensemble samples the allowed region of (φ,ψ)-space in the absence of any dominant stabilizing interactions.
To define a secondary chemical shift, one needs first to define a reference chemical shift. In the absence of methods able ab initio to predict structural effects on measured chemical shifts, the choice of reference chemical shift assumes an important role. According to Vila et al. [94], NMR-based chemical shift methods to date have not focused on statistical coil peptides, mainly because of the intrinsic difficulties associated with the characterization of unstructured states; i.e., the experimentally-determined (NMR) chemical shift values for statistical coil peptides are not associated with a unique set of canonical dihedral angles, making a theoretical description of non-structured states difficult to achieve. Studies of the factors that affect the chemical shift are very important, because NMR methods used to determine secondary structure (e.g., CSI and others discussed earlier) rely heavily on a comparison with the chemical shifts of the so-called statistical coil, which is frequently, but erroneously, referred to as a random coil [94].
A considerable amount of effort has gone into determining random coil chemical shifts, but the specific consequences of using a particular data set to determine protein secondary structures have not been investigated in detail. Over a selected set of well-characterized protein structures, it has been suggested that CSI-based secondary structure determination is 93% accurate in comparison to X-ray structure-based determinations [19]. Our analysis of a considerable amount of raw data from the BMRB and PDB shows that CSI estimates helical and sheet structures to an accuracy of only 90% and 79%, respectively. These results do not reflect the quality of the CSI method itself, but rather the sensitivity of the method to the choice of reference chemical shifts, and the large variation inherent in chemical shift data. Our results further suggest that secondary chemical shifts are more reliable for identifying helical regions of proteins than strand regions. Sharman et al. [95] have recently proposed that long-range effects from distant amino acids are one of the dominant factors in determining experimental chemical shifts in β-sheets. The absence of a good correlation for β-sheets in the data presented here is perhaps suggestive of this. Though rigorous experimental and statistical methods have been able to estimate random coil shifts more accurately in the last decade, our findings indicate that additional experimental and theoretical developments are mandatory for an explanation of the observed deviations. The present analysis forms a critical evaluation of the current status of the reliability of secondary chemical shifts as a direct refinement parameter in structure calculations. Though caution must be advised, since this work relies only on secondary chemical shifts, it nevertheless suggests the importance of pursuing a combined experimental, theoretical, and database-driven approach to secondary structure estimation that can provide a better understanding of the factors governing both the chemical shift, and its relationship to protein structure. From a practical point of view, one might want to know what is the best set of reference (random coil or statistical coil) chemical shifts (or combination of sets) to estimate the secondary or even tertiary structure in proteins. Evaluations of the different choices of reference set have shown clear discrepancies, and suggested which choices are best for specific sets of proteins [73], [80]. However, a complete understanding of the origin of these effects, and of how well a ‘secondary chemical shift’ can be defined for purposes of accurate estimation of secondary structure, remains a challenge.
2.7. Secondary chemical shifts in DNA and RNA
In contrast to the extensive development of empirical and semi-empirical chemical shift methods for proteins, these methods are limited for DNA and, in particular, for RNAs. Though a discussion on nucleic acids might sound anomalous in an article that focuses on proteins, from the point of view of secondary chemical shifts in biopolymers in general, this section makes it complete. Lam and co-workers have extensively contributed to the measurement and categorization of random coil and B-form DNA chemical shifts [96], [97], [98], [99], while work on RNA is essentially limited to work by Cromsigt et al. [100]. Chemical shift-structure relationships in DNA can provide a quick reference guide for resonance assignments based on conventional experiments, thus facilitating solution structure studies of DNAs. These results can also provide useful information for studying structure–chemical shift relationships, identifying unstructured or right-handed double helical regions, monitoring DNA–drug or DNA–protein binding, and investigating conformational details of special features in DNA structures [101], [102], [103], [104].
Chemical shift information in DNA contains a wealth of structural information that is seldom used extensively. Over the last few years, methods have been established to predict chemical shifts of DNAs in random coil form (single stranded) [96], [97], [99] and double-helical B-form [105], [106]. These methods are based on sets of reference chemical shift values and correction factors from experimental measurements, statistical analysis or semi-empirical calculations. Shielding or deshielding contributions from nearest neighbor and/or next-nearest neighbor nucleotides have been included in these prediction methods.
To automate these prediction methods, Lam has established a web server called ‘DSHIFT’ for predicting random coil or double-helical B-DNA chemical shifts of any specific sequence (http://www.chem.cuhk.edu.hk/DSHIFT,).
Random coil chemical shifts in DNA are more sensitive to the nearest neighboring residues (contradicting the conventional definition of a ‘random coil’), and therefore a pentamer or triplet model must be defined. In the case of a triplet model, for each residue (e.g., the base ‘C’), there are 16 possible chemical shift values. For random coil proton prediction, DSHIFT uses a pentamer model: . Here the prediction is based on the triplet chemical shift, , and a correction factor is invoked to account for the effects of second nearest neighbors using the expression:
| [4] |
Here, and are the 5′ and 3′ nearest neighbor thymine effects on the central residue (X) and and are the corresponding effects on X in the predicted sequence. Modifications for the terminal residues are accounted for separately in the prediction algorithm, as the 5′ and 3′ phosphate groups at the termini are absent [98], [105]. For random coil carbon chemical shifts, the prediction method is based on a trimer model, as only nearest neighbor effect has been found to be significant [97].
For prediction of double-helical (B-form) DNA, DSHIFT can use either the methods introduced by Altona et al., [105] or Wijmenga at al., [106]. In Altona’s method, proton chemical shift prediction is based on a trimer model in which an incremental scheme and statistical reference values from experimental results are used [105]. In Wijmenga’s method, the proton chemical shift of a specific nucleotide is predicted based on a set of calculated reference shift values (δref) plus the chemical shift effect induced by its own base (δib), as well as its 3′ (δ3′b) and 5′ (δ5′b) neighboring bases [106]. As noted by Lam [98], the prediction accuracy of the various methods depends mainly on DNA conformations. Since temperature and solution conditions affect stabilities of DNA structures, it is expected that these factors will also affect the prediction accuracy.
3. Empirical methods correlating averaged chemical shifts (ACS)
3.1. Basic concepts: averaged chemical shifts, protein secondary structure content, and protein structural class
3.1.1. Averaged chemical shifts
The averaged chemical shift (ACS) of a nucleus, i, is defined by:
| [5] |
where N is the total number of observed cross peaks (typically in a single bond-correlated spectrum, such as a heteronuclear single quantum correlation, HSQC) and ωk is the corresponding chemical shift of the kth resonance (referenced using recommended procedures [78]). Averaged values of chemical shifts of random coil proteins were also calculated from the respective amino acid sequences using recently published experimental values [74], [90].
3.1.2. Protein secondary structure content
Protein secondary structure content refers to the proportion of each secondary structure type constituting a given protein. Formally, it is defined as the ratio of the number of residues in a certain secondary structure to the number of total residues of a protein. According to the conventional classification by DSSP [107], there are eight secondary structure types, namely, α-helix,β-strand, β-bridge, three-turn helix, π-helix, hydrogen-bonded turn, bend, and random coil. Protein secondary structure provides fundamental information about proteins, and knowing a protein’s secondary structure content is often the first step towards more detailed knowledge of its structure and function.
Protein secondary structure content can be semi-empirically estimated using variants of spectroscopic methods, such as UV-Raman [108], CD [109], FTIR [110] and NMR [111]. However, generally speaking, these experiment-based approaches have been of questionable accuracy [112], [113]. For that reason, there have been many attempts to make ab initio predictions of secondary structure content [114], [115], [116], [117], [118], [119], [120], [121]. Among notable early attempts to do so are the multiple linear regression approach [122], [123], [124], [125], [126], [127], [128], [129], the artificial neural network approach [121], and the analytic vector decomposition approach [130], [131]. Of course, the validity of such approaches ultimately depends on the accuracy with which they predict the actual secondary structure contents of proteins, so experimental methods continue to play a significant role in these efforts.
3.1.3. Protein structural class
Classification and prediction of protein structure are essential goals of protein science, and the structural class is an important attribute used to characterize the overall folding type of a protein or its domains [132], [133], [134]. Nikashima et al. were the first investigators to suggest that protein structural class is correlated with protein secondary structural information and amino acid composition [132]. Subsequent efforts have primarily focused on designating structural classes based on amino acid composition [116], [117], [135], [136], [137], [138], [139], [140], from which folding pattern information can be obtained without addressing the complicated issue of three-dimensional structure [133], [134]. However, in the last decade, the designation of protein structural class based on secondary structure content has proven to be extremely useful from both experimental and theoretical points of view [133], [134], [140], [141], [142], [143], [144], [145], [146], [147], [148]. In the following section, we discuss a chemical shift-based structural classification method motivated by the success of secondary structure-based approaches.
3.2. Correlation between averaged chemical shift and protein structural class
This section summarizes the results of an empirical approach for estimating protein structural class directly from NMR spectra, prior to resonance assignment [149]. For a detailed discussion, see Ref. [149]. Briefly, the method seeks to correlate an empirical parameter, an averaged chemical shift (ACS) obtained by mining the BioMagResBank (BMRB) [38], with protein structural classes obtained from CATH [165,166] and SCOP [69,70,164]. This correlation permits an estimation of the classes of proteins of unknown structure based solely on the average of chemical shift values obtained from NMR.
3.2.1. Averaged chemical shifts are sensitive to protein structural class
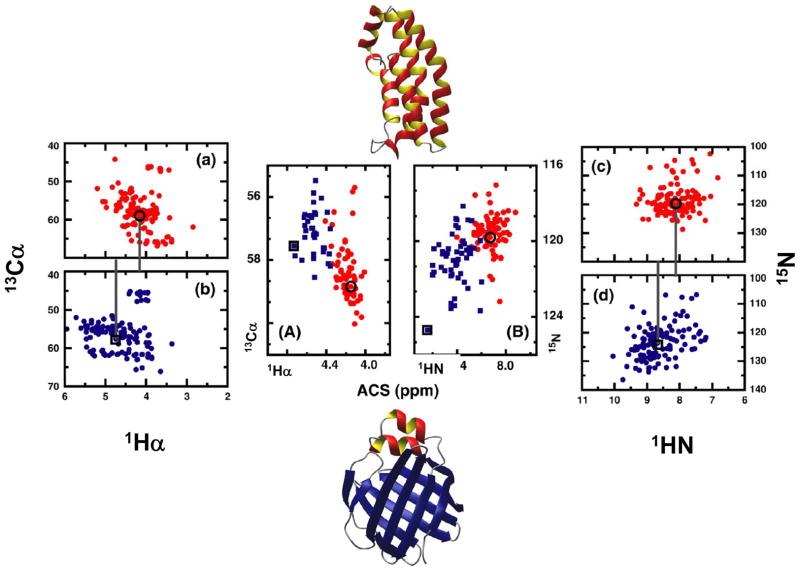
Figures 6A and 6B plot, respectively, the 13Cα versus 1Hα and 15N versus 1HN ACS values reported in Ref. [167]. Values indicated by red circles correspond to proteins deemed α-class according to CATH, and values indicated by blue squares correspond to molecules deemed β-class. As pointed out in Ref. [167], the figures are suggestive of a correlation between structural class and ACS. This is borne out by ACS values calculated from 13C-HSQC spectra (see Figures 6a–d) from histidine kinase (PDB code 1A0B, BMRB number 4857) [150], a predominantly α-helical protein, and from liver fatty acid binding protein (PDB code 1LFO, BMRB number 4098) [151], a predominantly β-sheet protein (the three-dimensional structures of these proteins are shown above and below Figures 6A and 6B, respectively). These values are indicated by circles for histidine kinase and squares for liver fatty acid binding protein. In both cases, the ACS values are reproduced in Figures 6A and 6B, where they are seen to lie within the appropriate cluster of α- or β-class proteins considered in Ref. [167].
Figure 6.
Representative examples to show that averaged chemical shift (ACS) is a structural parameter directly obtainable from NMR spectra. (a) and (c): simulated 13C and 15N-HSQC spectra of an α-helical protein (Histidine kinase, PDB code 1A0B, BMRB number 4857), respectively. (b) and (d): simulated 13C and 15N-HSQC spectra of a β-sheet protein (Liver fatty acid binding protein, PDB code 1LFO, BMRB number 4098). The ACS calculated from each spectrum is noted by a black circle (helical protein) and square (sheet protein). (A) and (B): representative examples of the ACS values calculated from 13Cα-1Hα and 15N-1HN correlations, respectively, for a set of proteins for which chemical shift information is obtained from BioMagResBank. The red circles and blue squares correspond to proteins that are classified as mainly-α and mainly–β, respectively, under the CATH classification scheme. ACS values from (a) and (b), and (c) and (d), are reproduced in (A) and (B), respectively. Reproduced with permission from Ref. [149].
3.2.2. Distribution of protein structural classes with respect to ACS values
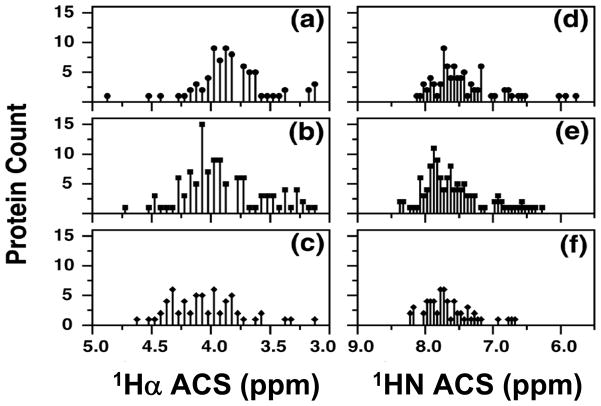
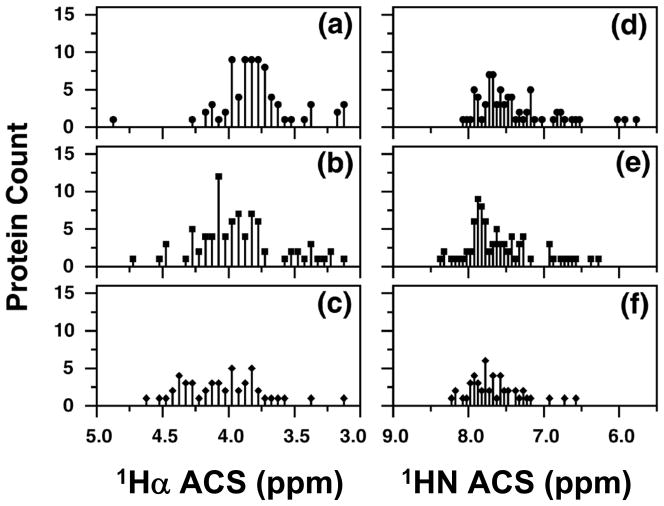
Figures 7 and 8 reproduce histograms of the protein distributions discussed in Ref. [167]. Figure 7 shows numbers of proteins, binned according to the ACS values of 1Hα (left panels) and 1HN (right panels), classified by SCOP as α (panels a and d), αβ (panels b and e) and β (panels c and f). Figure 8 shows the distributions resulting from classification by CATH. Statistical information on these distributions is summarized in Table 6. As noted in Ref. [167], because they are insufficiently resolved, the distributions based on 1HN ACS values (Figures 7,8 (right panels) and Table 6) are able to discriminate only α and “αβ/β” structural classes.
Figure 7.
ACS values vs. number of proteins in the three major structural classes defined according to the SCOP method. (a), (b), and (c) display the 1Hα ACS values for proteins that are mainly-α, mainly-β, and a mixture of α and β (αβ) (both α/β and α+β), respectively. (d), (e), and (f) display the corresponding 1HN values for mainly-α, mainly-β, and αβ (both α/β and α+β), respectively. Reproduced with permission from Ref. [149].
Figure 8.
ACS values vs. number of proteins in the three major structural classes defined according to the CATH method. (a), (b), and (c) display the 1Hα ACS values for proteins that are α, β, and αβ (both α/β and α+β), respectively. (d), (e), and (f) display the corresponding 1HN values for α, β, and αβ (both α/β and α+β), respectively. Reproduced with permission from Ref. [149].
Table 6.
Characterization of the statistical distribution of structural classes
| SCOPa Nucleus 1Hα | |||||
|---|---|---|---|---|---|
| Class | Totalb | Mean Chemical shift (ppm) | SDc | SDMd | 2*SDM |
| α | 88 | 3.83 | 0.34 | 0.040 | 0.072 |
| αβ | 122 | 3.94 | 0.52 | 0.047 | 0.093 |
| β | 61 | 4.05 | 0.30 | 0.038 | 0.076 |
| Nucleus 1HN | |||||
| α | 87 | 7.54 | 0.64 | 0.069 | 0.14 |
| αβ | 122 | 7.68 | 0.99 | 0.089 | 0.18 |
| β | 60 | 7.70 | 0.41 | 0.053 | 0.11 |
| CATHe Nucleus 1 Hα | |||||
| α | 77 | 3.79 | 0.29 | 0.033 | 0.066 |
| αβ | 83 | 3.93 | 0.32 | 0.035 | 0.070 |
| β | 49 | 4.05 | 0.30 | 0.043 | 0.086 |
| Nucleus 1HN | |||||
| α | 75 | 7.45 | 0.56 | 0.064 | 0.13 |
| αβ | 83 | 7.62 | 0.48 | 0.053 | 0.11 |
| β | 49 | 7.69 | 0.43 | 0.061 | 0.12 |
SCOP (Structural Classification of Proteins),
Total number of proteins,
SD Standard Deviation,
SDM Standard deviation about the mean,
and CATH (Class-Architecture-Topology-Homologous Superfamily) protein classification protocols.
3.2.3. Kolmogorov-Smirnov (K-S) tests
To check the statistical independence of the distributions presented in Figures 7, 8 and Table 6, Kolmogorov-Smirnov (K-S) tests were performed. Table 7 presents the results of these tests for all nuclei. As described in Ref. [167], two distributions are considered independent if the significance of the D statistic is less than or equal to 0.05. The comparisons for which this is the case are indicated by significance values in boldface type in Table 7. Only the separation of 1Hα ACS values according to SCOP-based classes leads to three independent distributions at a 5% level of significance.
Table 7.
Results of Kolmogorov-Smirnov test
| Classes Compared | SCOPa |
CATHb |
||
|---|---|---|---|---|
| K-S D Statisticc | Significanced | K-S D Statisticc | Significanced | |
|
1Hα | ||||
| α ↔ αβ | 0.24 | 0.0039 | 0.32 | 0.00042 |
| α ↔ β | 0.41 | 0.0000060 | 0.41 | 0.000042 |
| αβ ↔ β | 0.24 | 0.018 | 0.23 | 0.058 |
|
13Cα | ||||
| α ↔ αβ | 0.29 | 0.00030 | 0.29 | 0.0021 |
| α ↔ β | 0.41 | 0.0000090 | 0.34 | 0.0015 |
| αβ ↔ β | 0.18 | 0.15 | 0.21 | 0.11 |
|
1HN | ||||
| α ↔ αβ | 0.22 | 0.012 | 0.28 | 0.0029 |
| α ↔ β | 0.26 | 0.015 | 0.27 | 0.021 |
| αβ ↔ β | 0.11 | 0.65 | 0.092 | 0.94 |
|
15N | ||||
| α ↔ αβ | 0.082 | 0.88 | 0.10 | 0.79 |
| α ↔ β | 0.11 | 0.77 | 0.14 | 0.59 |
| αβ ↔ β | 0.13 | 0.47 | 0.13 | 0.65 |
Proteins classified using SCOP,
Proteins classified using CATH,
Maximum value of absolute difference between cumulative distribution functions,
Significance: values less than/equal to 0.05 are considered significant (numbers in bold print).
3.3. Empirical correlation between averaged chemical shifts and protein secondary structure content
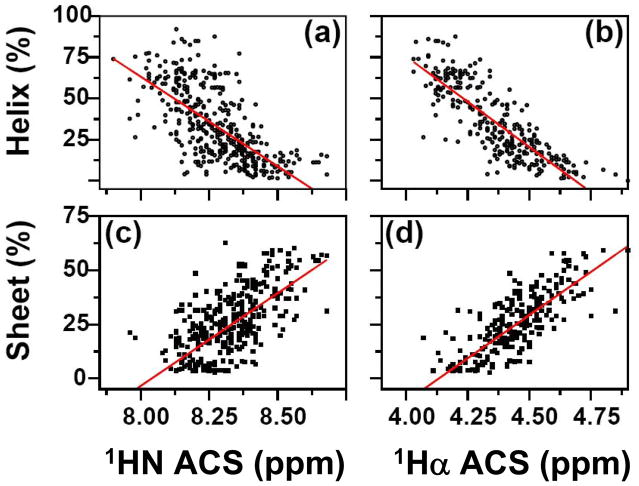
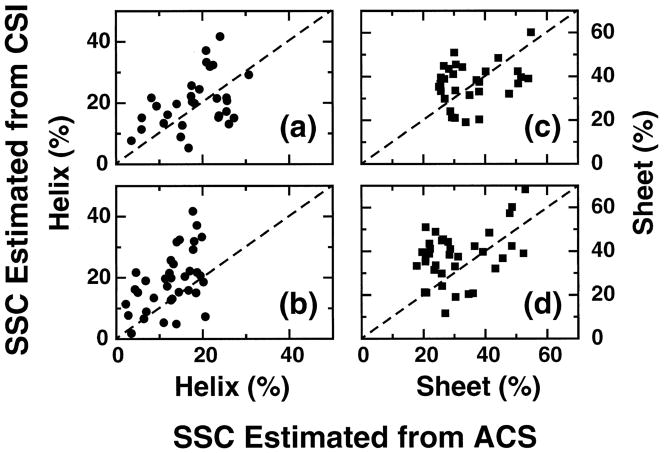
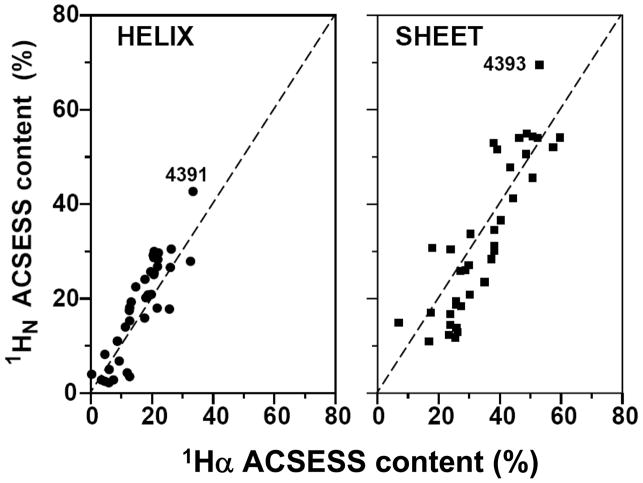
It is possible to take an educated ‘guess’ by looking at the chemical shift dispersion of an 15N-HSQC spectrum to say whether it contains predominantly helical or sheet secondary structure. This is because helical proteins generally have narrow spectral dispersion in the 1H dimension of the amide protein region, while proteins with β-sheets tend to be more dispersed. The averaged chemical shift method essentially quantifies this observation. It is based on the hypothesis that if one considers an NMR spectrum as a projection of the protein’s three-dimensional structure on a chemical shift dimension (dimensions), the distribution of the points represents some of the dominant features of the three-dimensional fold. For any distribution, the mean value—in this case the ‘averaged chemical shift’—is the first statistical quantity that distinguishes itself from other such quantities. Although the mean value in any given NMR spectrum can be calculated in a straightforward manner, there is no simple translation from this value to the three-dimensional structure. As a first step in the ‘reverse-engineering’ process one can resort to examining empirical relationships between a set of known three-dimensional structures and their respective chemical shift distributions.
In this section we address the correlation between protein secondary structure content and the average chemical shift (ACS) value for a particular type of nucleus within the protein. By using current NMR data processing software, it is fast and easy to obtain an experimentally-determined ACS value compared with obtaining complete resonance assignments. We have determined that the highest correlation with secondary structure content is found with the 1Hα ACS value, followed by the 1HN ACS. The empirical correlation that is derived from these relationships has been named ACSESS (averaged chemical shift to estimate secondary structure content. The application of ACSESS to determine secondary structure (helix and sheet) content under conditions where it is often difficult to obtain structural information, such as denaturing conditions, is also demonstrated. Predictions of secondary structure content obtained using ACSESS are better than those obtained using methods that rely on primary sequence, because the latter do not provide any information about conformational changes that result from different solvent conditions. Estimating changes in secondary structure content is relevant to studies of proteins, such as prions, that undergo conformational rearrangements, and to following major conformational changes of proteins in the presence of ligands or nucleic acids. It is emphasized, however, that ACSESS does not provide an alternative to other conventional NMR methods for secondary structure determination, such as the Chemical Shift Index (CSI) [19], [54]; it only provides information about overall secondary structure content prior to complete structural analysis, or in cases where it is difficult, if not impossible, to obtain such information by other means. Thus, ACSESS has several important potential applications in proteomics and protein folding studies.
3.3.1. Linear Correlations between ACS and SSC
The empirical correlation between averaged chemical shift and secondary structure content is referred as “ACSESS”. Figures 9a–d show plots of the ACS values of HN and Hα nuclei versus helix and sheet content. A total of 426 proteins was used for both 1HN (Fig. 9a and c) and 1Hα (Fig. 9b and d) to establish a correlation. Linear-regression analyses of the data in Figure 9 (helix and sheet structure content vs. ACS) are summarized in Table 6 (SSC = Slope × ACS + Intercept). Only ACS values corresponding to HN and 1Hα nuclei were considered, since values associated with these nuclei are in general much more indicative of overall secondary structure content than those associated with the heavy backbone atoms [111], [152]. BMRB is the first public database to collect chemical shift information from a large number of proteins. Though highly useful, BMRB is new by comparison with three-dimensional structural databases, such as the PDB, and currently lacks a rigorous strategy for quality control. RefDB is an even newer database, assembled by Zhang et al. [153], in which chemical shift information obtained from BMRB is uniformly referenced, and unassigned or missing resonances are predicted using other empirical correlations. In addition to those obtained using BMRB data, we obtained similar correlations with secondary structure content using RefDB data (similar to Fig. 9). Table 8 lists the results of the linear correlation for both BMRB and RefDB chemical shift values. For BMRB-based chemical shifts, the coefficients of correlation between HN ACS and helix or sheet content are −0.67 and +0.71, respectively, while the corresponding Hα values are −0.84 and +0.84. It can be seen from Table 8 that performing the analysis on the same set of proteins using RefDB-based information produces values in close agreement with these BMRB-derived values; omissions and nonstandard referencing in BMRB evidently have little impact on correlations between ACS and SSC. On the other hand, comparison of these with our earlier results [152] shows that increasing the number of proteins in the data set significantly improves the correlation. For example, the coefficients of correlation between ACS and sheet content are seen to increase from 0.75 to 0.84 for Hα, and from 0.66 to 0.71 for HN. We note that the intercept values do not have any physical meaning, as this empirical approach is intended to show a linear correlation over a subset of chemical shifts of folded proteins only. The present results are consistent with our earlier findings that, for both Hα and HN, the relationship between ACS and SSC are characterized by a positive correlation coefficient for sheet content and a negative coefficient for helix content. A similar correlation for the heteronuclei (13Cα and 15N) was also performed [152]. Correlation coefficients for the plots of 15N and 13Cα versus percent sheet content are 0.44 for both, while the coefficients obtained in the plots versus percent helix content are 0.40 and 0.58, respectively. Although 13Cα ACS values show a wider dispersion with respect to helical content than the corresponding 15N data, the correlation coefficients for the plots of heteronuclei are equally poor [152]. Overall, the best correlations were obtained with the 1HN and 1Hα data.
Figure 9.
Plots of the averaged chemical shift (ACS) values from experimental data versus the secondary structure content (SSC) estimated from three-dimensional structures. (a) and (c) show percent helix (circles) and sheet (squares) versus ACS for HN, whereas (b) and (d) show the corresponding plots for Hα. The continuous lines show the a linear regression analysis of the data. Reproduced with permission from Ref. [111].
Table 8.
Linear Correlation of ACS to Secondary Structure Content
| Nucleus | a Helix (%) | a Sheet (%) | ||||
|---|---|---|---|---|---|---|
| b CC | Slope | Intercept | b CC | Slope | Intercept | |
| HN | −0.67 (−0.68) | −108.7±5.7 (−109.3±5.8) | 933.03±47.5 (938.0±48.7) | 0.71 (0.70) | 85.73±4.0 (85.3±4.2) | −689.3±33.8 (−686.2±35.4) |
| 1Hα | −0.84 (−0.84) | −109.7±4.0 (−108.6±4.0) | 514.54±17.5 (509.3±17.6) | 0.84 (0.84) | 79.81±2.9 (79.5±2.9) | −329.8±12.7 (−328.5±12.9) |
Secondary structures defined based on PROMOTIF.
CC: Correlation coefficient from linear regression analysis.
Slope and intercept are defined based on a linear equation: SSC = Slope×ACS (ppm) + Intercept.
Each cell contains the analyses of chemical shift information from BMRB (RefDB).
A notable feature of these results is that the slopes of the lines for the ACS values versus helix and sheet content are opposite in sign (most clearly seen in panels a and c of Figures 6 and 9). The change in the sign of the slope indicates that changes in ACS values allow differentiation of increasing or decreasing helical or sheet secondary structural elements upon changes in environment. The ACS values increase with an increase in the total helical content and decrease with an increase in the total sheet content.
The statistical analysis of the correlation between ACS and SSC is relatively good for the 1Hα ACS values (84%), while a moderate correlation (67%) is obtained with the 1HN ACS values. As the number of proteins that can be added into the correlations of ACS with secondary structure increases, the correlation coefficients should improve significantly. However, certain factors may result in lowering the correlation coefficient. ACS values were based on the total number of cross-peaks that were observed, not on the total number of residues in the protein. For example, an 15N-HSQC spectrum will not contain resonances from a proline residue, which will consequently not be included in the ACS value, though it is present in the sequence. Significant contributions in lowering the correlation are expected from the residues that are present in the turns that will contribute to the ACS value as a sheet or helix. For example, residues that are part of a β-turn will be considered as β-sheet when the average values are calculated. The distribution of chemical shifts for each of the amino acids found in the BMRB database suggests that no particular amino acid dominates the ACS values; hence, the chemical shifts for a particular type of amino acid are not expected to bias the correlation. Moreover, Sharman et al. [95] have used rigorous statistical analyses of 1Hα chemical shifts to show that there is no correlation between amino acid type and propensity to fall within helical or sheet regions. However, it is possible that certain proteins will contain a large number of one type of residue (or a preponderance of a few types of residues) that may skew the ACS value. The relatively low correlation coefficients (0.64–0.8) for the ACS versus SSC correlations may result from these and other factors.
3.4. Applications of empirical correlations of ACS
3.4.1. Identification of the protein class from ACS
Rigorous statistical analysis of the data clearly suggests that only the 1Hα ACS values are capable of distinguishing the three different structural classes of the proteins. Based on the results of K-S test for 1Hα chemical shifts, it is possible to define the range of 1HαACS values corresponding to each class. For protein structural classes, α, αβ, and β, defined by SCOP, the centers of the ACS values are 3.83 ± 0.072, 3.94 ± 0.093, and 4.05 ±0.076 ppm, respectively. The corresponding values for the CATH-classified proteins are 3.79 ± 0.066, 3.93 ± 0.070, and 4.05 ± 0.086 ppm, respectively. Following this criteria, the results for a total of 37 proteins, predicted using both CATH- and SCOP-derived empirical relations, are summarized in Table 9. Only two proteins could be classified using the CATH-, but not the SCOP-based, relation (noted as NP, no prediction) and there is no cross prediction between α and β classes.
Table 9.
Prediction of structural class from NMR data for proteins of undetermined three-dimensional structure
| BMRBa | ACS (1Hα)b | Protein Name | Structural Class (using CATH-based correlation)c | Structural Class (using SCOP-based correlation)d |
|---|---|---|---|---|
| 4664 | 3.818 | Lipocalin Q83 | α | α |
| 4688 | 3.899 | L18 | αβ | α/αβ |
| 4698 | 3.846 | Transforming Growth Factor β type II receptor | α | α/αβ |
| 4722 | 3.823 | Shikimate Kinase | α | α |
| 4752 | 3.819 | Gpnu1-E68 | α | α |
| 4771 | 3.726 | Tola3 | α | NP |
| 4791 | 3.808 | HCV NS3 RNA helicase | α | α |
| 4792 | 3.778 | ParD dimer | α | α |
| 4829 | 3.841 | Interleukin enhancer binding factor | α | α |
| 4834 | 3.766 | S. aureus peptide deformylase | α | α |
| 4908 | 3.769 | α′-domain of ERp57 | α | α |
| 5014 | 3.724 | MyBP-C cC5 | α | NP |
| 5040 | 3.778 | I1(I29T) monomer | α | α |
| 5093 | 3.881 | RbfADelta25 | αβ | α/αβ |
| 5107 | 3.826 | Sensor & Substrate Binding Domain from Lon (La) Protease | α | α |
| 5316 | 3.781 | Gag | α | α |
| 4113 | 3.931 | Vaccinia Glutaredoxin-1 | αβ | αβ |
| 4132 | 4.015 | Human ubiquitin-conjugating enzyme | β | αβ/β |
| 4719 | 3.922 | Ras binding domain of rat AF6 | αβ | αβ |
| 4802 | 3.968 | N-terminal domain of H-NS | αβ/β | αβ |
| 4881 | 3.983 | Azotobacter vinelandii C69A holoflavodoxin II | αβ | αβ |
| 4901 | 3.991 | p62 N-terminal domain | αβ | αβ |
| 4940 | 3.933 | Antennal Specific Protein 1 | αβ | αβ |
| 4965 | 3.925 | L11 | αβ | αβ |
| 5030 | 3.937 | Honeybee antennal specific Protein 2 | αβ | αβ |
| 5093 | 3.881 | RbfADelta25 | αβ | αβ |
| 4302 | 4.010 | Protein disulfide isomerase a′ domain | β | αβ/β |
| 4720 | 4.066 | Inhibitor-2 monomer | β | β |
| 4870 | 4.094 | region 4.2 of sigma70 of E. coli RNA polymerase holoenzyme | β | β |
| 4881 | 3.983 | Azotobacter vinelandii C69A holoflavodoxin II | β | β |
| 4901 | 3.991 | p62 N-terminal domain | β | β |
| 4913 | 4.046 | cAMP-regulated phosphoprotein-19 monomer | β | β |
| 4929 | 4.090 | Tctex1 dimer | β | β |
| 4956 | 4.013 | YajQ from E. coli | β | αβ/β |
| 4973 | 4.100 | Saratin | β | β |
| 4999 | 3.979 | Nucleocapsid binding domain of the sendai virus phosphoprotein | β | β |
| 5049 | 4.053 | Extracellular domain of subunit 2 of the human receptor | β | β |
BioMagResBank (BMRB) accession number (http://www.bmrb.wisc.edu/),
Averaged chemical shift (ACS) calculated for the 1Hα nuclei,
Structural class estimation based on the empirical distribution obtained by CATH classification,
Structural class estimation based on the empirical distribution obtained by SCOP classification.
3.4.2. Estimation of secondary structure content from ACS
To determine the effectiveness of the ACS in estimating the SSC, we have used an independent set of proteins that are not part of the derived correlations. A set of 36 proteins obtained from the BMRB for which complete assignments of the backbone atoms are known, but the structures have not yet been determined, were used to estimate SSC by using the empirical correlation between SSC and 1Hα or 1HN ACS values. SSC was also calculated using the consensus chemical shift indices using the program PSSI using all the backbone atoms. In order to evaluate the secondary structure content for a set of proteins, the program Probability-based protein secondary structure identification (PSSI) was used [59] (discussed in section II.D.2). In this method, chemical shift indices (CSI) of the set of backbone atoms are used to define the probability with which the secondary structure (sheet or helix) is assigned. Secondary structure content in percentage is then calculated with respect to the total number of residues in the sequence. The list of all the proteins and their estimated SSC, using the correlation and CSI based methods are given in Table 10. There is an overall agreement between the SSC estimated between these two methods (Figure 10). Larger deviations were observed in the 1HN ACS values compared to the 1Hα ACS values. To compare the predictions from 1HN and 1Hα ACS values, Figure 11 shows the comparison. For example, the BMRB numbers 4391 (candoxin) [154] and 4393 (N-terminal domain of human spectrin including one structural domain) [155], are predicted to contain predominantly helical and sheet secondary structures, respectively. Figure 4 shows a comparison of the estimates of helical (left panel) and sheet (right panel) content for these proteins derived from either the 1HN or 1Hα ACS values Ideally, both ACS values should provide exactly the same values, within experimental error.
Table 10.
Estimated SSC using ACS and CSI based methods for proteins with no three-dimensional structural information.
| BMRB | Protein Name | Helix (%) from |
Sheet (%) from |
||||
|---|---|---|---|---|---|---|---|
| a ACS (1Hα) | a ACS (1HN) | b CSI | a ACS (1Hα) | a ACS (1HN) | b CSI | ||
| 4840 | Adenylate kinase | 15.31 | 12.65 | 12.70 | 38.11 | 34.58 | 20.40 |
| 4834 | S. aureus peptide deformylase | 25.56 | 18.69 | 21.70 | 25.54 | 20.70 | 36.50 |
| 4825 | Recombinant RC-RNase 2 | 27.28 | 18.45 | 15.10 | 25.70 | 19.60 | 39.60 |
| 4821 | DcuS | 17.62 | 15.87 | 20.40 | 33.70 | 30.37 | 19.10 |
| 4795 | Human D187N gelsolin domain 2 | 20.83 | 18.67 | 37.10 | 28.73 | 26.01 | 24.10 |
| 4794 | Human wild type gelsolin domain 2 | 25.69 | 19.45 | 20.70 | 25.06 | 20.62 | 35.30 |
| 4787 | Apical Membrane Antigen 1 | 26.10 | 12.95 | 13.10 | 29.25 | 20.36 | 21.30 |
| 4784 | Tyrosine repressor | n.a | 6.38 | 6.60 | n.a | 47.90 | 57.40 |
| 4776 | Sud dimer | 14.03 | 11.31 | 19.70 | 40.21 | 36.56 | 42.30 |
| 4771 | Tola3 | 8.18 | 4.57 | 21.70 | 50.72 | 45.64 | 36.80 |
| 4752 | gpnu1-E68 | 11.92 | 4.32 | 16.20 | 51.62 | 39.25 | 39.70 |
| 4735 | Olfactory marker protein | 23.33 | 12.27 | 21.50 | 29.68 | 22.13 | 41.10 |
| 4722 | Shikimate Kinase | 9.34 | 6.80 | 19.00 | 47.78 | 43.28 | 32.10 |
| 4716 | Auxilin | 3.53 | 2.78 | 7.70 | 54.01 | 52.33 | 39.00 |
| 4712 | Newt acidic FGF | 16.76 | 10.94 | 5.30 | 30.54 | 26.34 | 45.50 |
| 4711 | RNA-binding protein | n.a | 13.99 | 31.70 | n.a | 36.02 | 20.80 |
| 4698 | Transforming Growth Factor Beta type II receptor | n.a | 13.86 | 4.90 | n.a | 28.66 | 41.00 |
| 4688 | L18 | 22.46 | 14.66 | 32.40 | 34.98 | 23.48 | 31.50 |
| 4670 | PIN1At | 30.71 | 17.80 | 29.20 | 25.68 | 17.80 | 33.30 |
| 4664 | Lipocalin Q83 | 25.54 | 11.77 | 17.20 | 30.01 | 20.71 | 51.00 |
| 4579 | FYVE domain of EEA1 | n.a | 20.16 | 18.60 | n.a | 27.05 | 11.60 |
| 4567 | Catalytic domain of yUBC1 | 19.30 | 13.23 | 24.50 | 37.21 | 28.38 | 38.40 |
| 4558 | YopH-NT monomer | 17.48 | 12.64 | 25.70 | 38.13 | 31.21 | 37.50 |
| 4463 | Ras-binding domain of Byr2 | 21.74 | 18.02 | 31.90 | 30.37 | 23.95 | 33.60 |
| 4447 | p23fyp | 20.93 | 19.80 | 33.30 | 26.97 | 25.85 | 29.80 |
| 4353 | p13 C-terminal domain | 23.81 | 16.69 | 15.90 | 26.84 | 21.82 | 38.90 |
| 4335 | calerythrin | 5.88 | 2.21 | 11.40 | 54.90 | 48.68 | 60.20 |
| 4313 | E2 | 24.09 | 17.73 | 41.70 | 30.19 | 20.94 | 21.10 |
| 4294 | Human MBF1(57–148) core domain | 5.96 | 4.99 | 15.20 | 50.59 | 48.55 | 42.40 |
| 4271 | Calcium binding protein from Entamoeba Histolytica | 11.00 | 8.70 | 13.40 | 44.27 | 41.27 | 48.50 |
| 4239 | f29-SSB bacteriophage | 23.69 | 14.46 | 15.30 | 28.27 | 21.90 | 43.50 |
| 4147 | Cold Shock domain | 14.95 | 6.93 | 8.90 | 32.65 | 27.95 | 44.30 |
| 4136 | E. coli multidrug resistance protein E | n.a | 3.46 | 1.80 | n.a | 52.96 | 68.20 |
| 4132 | Human ubiquitin-conjugating enzyme | 18.15 | 12.68 | 19.90 | 38.07 | 30.17 | 33.10 |
| 4027 | S.aureus DHFR(F98Y)-NADPH-TMP ternary complex | 17.36 | 17.00 | 22.20 | 26.64 | 25.96 | 44.90 |
| 1583 | Micrococcal nuclease | n.a | 20.59 | 7.30 | n.a | 23.89 | 48.90 |
Figure 10.
Comparison of helical and sheet content percent calculated using 1Hα or 1HN ACS values to that obtained using a consensus chemical shift index based method for a set of proteins for which no three dimensional structures are available. (a) and (b) correspond to the helical content using the 1Hα and 1HN ACS values, respectively, while (c) and (d) are the corresponding sheet content using the same ACS values. The dashed lines correspond to a perfect correlation between these two methods.
Figure 11.
Comparison of helical (LEFT) and sheet (RIGHT) content calculated using ACSESS with either 1Hα or 1HN ACS values for a set of proteins for which no three dimensional structure are available. The numbers for two proteins are their identification codes in the BMRB database (see Table 10).
3.4.3. Averaged chemical shifts and protein folding
The utility of ACSESS as a tool to identify what structural changes occur in proteins under denaturing conditions has been demonstrated for ubiquitin. Chemical shift data acquired under a variety of conditions are available for this protein (see Table 11). Ubiquitin belongs to the αβ class according to CATH, and both chemical shift information and structures are available for three variants: multiple mutant (BMRB 4663, pdb 1C3T) [156], yeast (4769, 1UBI) [157] and core mutant (4493, 1UD7) [158]. Chemical shift information for the denatured state (BMRB 4375) is also available [159]. ACSESS- predicted sheet and helix content obtained using both 1HN and 1Hα ACS values from the folded forms of ubiquitin are in close agreement with secondary structure estimates obtained from their three-dimensional structures using PROMOTIF (Table 11). In the case of denatured ubiquitin, the ACSESS method estimates a loss of helical structure of approximately 6% and a gain in sheet content of the same amount, suggesting that even in the denatured state, significant residual secondary structure is present in ubiquitin.
Table 11.
Secondary structure of ubiquitin predicted by ACSESS
| a BMRB | State | b ACS (ppm) | c ACSESS (%) | d PDB | e PROMOTIF (%) | ||
|---|---|---|---|---|---|---|---|
| Helical | Sheet | Helical | Sheet | ||||
| 4663 | Mutant, multiple | 8.38 (1HN) | 22.3±1.6 | 23.0±1.6 | 1C3T | 27.6 | 21.1 |
| 4.29 (1Hα) | 24.9±1.4 | 20.4±1.0 | |||||
| 4769 | Yeast | 8.37 (1HN) | 21.8±1.6 | 23.9±1.7 | 1UBI | 31.6 | 23.7 |
| 4.54 (1Hα) | 27.6±1.5 | 16.2±0.8 | |||||
| 4493 | Core Mutant | 8.40 (1HN) | 23.7±1.7 | 20.9±1.5 | 1UD7 | 27.6 | 23.7 |
| 4.48 (1Hα) | 23.7±1.1 | 22.3±1.1 | |||||
| 4375 | denatured | 8.34 (1HN) | 20.4±1.5 | 26.0±1.8 | none | ||
| 4.37 (1Hα) | 16.7±0.9 | 33.1±1.7 | |||||
BMRB id number from http://www.bmrb.wisc.edu/
ACS: Averaged chemical shift calculated form the BMRB.
ACSESS: Averaged chemical shift to estimate secondary structure
PDB: Protein data back structure coordinate files from www.rcsb.org.
PROMOTIF estimation of secondary structure content from 3D structure.
We have demonstrated that ACSESS provides information about the denatured state of ubiquitin. Our results show that ubiquitin retains significant residual helical and sheet structure in denaturing solvents, and that the β-strand content increases relative to that of the folded state. This increase in sheet content can be attributed to the presence of additional turns in an extended conformation. The retention of helical structure, though reduced, could be due to retention of local secondary structural elements that are no longer folded into a three-dimensional conformation. This idea is consistent with the original paper by Peti et al. [159] that reported the chemical shifts of denatured ubiquitin, and compared them with chemical shifts of other denatured proteins, by assuming that all interactions in the unfolded state are local.
3.5. Some important aspects of using ACS to obtain low-resolution structural information
NMR spectroscopy plays a vital role in determining the structures of proteins in the solution state. In spite of advancement in the field during the past decade, determining the complete three-dimensional structure of any given protein remains a time-consuming proposition. Though the information content of a complete structure at atomic resolution is indisputable, in recent times several groups have begun exploring alternative methods that are faster than conventional experiments [160], [161]. Prior to collecting several days’ worth of NMR spectra for structure determination, other biophysical methods are generally adopted to infer secondary structural information about the protein of interest. In particular, circular dichroism (CD) spectroscopy is extensively used to estimate the secondary structure content of medium-sized proteins. In CD spectroscopy, deconvolution of the experimental molar ellipticity at 222 nm is used to estimate secondary structure content. In the case of NMR, chemical shifts have been used as regular indicators of a particular secondary structure. For example, an 1Hα resonance that is shifted upfield with respect to the corresponding random coil value is considered to be α-helical, while one shifted downfield to be β-strand. This is a widely accepted procedure, and a large number of NMR studies have shown that such correlation is valid [5], [17]. However, NMR spectral information has seldom been used to obtain relatively low-resolution structural information, such as secondary structure content. In some cases, the results of CD are used to determine whether it is feasible to obtain complete, three-dimensional structural information for a particular protein, using NMR. This suggests the critical importance of evaluating whether data obtained from NMR itself can be used to estimate secondary structure content. Lee and Cao have addressed this question extensively in their comprehensive study [162], and have shown that the correlation between NMR- and CD-based secondary structure estimation is poor. Further, while CD spectroscopy is more suitable for studying relatively small proteins and polypeptides, the characterization of larger molecules requires NMR.
Computational methods often play a primary role in initial predictions of protein structure; for example, in predictions of protein structural class. These methods are typically invoked even before a protein is expressed or extracted for any biophysical characterization. Secondary structure estimations from CD are often inconsistent with such computational predictions from NMR. On the other hand, to date, estimations from NMR have required the time-consuming process of resonance assignment. A method such as that proposed here could essentially fulfill the need for an empirical, NMR-based estimator of protein structural class that is both accurate and efficient.
The results discussed above show that 1Hα and 1HN ACS values clearly distinguish the three different protein classes, α, mixed αβ, and β, when the proteins are classified either by CATH or SCOP, and can be used in estimating secondary structure content. The empirical correlations provide a way to determine directly the structural information of proteins in the absence of resonance assignments. They can be easily incorporated into any commercial or academic software package that employs manual or automated peak picking routines to reduce an HSQC spectrum into a single ACS value. ACS is expressed in the same unit as chemical shift (ppm). Instead of using the absolute chemical shift values to determine the averages, we have also explored definitions such as chemical shift index (CSI), which determines the relative change in the chemical shift with respect to the corresponding random coil value. CSI may better distinguish proteins that are comprised primarily of either helices or sheets; αβ proteins cannot be identified by this method, because the values of α and β segments are opposite in sign, and therefore cancel each other.
Determination of the structural classes of proteins with no available experimental three-dimensional structure information (from NMR or X-ray), using 1Hα ACS values, provides an internal test of the reliability factor (Tables 9–11). The secondary structures of these proteins were also estimated using prediction algorithms that utilize only amino acid sequences. For many of the proteins, the sequence-based class prediction approach provided similar results for the mainly-α class, while larger differences were observed for mainly-β class proteins. However, considering the variability and confidence limits associated with such predictions (http://cubic.bioc.columbia.edu/eva/ and references therein), it is difficult to define a suitable control set for comparison. In some cases, using the sequence-based prediction method (http://www.bork.embl-heidelberg.de/SSCP/) [130], [131], we have observed large variations in the estimation of sheet and helical classes for the same amino acid sequence (data not shown).
In general, the quality of structural predictions based on specific algorithms is examined either by redistribution test or jack-knife test [163]. However, in the correlations presented here, neither of these methods was considered, for the following reasons. First, our methods are not algorithm-based; our results are strictly the outcome of an empirical correlation between known protein structural classes or SSC and averaged chemical shifts. Second, in self-consistency tests [163], it is necessary to define a training set of proteins that obey a particular criterion; for example, the resolution of three-dimensional structure. Though it is possible to define such criteria for protein classes, use of chemical shift information as the test criterion must be considered premature, as there is currently no consensus definition of the “accuracy” of such information [19].
Although we have shown that ACS values can be used to identify directly the structural classes or SSC of proteins, thereby providing a first, low-resolution structural estimate from experiments, critical questions still remain. For example, what is the reliability of the estimates? As the number of proteins that we add into our correlations of ACS with protein class or SSC increases, one can expect the reliability of the method to improve. In the empirical correlation derived between secondary structure content and ACS values, we have determined a reliability factor > 84% when 1Hα nuclei are used. Notwithstanding the limited number of proteins in the current study, and that we have defined the relative regions of ACS values demarcating the structural classes in a conservative manner, we suggest the reliability of this method is about 80%.
Another remaining question is whether it is possible that certain amino acids bias the current estimates, since the method is based on an average of the chemical shifts. The distribution of chemical shifts for each of the amino acids found in the BMRB database suggests that no particular amino acid dominates the ACS values. In a recent paper, Sharman et al. [95] have used rigorous statistical analyses of 1Hα chemical shifts to show that there is no correlation between amino acid type and propensity to fall within helical or sheet regions. The exact nature of the chemical shift dependence on secondary structure for a specific amino acid residue remains to be determined [95], [164]. In addition, long range and context-dependent effects on protein structural class definition are still not clearly understood [164], and may also play important roles in influencing chemical shifts.
As the estimation of structural class from NMR is directly influenced by the quality of the data used, the method is most useful in cases in which the resolution of the corresponding HSQC spectrum is excellent. Experiments based on transverse relaxation optimized spectroscopy (TROSY) [165] provide an additional advantage in applicability to large proteins. From a practical point of view, the method would be most appropriate if a sufficient number of individual cross-peaks are observed in an HSQC spectrum. Further, since calculated ACS values are based on the total number of residues in a protein, and not on the total number of crosspeaks observed, we recommend that a minimum of 70% of the total number of peaks expected be present in a given spectrum for determination of a reliable ACS value. As a final point, all amino acid residues have 1Hα resonances (glycine has two), so these will be fully represented in any calculation of the 1Hα ACS. In contrast, proline residues lack an amide proton resonance, and consequently are not observed in 15N-HSQC spectra; an abundance of proline-rich proteins in a data set could conceivably lead to an underestimate of amide ACS values.
It must be emphasized that ACS-based methods do not provide an alternative to conventional NMR-based experiments, and should only be considered initial predictors of protein class or secondary structure content. ACS methods might provide a novel technique for monitoring protein structural changes in real time, such as in protein folding experiments. Such methods might also be used to detect major structural changes that occur upon protein-protein, protein-DNA/RNA, and other complex formations, to provide some direct experimental structural information in situations in which other techniques are incapable of doing so (e.g., in studies of large and/or highly disordered proteins), and to facilitate initial protein fold identification in high throughput proteomics applications.
4. Other empirical correlations of chemical shift
Methods to elucidate empirical relationships between chemical shift and protein structure have been under development for decades. Examples include magnetic anisotropy [166] and methods that investigate electrostatic [167] and aromatic ring current effects [168]. In addition to methods focused on estimating the secondary structure of individual residues from a secondary structure index, and secondary structure content or structural classes from chemical shifts, a few empirical correlations have been developed to address specific features of protein structure, such as the redox state of cystines [11], [169] and Xaa-Pro peptide bond conformations [170]. In this section, we briefly review some of these methods.
4.1. Semi-empirical methods for chemical shift estimation from 3D structure
One of the earliest methods extensively to use the empirical relationship between NMR chemical shifts and protein structure is TALOS, developed by Cornilescu et al., [5]. This method is based on the observation that homologous proteins have similar secondary chemical shifts, because these correlate with local protein conformation. This relation provides a basis for searching a database for triplets of adjacent residues with secondary chemical shifts and sequence similarity that provide the best match to the query triplet of interest. Tests carried out using proteins of known structure indicate that the root-mean-square difference (rmsd) between the output of TALOS and the X-ray derived backbone angles is about 15°, and has an error rate of ~3%. TALOS is freely available (http://spin.niddk.nih.gov/bax/software/TALOS/index.html).
A range of semi-empirical methods is available to predict protein chemical shifts from three-dimensional structure and dynamics. These include: (a) SHIFTS: The first version of this program was developed by Case and co-workers [171], and has seen several subsequent improvements [18], [19], [171], [172], including a most recent version [173]. SHIFTS is available from the authors’ group page (http://www.scripps.edu/mb/case/qshifts/qshifts.htm). (b) SHIFTCALC: this method was developed by Williamson and his group, with details presented in a number of papers in the 1990s [3], [4], [174], [175], [176], [177], [178], [179]. Source code and a web-server for SHIFTCALC are available (http://nmr.group.shef.ac.uk/NMR/mainpage.html). (c) SHIFTX and SHIFTY: Wishart s group provides a wide range of software tools for correlating chemical shift with protein structure. These include: SHIFTX to predict 1H, 13C, and 15N protein chemical shifts from 3D structure [180], [181] and SHIFTY to predict protein chemical shifts using only amino acid sequence [182]. In particular, Neal et al. [180], [181] have shown that accuracy for predicting chemical shifts (including amide proton shifts) can be improved by combining empirical formulas for spatial interactions with ‘hyper-surfaces’ representing local covalent interactions. (d) PROSHIFT: this neural network-based method, developed by Meiler [183], predicts the 1H, 13C, and 15N chemical shifts of proteins from their three-dimensional structure as a function experimental conditions as input parameters. A webserver for PROSHIFT is available from Meiler’s group (http://www.meilerlab.org/view.php). A more recent program, Random Coil Index (RCI), predicts protein flexibility from backbone chemical shifts (13Cα, 13CO, 13Cβ, 15N, 1Hα), and estimates values of model-free order parameters as well as per-residue RMSDs of NMR and MD ensembles [184], [185]. All these programs are either available to download or on a webserver at Wishart’s group (http://redpoll.pharmacy.ualberta.ca/).
Figure 12 shows a straightforward comparison of the experimental chemical shifts of protein G (BMRB 5875), represented by filled circles, with chemical shifts calculated using SHIFTX (open circles), SHIFTS (filled triangles) and PROSHIFT (open triangles). A dashed line connects the experimental points to show a visual trend. Panels (a), (b), (c) and (d) show plots of chemical shift values of the nuclei, 1HN, 15N, 13Cα and 1Hα, respectively. We were unable to perform a similar calculation with SHIFTCALC due to technical issues with the webserver. Overall, we find that all the calculations follow the experimental values, but exhibit differences with respect to specific nuclei and residues. This area sees continuing development by several groups in recent time [173], [186].
Figure 12.
Comparison of semi-empirical methods to calculate the chemical shifts of an example proteins, protein G. Panels (a), (b), (c) and (d) show the plots of chemical shifts values of the nuclei, 1HN, 15N, 13Cα and 1Hα, respectively. Experimental chemical shifts of protein G (bmr5875) in filled circles to that chemical shifts calculated using SHIFTX (open circles), SHIFTS (filled triangles) and PROSHIFT (open triangles). A dashed line is connected through a experimental points to show a visual trend.
4.2. Prediction of redox states of cysteines from chemical shifts
Disulfide bonds play a pivotal role in protein structure, function, folding, and stability. The importance of disulfide bonds has been extensively studied, but invariably involves either breaking or forming a disulfide bond. Further, it is not the disulfide bond but the effect of the disulfide bond on the rest of the structure that has been studied. Two groups have developed empirical relationship to predict the redox state; Sharma and Rajarathnam provided the first such correlation [11] and recently Wang et al., [63], [169] have developed a two-dimensional cluster approach for a similar purpose.
These results in general show that that the Cβ shift is extremely sensitive to the redox state, and can predict the disulfide-bonded state. Further, chemical shifts in both states occupy distinct groups in a XY plots of Cα, Cβ chemical shifts. The redox state chemical shifts of cysteines also sensitive to the secondary structural state of the protein. The results of Sharma and Rajarathnam are summarized in Table 12. The rules to define the empirical state with confirmed chemical shifts assignments are given in the original reference [11]. Wang et al., [169] have performed a two-dimensional cluster analysis, while the earlier method looked only Cα, Cβ correlations. This analysis showed that different clusters of (Cα, Cβ), (C′, Cβ), (HN, Cβ) and (Hα, Cβ) are helpful in distinguishing the redox state of cysteine residues. Similar to the first approach, the authors derived rules using a score matrix to predict the redox state of cysteines using their chemical shifts. The score matrix predicts the redox state of cysteine residues in proteins with 90% accuracy. Table 12 also lists the summary of the results of the most sensitive nuclei for redox state, Cβ. Table 12 shows that the results from the two methods are similar.
Table 12. Distribution of Cβ chemical shifts in oxidized and reduced cysteinea.
| Method Ia |
Method IIb |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cβ (S-S) Oxidized |
Cβ (C-H) Reduced |
Cβ (S-S) Oxidized |
Cβ (C-H) Reduced |
|||||
| α-helix | β-strand | α-helix | β-strand | α-helix | β-strand | α-helix | β-strand | |
| Number of chemical shifts | 33 | 49 | 39 | 35 | 67 | 79 | 90 | 86 |
| Minimum (ppm) | 32.8 | 35.9 | 23.8 | 25.1 | 32.6 | 34.5 | 24.1 | 25.9 |
| Maximum (ppm) | 47.4 | 50.9 | 28.8 | 33.3 | 44.6 | 51.2 | 43.7 | 35.1 |
| Range (ppm) | 14.6 | 15.0 | 5.0 | 8.2 | 12.0 | 16.7 | 19.6 | 9.2 |
| Median (ppm) | 38.8 | 43.2 | 26.6 | 30.2 | 38.9 | 43.0 | 27.0 | 30.4 |
| Mean (ppm) | 38.4 | 43.0 | 26.5 | 29.7 | 38.6 | 43.0 | 27.6 | 30.5 |
| Std. Dev (ppm) | 3.2 | 4.2 | 1.1 | 2.0 | 3.2 | 4.1 | 2.7 | 2.1 |
4.3. Prediction of Xaa-Pro peptide bond conformations
In peptides and proteins, the planar peptide bond occurs predominantly in the trans conformation [187]. In general the cis form is energetically less favorable due to the steric repulsion of the Cα/Hα atoms of the two sequential amino acids. However, in peptide bonds preceding prolines (Xaa-Pro), the Cδ/Hδ in the pyrolidine ring and the Cα/Hα atoms of the preceding residue experience a comparable repulsion and the energy difference between the cis and the trans conformation is reduced. Therefore an appreciable fraction of the Xaa-Pro peptide bonds occur in the cis form. A survey of a non-redundant database of 571 high resolution protein structures found 5.2% of all Xaa-Pro peptide bonds occur in the cis conformation, as compared to only 0.03% of all Xaa-nonPro peptide bonds [188], [189]. Earlier studies on small peptides containing prolines observed signature features of the cis conformation include an upfield change in the 13Cγ chemical shift and a downfield change in the 13Cβ chemical shift [190]. Therefore, the chemical shift difference between them, Δβγ… (δ [13Cβ]− δ[13Cγ]) is expected to be an indicator for cis or trans conformation [191]. These observations lead Schubert et al [170] to develop a chemical shift based empirical relationship. This method, also referred as POP (Prediction of Proline) conformation 304 protein entries in the BMRB, representing an overall number of 1033 prolines for the analysis.
The chemical shift difference Δβγ is a reference-independent indicator of the Xaa-Pro peptide bond conformation. Based on a statistical analysis of the 13C chemical shifts, a software tool was created to predict the probabilities for cis or transconformations of Xaa-Pro peptide bonds. Using this approach, the conformation at a given Xaa-Pro bond can be identified in a simple NOE-independent way immediately after obtaining its NMR resonance assignments. Table 13 lists of the results of the analysis [170]. Distribution of Δβγ were fitted a single Gaussian and the fitted parameters (average, variance and standard deviation) are used for the prediction (also listed in Table 13)). For Δβγ ιn the range from 0.0 ppm to 4.8 ppm the peptide bond conformation is predicted to be 100% trans, whereas from 9.15 ppm to 14.4 ppm it is 100% cis. In the range from 4.8 ppm to 9.15 ppm, the prediction is ambiguous and only probabilities can be given for both conformers and the results must be confirmed using the conventional NOE-based method [21].
Table 13.
Summary of the empirical analysis for the prediction of Xaa-Pro peptide bond conformationa
| Cβ resonances (ppm) |
Cγ resonances (ppm) |
Δβγ (ppm) |
||||
|---|---|---|---|---|---|---|
| cis | trans | cis | trans | cis | trans | |
| Average Value | 31.75 | 34.16 | 27.26 | 24.52 | 4.51 | 9.64 |
| Minimum | 26.3 | 30.74 | 19.31 | 22.1 | ||
| Maximum | 35.83 | 36.23 | 33.39 | 27.01 | ||
| Standard Deviation | 0.98 | 1.15 | 1.05 | 1.09 | 1.17 | 1.27 |
Table adopted from [170].
5. Summary
Progress in the structural biology of proteins comes from both experimental and theoretical efforts. Computational methods are capable of delivering fast structural information, ranging from low-resolution protein structural class definition to high-quality information based on homology modeling. Experimental methods that concentrate on obtaining high-resolution information are hampered by inherent time cost, and lack the capacity to provide low-resolution structural information expediently. NMR spectroscopy is a powerful tool for obtaining high-resolution structural and dynamical details of molecules in the solution state. In order to explore new experimental methods for the fast identification of protein structures using NMR, we have presented the degree to which chemical shifts of a particular nuclear species in the protein backbone can be used as a low-resolution structural parameter that correlates with a variety of protein structural parameters.
Acknowledgments
Thanks to Anaika Sibley and Dr. Monique Cosman for some of the initial contributions on the project. This work funded in part by Student Employee Graduate Research Fellowship (SEGRF) for SPM and NIH Grant #GM077520 for VVK.
Glossary of abbreviations
- ACS
Averaged Chemical Shift
- ACSESS
Averaged chemical shifts to secondary structure
- BMRB
BioMagResBank
- CATH
Class, Architecture, topology and homologous super family
- CD
Circular Dichroism
- COSY
Correlated Spectroscopy
- CSI
Chemical shift index
- CSP
Chemical shift pattern
- DEFINE
Determine the secondary and first level supersecondary structure
- DSSP
Database of secondary structure Predictions
- HSQC
Heteronucelar single quantum correlation
- K-S test
Kolmogorov-Smirnov test
- PDB
Protein data bank
- POP
Prediction of Proline
- RCSB
Research collaboratory for structural bioinformatics
- RefDB
Referenced database
- SCOP
Structural classification of proteins
- SSC
Secondary structure content
- STRIDE
Secondary structure assignment from atomic coordinates
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Proctor WG, Yu FC. The dependence of a nuclear magnetic resonance frequency upon chemical compound. Phy Rev. 1950;77:717. [Google Scholar]
- 2.Arnold JT, DSS, Packard ME. Chemical effects on nuclear-induction signals from organic compounds. J Chem Phys. 1951;19:507. [Google Scholar]
- 3.Asakura T, Iwadate M, Demura M, Williamson MP. Structural analysis of silk with C-13 NMR chemical shift contour plots. Int J Biol Macromol. 1999;24:167–171. doi: 10.1016/s0141-8130(98)00082-8. [DOI] [PubMed] [Google Scholar]
- 4.Asakura T, Taoka K, Demura M, Williamson MP. The Relationship Between Amide Proton Chemical Shifts and Secondary Structure in Proteins. J Biomol NMR. 1995;6:227–236. doi: 10.1007/BF00197804. [DOI] [PubMed] [Google Scholar]
- 5.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 6.Dalgarno DC, Levine BA, Williams RJ. Structural information from NMR secondary chemical shifts of peptide alpha C-H protons in proteins. Biosci Rep. 1983;3:443–52. doi: 10.1007/BF01121955. [DOI] [PubMed] [Google Scholar]
- 7.Laws DD, Dedios AC, Oldfield E. NMR Chemical Shifts and Structure Refinement in Proteins. J Biomol NMR. 1993;3:607–612. doi: 10.1007/BF00174614. [DOI] [PubMed] [Google Scholar]
- 8.Oldfield E. Chemical Shifts and Three-Dimensional Protein Structures. J Biomol NMR. 1995;5:217–225. doi: 10.1007/BF00211749. [DOI] [PubMed] [Google Scholar]
- 9.Osapay K, Case DA. A New Analysis of Proton Chemical Shifts in Proteins. J Am Chem Soc. 1991;113:9436–9444. [Google Scholar]
- 10.Pastore A, Saudek V. The Relationship Between Chemical Shift and Secondary Structure in Proteins. J Magn Reson. 1990;90:165–176. [Google Scholar]
- 11.Sharma D, Rajarathnam K. C-13 NMR chemical shifts can predict disulfide bond formation. J Biomol NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 12.Williamson MP. Secondary-Structure Dependent Chemical Shifts in Proteins. Biopolymers. 1990;29:1428–1431. doi: 10.1002/bip.360291009. [DOI] [PubMed] [Google Scholar]
- 13.Wishart DS, Sykes BD, Richards FM. Simple Techniques For the Quantification of Protein Secondary Structure By H-1 NMR Spectroscopy. FEBD Lett. 1991;293:72–80. doi: 10.1016/0014-5793(91)81155-2. [DOI] [PubMed] [Google Scholar]
- 14.Szilagyi L. Chemical Shifts in Proteins Come of Age. Prog Nucl Magn Reson Spectros. 1995;27:325–443. [Google Scholar]
- 15.Ando I, Kuroki S, Kurosu H, Yamanobe T. NMR chemical shift calculations and structural characterizations of polymers. Prog Nucl Magn Reson Spectros. 2001;39:79–133. [Google Scholar]
- 16.Case DA. The use of chemical shifts and their anisotropies in biomolecular structure determination. Curr Opin Struct Biol. 1998;8:624–630. doi: 10.1016/s0959-440x(98)80155-3. [DOI] [PubMed] [Google Scholar]
- 17.Case DA. Interpretation of chemical shifts and coupling constants in macromolecules. Curr Opin Struct Biol. 2000;10:197–203. doi: 10.1016/s0959-440x(00)00068-3. [DOI] [PubMed] [Google Scholar]
- 18.Case DA, Dyson HJ, Wright PE. Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies of peptides and proteins. Meth Enzym. 1994;239:392–416. doi: 10.1016/s0076-6879(94)39015-0. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Meth Enzym. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Nip AM. Protein chemical shift analysis: a practical guide. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 1998;76:153–163. doi: 10.1139/bcb-76-2-3-153. [DOI] [PubMed] [Google Scholar]
- 21.Wüthrich K. NMR of proteins and nucleic acids. Wiley; New York: 1986. [Google Scholar]
- 22.Bailey-Kellogg C, Widge A, Kelley JJ, Berardi MJ, Bushweller JH, Donald BR. The NOESY JIGSAW: Automated protein secondary structure and main-chain assignment from sparse, unassigned NMR data. J Comput Biology. 2000;7:537–558. doi: 10.1089/106652700750050934. [DOI] [PubMed] [Google Scholar]
- 23.Croft D, Kemmink J, Neidig KP, Oschkinat H. Tools for the automated assignment of high-resolution three-dimensional protein NMR spectra based on pattern recognition techniques. J Biomol NMR. 1997;10:207–219. doi: 10.1023/A:1018329420659. [DOI] [PubMed] [Google Scholar]
- 24.Koradi R, Billeter M, Engeli M, Guntert P, Wuthrich K. Automated peak picking and peak integration in macromolecular NMR spectra using AUTOPSY. J Magn Reson. 1998;135:288–297. doi: 10.1006/jmre.1998.1570. [DOI] [PubMed] [Google Scholar]
- 25.Li KB, Sanctuary BC. Automated resonance assignment of proteins using heteronuclear 3D NMR. Backbone spin systems extraction and creation of polypeptides. J Chem Inform Comput Sci. 1997;37:359–366. doi: 10.1021/ci960372k. [DOI] [PubMed] [Google Scholar]
- 26.Li KB, Sanctuary BC. Automated resonance assignment of proteins using heteronuclear 3D NMR.2. Side chain and sequence-specific assignment. J Chem Inform Comput Sci. 1997;37:467–477. doi: 10.1021/ci960372k. [DOI] [PubMed] [Google Scholar]
- 27.Moseley HNB, Montelione GT. Automated analysis of NMR assignments and structures for proteins. Curr Opin Struct Biol. 1999;9:635–642. doi: 10.1016/s0959-440x(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 28.Shimotakahara S, Rios CB, Laity JH, Zimmerman DE, Scheraga HA, Montelione GT. NMR structural analysis of an analog of an intermediate formed in the rate-determining step of one pathway in the oxidative folding of bovine pancreatic ribonuclease A: Automated analysis of H-1, C-13, and N-15 resonance assignments for wild-type and [C65S, C72S] mutant forms. Biochemistry. 1997;36:6915–6929. doi: 10.1021/bi963024k. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman DE, Montelione GT. Automated Analysis of Nuclear Magnetic Resonance Assignments For Proteins. Curr Opin Struct Biol. 1995;5:664–673. doi: 10.1016/0959-440x(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 30.Ab E, Atkinson AR, Banci L, Bertini I, Ciofi-Baffoni S, Brunner K, Diercks T, Dotsch V, Engelke F, Folkers GE, Griesinger C, Gronwald W, Gunther U, Habeck M, de Jong RN, Kalbitzer HR, Kieffer B, Leeflang BR, Loss S, Luchinat C, Marquardsen T, Moskau D, Neidig KP, Nilges M, Piccioli M, Pierattelli R, Rieping W, Schippmann T, Schwalbe H, Trave G, Trenner J, Wohnert J, Zweckstetter M, Kaptein R. NMR in the SPINE Structural Proteomics project. Acta Crystallogr D Biol Crystallogr. 2006;62:1150–61. doi: 10.1107/S0907444906032070. [DOI] [PubMed] [Google Scholar]
- 31.Yee A, Gutmanas A, Arrowsmith CH. Solution NMR in structural genomics. Curr Opin Struct Biol. 2006;16:611–7. doi: 10.1016/j.sbi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Taylor WR. A ‘periodic table’ for protein structures. Nature. 2002;416:657–60. doi: 10.1038/416657a. [DOI] [PubMed] [Google Scholar]
- 33.Chou KC. Progress in protein structural class prediction and its impact to bioinformatics and proteomics. Curr Protein Pept Sci. 2005;6:423–36. doi: 10.2174/138920305774329368. [DOI] [PubMed] [Google Scholar]
- 34.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucl Acid Res. 2007;35:D301–3. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nat Struct Biol. 2000;7(Suppl):957–9. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 36.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, Green RK, Flippen-Anderson JL, Westbrook J, Berman HM, Bourne PE. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucl Acid Res. 2005;33:D233–7. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucl Acid Res. 2006;34:D302–5. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J Biomol NMR. 1991;1:217–36. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- 39.Laws DD, de Dios AC, Oldfield E. NMR chemical shifts and structure refinement in proteins. J Biomol NMR. 1993;3:607–12. doi: 10.1007/BF00174614. [DOI] [PubMed] [Google Scholar]
- 40.de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260:1491–6. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 41.deDios AC. Ab initio calculations of the NMR chemical shift. Prog Nucl Magn Reson Spectros. 1996;29:229–278. [Google Scholar]
- 42.Vila JA, Scheraga HA. Factors affecting the use of (13)C(alpha) chemical shifts to determine, refine, and validate protein structures. Proteins. 2007 doi: 10.1002/prot.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A. 2007;104:9615–20. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vila JA, Arnautova YA, Scheraga HA. Use of 13C(alpha) chemical shifts for accurate determination of beta-sheet structures in solution. Proc Natl Acad Sci U S A. 2008;105:1891–6. doi: 10.1073/pnas.0711022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–90. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardi A, Wagner G, Wuthrich K. Protein conformation and proton nuclear-magnetic-resonance chemical shifts. Eur J Biochem. 1983;137:445–54. doi: 10.1111/j.1432-1033.1983.tb07848.x. [DOI] [PubMed] [Google Scholar]
- 47.Markley JL, Meadows DH, Jardetzky O. Nuclear magnetic resonance studies of helix-coil transitions in polyamino acids. J Mol Biol. 1967;27:25–40. doi: 10.1016/0022-2836(67)90349-x. [DOI] [PubMed] [Google Scholar]
- 48.Delepierre M, Dobson CM, Poulsen FM. Studies of beta-sheet structure in lysozyme by proton nuclear magnetic resonance. Assignments and analysis of spin-spin coupling constants. Biochemistry. 1982;21:4756–61. doi: 10.1021/bi00262a036. [DOI] [PubMed] [Google Scholar]
- 49.Rico M, Nieto JL, Santoro J, Bermejo FJ, Herranz J, Gallego E. Low-temperature 1H-NMR evidence of the folding of isolated ribonuclease S-peptide. FEBS Lett. 1983;162:314–9. doi: 10.1016/0014-5793(83)80779-0. [DOI] [PubMed] [Google Scholar]
- 50.Zuiderweg ER, Kaptein R, Wuthrich K. Sequence-specific resonance assignments in the 1H nuclear-magnetic-resonance spectrum of the lac repressor DNA-binding domain 1–51 from Escherichia coli by two-dimensional spectroscopy. Eur J Biochem. 1983;137:279–92. doi: 10.1111/j.1432-1033.1983.tb07827.x. [DOI] [PubMed] [Google Scholar]
- 51.Jimenez MA, Nieto JL, Herranz J, Rico M, Santoro J. 1H NMR and CD evidence of the folding of the isolated ribonuclease 50–61 fragment. FEBS Lett. 1987;221:320–4. doi: 10.1016/0014-5793(87)80948-1. [DOI] [PubMed] [Google Scholar]
- 52.Hutchinson EG, Thornton JM. Promotif - a Program to Identify and Analyze Structural Motifs in Proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreau VH, Valente AP, Almeida FCL. Prediction of the amount of secondary structure of proteins using unassigned NMR spectra: A tool for target selection in structural proteomics. Genetics and Molecular Biology. 2006;29:762–770. [Google Scholar]
- 54.Wishart DS, Sykes BD, Richards FM. The Chemical Shift Index - a Fast and Simple Method For the Assignment of Protein Secondary Structure Through NMR Spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 55.Wishart DS, Sykes BD. The C-13 Chemical-Shift Index - a Simple Method For the Identification of Protein Secondary Structure Using C-13 Chemical-Shift Data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 56.Wishart DS, Sykes BD. Chemical shifts a tool for structure determination. Meth Enzym. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 57.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 58.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:605–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 59.Wang YJ, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung LH, Samudrala R. Accurate and automated classification of protein secondary structure with PsiCSI. Protein Sci. 2003;12:288–95. doi: 10.1110/ps.0222303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labudde D, Leitner D, Kruger M, Oschkinat H. Prediction algorithm for amino acid types with their secondary structure in proteins (PLATON) using chemical shifts. J Biomol NMR. 2003;25:41–53. doi: 10.1023/a:1021952400388. [DOI] [PubMed] [Google Scholar]
- 62.Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 63.Wang CC, Chen JH, Lai WC, Chuang WJ. 2DCSi: identification of protein secondary structure and redox state using 2D cluster analysis of NMR chemical shifts. J Biomol NMR. 2007;38:57–63. doi: 10.1007/s10858-007-9146-x. [DOI] [PubMed] [Google Scholar]
- 64.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 65.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 66.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 67.Achari A, Hale SP, Howard AJ, Clore GM, Gronenborn AM, Hardman KD, Whitlow M. 1.67-A X-ray structure of the B2 immunoglobulin-binding domain of streptococcal protein G and comparison to the NMR structure of the B1 domain. Biochemistry. 1992;31:10449–57. doi: 10.1021/bi00158a006. [DOI] [PubMed] [Google Scholar]
- 68.Frank MK, Clore GM, Gronenborn AM. Structural and dynamic characterization of the urea denatured state of the immunoglobulin binding domain of streptococcal protein G by multidimensional heteronuclear NMR spectroscopy. Protein Sci. 1995;4:2605–15. doi: 10.1002/pro.5560041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallagher T, Alexander P, Bryan P, Gilliland GL. Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Biochemistry. 1994;33:4721–9. [PubMed] [Google Scholar]
- 70.Orban J, Alexander P, Bryan P. Sequence-specific 1H NMR assignments and secondary structure of the streptococcal protein G B2-domain. Biochemistry. 1992;31:3604–11. doi: 10.1021/bi00129a008. [DOI] [PubMed] [Google Scholar]
- 71.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucl Acid Res. 2004;32:D226–9. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucl Acid Res. 2000;28:257–9. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mielke SP, Krishnan VV. An evaluation of chemical shift index-based secondary structure determination in proteins: influence of random coil chemical shifts. J Biomol NMR. 2004;30:143–53. doi: 10.1023/b:jnmr.0000048940.51331.49. [DOI] [PubMed] [Google Scholar]
- 74.Schwarzinger S, Kroon GJA, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in NMRView. J Biomol NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 75.Lukin JA, Gove AP, Talukdar SN, Ho C. Automated Probabilistic Method For Assigning Backbone Resonances of (C-13,N-15)-Labeled Proteins. J Biomol NMR. 1997;9:151–166. doi: 10.1023/a:1018602220061. [DOI] [PubMed] [Google Scholar]
- 76.Braun D, Wider G, Wuthrich K. Sequence-Corrected N-15 Random Coil Chemical Shifts. J Am Chem Soc. 1994;116:8466–8469. [Google Scholar]
- 77.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. H-1, C-13 and N-15 Random Coil NMR Chemical Shifts of the Common Amino Acids.1. Investigations of Nearest-Neighbor Effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 78.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. H-1, C-13 and N-15 Chemical Shift Referencing in Biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 79.Wang YJ, Jardetzky O. Investigation of the neighboring residue effects on protein chemical shifts. J Am Chem Soc. 2002;124:14075–14084. doi: 10.1021/ja026811f. [DOI] [PubMed] [Google Scholar]
- 80.Wang L, Eghbalnia HR, Markley JL. Probabilistic approach to determining unbiased random-coil carbon-13 chemical shift values from the protein chemical shift database. J Biomol NMR. 2006;35:155–65. doi: 10.1007/s10858-006-9022-0. [DOI] [PubMed] [Google Scholar]
- 81.Plaxco KW, Morton CJ, Grimshaw SB, Jones JA, Pitkeathly M, Campbell ID, Dobson CM. The effects of guanidine hydrochloride on the ‘random coil’ conformations and NMR chemical shifts of the peptide series GGXGG. J Biomol NMR. 1997;10:221–230. doi: 10.1023/A:1018340217891. [DOI] [PubMed] [Google Scholar]
- 82.Bernstein FC, Koetzle TF, Williams GJ, Meyer EE, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–42. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 83.Kabsch W, Sander C. A dictionary of protein secondary structure. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 84.Richards FM, Kundrot CE. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins. 1988;3:71–84. doi: 10.1002/prot.340030202. [DOI] [PubMed] [Google Scholar]
- 85.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–79. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 86.Cuff JA, Barton GJ. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–19. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 87.Figureau A, Soto MA, Toha J. Secondary structure of proteins and three-dimensional pattern recognition. J Theor Biol. 1999;201:103–111. doi: 10.1006/jtbi.1999.1016. [DOI] [PubMed] [Google Scholar]
- 88.Figureau A, Soto MA, Toha J. A pentapeptide-based method for protein secondary structure prediction. Protein Eng. 2003;16:103–107. doi: 10.1093/proeng/gzg019. [DOI] [PubMed] [Google Scholar]
- 89.Flory PJ. Statistical mechanics of chain molecules. Interscience Publishers; New York: 1969. [Google Scholar]
- 90.Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 91.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 92.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 93.Shortle D. The denatured state (the other half of the folding equation) and its role in protein stability. Faseb J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- 94.Vila JA, Ripoll DR, Baldoni HA, Scheraga HA. Unblocked statistical-coil tetrapeptides and pentapeptides in aqueous solution: A theoretical study. J Biomol NMR. 2002;24:245–262. doi: 10.1023/a:1021633403715. [DOI] [PubMed] [Google Scholar]
- 95.Sharman GJ, Griffiths-Jones SR, Jourdan M, Searle MS. Effects of amino acid phi, psi propensities and secondary structure interactions in modulating H alpha chemical shifts in peptide and protein beta-sheet. J Am Chem Soc. 2001;123:12318–12324. doi: 10.1021/ja0116369. [DOI] [PubMed] [Google Scholar]
- 96.Ho CN, Lam SL. Random coil phosphorus chemical shift of deoxyribonucleic acids. J Magn Reson. 2004;171:193–200. doi: 10.1016/j.jmr.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 97.Kwok CW, Ho CN, Chi LM, Lam SL. Random coil carbon chemical shifts of deoxyribonucleic acids. J Magn Reson. 2004;166:11–8. doi: 10.1016/j.jmr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 98.Lam SL. DSHIFT: a web server for predicting DNA chemical shifts. Nucl Acid Res. 2007 doi: 10.1093/nar/gkm320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lam SL, Ip LN, Cui X, Ho CN. Random coil proton chemical shifts of deoxyribonucleic acids. J Biomol NMR. 2002;24:329–37. doi: 10.1023/a:1021671531438. [DOI] [PubMed] [Google Scholar]
- 100.Cromsigt JA, Hilbers CW, Wijmenga SS. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J Biomol NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- 101.Boudreau EA, Pelczer I, Borer PN, Heffron GJ, LaPlante SR. Changes in drug 13C NMR chemical shifts as a tool for monitoring interactions with DNA. Biophys Chem. 2004;109:333–44. doi: 10.1016/j.bpc.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 102.Buchko GW, Tung CS, McAteer K, Isern NG, Spicer LD, Kennedy MA. DNA-XPA interactions: a (31)P NMR and molecular modeling study of dCCAATAACC association with the minimal DNA-binding domain (M98-F219) of the nucleotide excision repair protein XPA. Nucl Acid Res. 2001;29:2635–43. doi: 10.1093/nar/29.12.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.LaPlante SR, Borer PN. Changes in 13C NMR chemical shifts of DNA as a tool for monitoring drug interactions. Biophys Chem. 2001;90:219–32. doi: 10.1016/s0301-4622(01)00143-0. [DOI] [PubMed] [Google Scholar]
- 104.Zeeb M, Balbach J. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 2003;12:112–23. doi: 10.1110/ps.0219703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Altona C, Faber DH, Hoekzema A. Double-helical DNA H-1 chemical shifts: an accurate and balanced predictive empirical scheme. Magn Reson Chem. 2000;38:95–107. [Google Scholar]
- 106.Wijmenga SS, Kruithof M, Hilbers CW. Analysis of H-1 chemical shifts in DNA: Assessment of the reliability of H-1 chemical shift calculations for use in structure refinement. J Biomol NMR. 1997;10:337–350. doi: 10.1023/A:1018348123074. [DOI] [PubMed] [Google Scholar]
- 107.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 108.Chi Z, Chen XG, Holtz JS, Asher SA. UV resonance Raman-selective amide vibrational enhancement: quantitative methodology for determining protein secondary structure. Biochemistry. 1998;37:2854–64. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 109.Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci. 2000;1:349–84. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 110.Hering JA, Innocent PR, Haris PI. Beyond average protein secondary structure content prediction using FTIR spectroscopy. Appl Bioinformatics. 2004;3:9–20. doi: 10.2165/00822942-200403010-00003. [DOI] [PubMed] [Google Scholar]
- 111.Mielke SP, Krishnan VV. Estimation of protein secondary structure content directly from NMR spectra using an improved empirical correlation with averaged chemical shift. J Struct Funct Genomics. 6(2005):281–5. doi: 10.1007/s10969-005-9002-8. [DOI] [PubMed] [Google Scholar]
- 112.Pancoska P, Bitto E, Janota V, Urbanova M, Gupta VP, Keiderling TA. Comparison of and limits of accuracy for statistical analyses of vibrational and electronic circular dichroism spectra in terms of correlations to and predictions of protein secondary structure. Protein Sci. 1995;4:1384–401. doi: 10.1002/pro.5560040713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 114.Lee S, Lee BC, Kim D. Prediction of protein secondary structure content using amino acid composition and evolutionary information. Proteins. 2006;62:1107–14. doi: 10.1002/prot.20821. [DOI] [PubMed] [Google Scholar]
- 115.Zhang CT, Zhang R. Q9, a content-balancing accuracy index to evaluate algorithms of protein secondary structure prediction. Int J Biochem Cell Biol. 2003;35:1256–62. doi: 10.1016/s1357-2725(02)00370-9. [DOI] [PubMed] [Google Scholar]
- 116.Cai YD, Liu XJ, Chou KC. Prediction of protein secondary structure content by artificial neural network. J Comput Chem. 2003;24:727–31. doi: 10.1002/jcc.10222. [DOI] [PubMed] [Google Scholar]
- 117.Cai YD, Liu XJ, Xu XB, Chou KC. Artificial neural network method for predicting protein secondary structure content. Comput Chem. 2002;26:347–50. doi: 10.1016/s0097-8485(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 118.Liu W, Chou KC. Prediction of protein secondary structure content. Protein Eng. 1999;12:1041–50. doi: 10.1093/protein/12.12.1041. [DOI] [PubMed] [Google Scholar]
- 119.Chou KC. Using pair-coupled amino acid composition to predict protein secondary structure content. J Protein Chem. 1999;18:473–80. doi: 10.1023/a:1020696810938. [DOI] [PubMed] [Google Scholar]
- 120.Clementi M, Clementi S, Cruciani G, Pastor M, Davis AM, Flower DR. Robust multivariate statistics and the prediction of protein secondary structure content. Protein Eng. 1997;10:747–9. doi: 10.1093/protein/10.7.747. [DOI] [PubMed] [Google Scholar]
- 121.Muskal SM, Kim SH. Predicting protein secondary structure content. A tandem neural network approach. J Mol Biol. 1992;225:713–27. doi: 10.1016/0022-2836(92)90396-2. [DOI] [PubMed] [Google Scholar]
- 122.Krigbaum WR, Knutton SP. Prediction of the amount of secondary structure in a globular protein from its aminoacid composition. Proc Natl Acad Sci U S A. 1973;70:2809–13. doi: 10.1073/pnas.70.10.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang CT, Zhang R. A graphic approach to evaluate algorithms of secondary structure prediction. J Biomol Struct Dyn. 2000;17:829–42. doi: 10.1080/07391102.2000.10506572. [DOI] [PubMed] [Google Scholar]
- 124.Zhang CT, Zhang R. S curve, a graphic representation of protein secondary structure sequence and its applications. Biopolymers. 2000;53:539–49. doi: 10.1002/(SICI)1097-0282(200006)53:7<539::AID-BIP2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 125.Zhang CT, Zhang R. A new criterion to classify globular proteins based on their secondary structure contents. Bioinformatics. 1998;14:857–65. doi: 10.1093/bioinformatics/14.10.857. [DOI] [PubMed] [Google Scholar]
- 126.Zhang CT, Zhang R. A new quantitative criterion to distinguish between alpha/beta and alpha+beta proteins (domains) FEBS Lett. 1998;440:153–7. doi: 10.1016/s0014-5793(98)01433-1. [DOI] [PubMed] [Google Scholar]
- 127.Zhang CT, Zhang Z, He Z. Prediction of the secondary structure contents of globular proteins based on three structural classes. J Protein Chem. 1998;17:261–72. doi: 10.1023/a:1022588803017. [DOI] [PubMed] [Google Scholar]
- 128.Pan XM. Multiple linear regression for protein secondary structure prediction. Proteins. 2001;43:256–9. doi: 10.1002/prot.1036. [DOI] [PubMed] [Google Scholar]
- 129.Pilizota T, Lucic B, Trinajstic N. Use of variable selection in modeling the secondary structural content of proteins from their composition of amino acid residues. J Chem Inform Comput Sci. 2004;44:113–21. doi: 10.1021/ci034037p. [DOI] [PubMed] [Google Scholar]
- 130.Eisenhaber F, Frommel C, Argos P. Prediction of secondary structural content of proteins from their amino acid composition alone. II. The paradox with secondary structural class. Proteins. 1996;25:169–79. doi: 10.1002/(SICI)1097-0134(199606)25:2<169::AID-PROT3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 131.Eisenhaber F, Imperiale F, Argos P, Frommel C. Prediction of secondary structural content of proteins from their amino acid composition alone. I. New analytic vector decomposition methods. Proteins. 1996;25:157–68. doi: 10.1002/(SICI)1097-0134(199606)25:2<157::AID-PROT2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 132.Nakashima H, Nishikawa K, Ooi T. The folding type of a protein is relevant to the amino acid composition. J Biochem (Tokyo) 1986;99:153–62. doi: 10.1093/oxfordjournals.jbchem.a135454. [DOI] [PubMed] [Google Scholar]
- 133.Klein P. Prediction of protein structural class by discriminant analysis. Biochim Biophys Acta. 1986;874:205–15. doi: 10.1016/0167-4838(86)90119-6. [DOI] [PubMed] [Google Scholar]
- 134.Klein P, Delisi C. Prediction of protein structural class from the amino acid sequence. Biopolymers. 1986;25:1659–72. doi: 10.1002/bip.360250909. [DOI] [PubMed] [Google Scholar]
- 135.Bahar I, Atilgan AR, Jernigan RL, Erman B. Understanding the recognition of protein structural classes by amino acid composition. Proteins. 1997;29:172–85. [PubMed] [Google Scholar]
- 136.Chou JJ, Zhang CT. A joint prediction of the folding types of 1490 human proteins from their genetic codons. J Theor Biol. 1993;161:251–62. doi: 10.1006/jtbi.1993.1053. [DOI] [PubMed] [Google Scholar]
- 137.Zhang CT, Chou KC. An analysis of protein folding type prediction by seed-propagated sampling and jackknife test. J Protein Chem. 1995;14:583–93. doi: 10.1007/BF01886884. [DOI] [PubMed] [Google Scholar]
- 138.Zhang CT, Chou KC. Monte Carlo simulation studies on the prediction of protein folding types from amino acid composition. II. Correlative effect. J Protein Chem. 1995;14:251–8. doi: 10.1007/BF01886766. [DOI] [PubMed] [Google Scholar]
- 139.Zhang CT, Chou KC, Maggiora GM. Predicting protein structural classes from amino acid composition: application of fuzzy clustering. Protein Eng. 1995;8:425–35. doi: 10.1093/protein/8.5.425. [DOI] [PubMed] [Google Scholar]
- 140.Chou KC. A novel approach to predicting protein structural classes in a (20–1)-D amino acid composition space. Proteins. 1995;21:319–44. doi: 10.1002/prot.340210406. [DOI] [PubMed] [Google Scholar]
- 141.Metfessel BA, Saurugger PN, Connelly DP, Rich SS. Cross-validation of protein structural class prediction using statistical clustering and neural networks. Protein Sci. 1993;2:1171–82. doi: 10.1002/pro.5560020712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang CT, Chou KC. An optimization approach to predicting protein structural class from amino acid composition. Protein Sci. 1992;1:401–8. doi: 10.1002/pro.5560010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou G, Xu X, Zhang CT. A weighting method for predicting protein structural class from amino acid composition. Eur J Biochem. 1992;210:747–9. doi: 10.1111/j.1432-1033.1992.tb17476.x. [DOI] [PubMed] [Google Scholar]
- 144.Boberg J, Salakoski T, Vihinen M. Accurate prediction of protein secondary structural class with fuzzy structural vectors. Protein Eng. 1995;8:505–12. doi: 10.1093/protein/8.6.505. [DOI] [PubMed] [Google Scholar]
- 145.Cai YD, Liu XJ, Xu Xb X, Zhou GP. Support Vector Machines for predicting protein structural class. BMC Bioinformatics. 2001;2:3. doi: 10.1186/1471-2105-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang ZX. The prediction accuracy for protein structural class by the component- coupled method is around 60% Proteins. 2001;43:339–40. doi: 10.1002/prot.1046. [DOI] [PubMed] [Google Scholar]
- 147.Wang ZX, Yuan Z. How good is prediction of protein structural class by the component- coupled method? Proteins. 2000;38:165–75. doi: 10.1002/(sici)1097-0134(20000201)38:2<165::aid-prot5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 148.Zhou GP. An intriguing controversy over protein structural class prediction. J Protein Chem. 1998;17:729–38. doi: 10.1023/a:1020713915365. [DOI] [PubMed] [Google Scholar]
- 149.Mielke SP, Krishnan VV. Protein structural class identification directly from NMR spectra using averaged chemical shifts. Bioinformatics. 2003;19:2054–64. doi: 10.1093/bioinformatics/btg280. [DOI] [PubMed] [Google Scholar]
- 150.Ikegami T, Okada T, Ohki I, Hirayama J, Mizuno T, Shirakawa M. Solution structure and dynamic character of the histidine-containing phosphotransfer domain of anaerobic sensor kinase ArcB from Escherichia coli. Biochemistry. 2001;40:375–86. doi: 10.1021/bi001619g. [DOI] [PubMed] [Google Scholar]
- 151.Wang H, He Y, Hsu KT, Magliocca JF, Storch J, Stark RE. 1H, 15N and 13C resonance assignments and secondary structure of apo liver fatty acid-binding protein. J Biomol NMR. 1998;12:197–9. doi: 10.1023/a:1008296530125. [DOI] [PubMed] [Google Scholar]
- 152.Sibley AB, Cosman M, Krishnan VV. An empirical correlation between secondary structure content and averaged chemical shifts in proteins. Biophys J. 2003;84:1223–7. doi: 10.1016/S0006-3495(03)74937-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang HY, Neal S, Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 154.Parvathy VR, Chary KVR, Kini RM, Govil G. Sequence-specific C-13 NMR assignments in a neurotoxin (candoxin) from Bungarus candidus. Magn Reson Chem. 2001;39:577–580. [Google Scholar]
- 155.Park S, Liao XB, Johnson ME, Fung LWM. H-1, N-15, and C-13 NMR backbone assignments of the N-terminal region of human erythrocyte alpha spectrin including one structural domain. J Biomol NMR. 1999;15:345–346. doi: 10.1023/a:1008339507922. [DOI] [PubMed] [Google Scholar]
- 156.Lazar GA, Johnson EC, Desjarlais JR, Handel TM. Rotamer strain as a determinant of protein structural specificity. Protein Sci. 1999;8:2598–2610. doi: 10.1110/ps.8.12.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hamilton KS, Ellison MJ, Shaw GS. Letter to the Editor: H-1, N-15 and C-13 resonance assignments for the catalytic domain of the yeast E2, UBC1. J Biomol NMR. 2000;16:351–352. doi: 10.1023/a:1008324602437. [DOI] [PubMed] [Google Scholar]
- 158.Johnson EC, Lazar GA, Desjarlais JR, Handel TM. Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin. Structure Fold Des. 1999;7:967–976. doi: 10.1016/s0969-2126(99)80123-3. [DOI] [PubMed] [Google Scholar]
- 159.Peti W, Smith LJ, Redfield C, Schwalbe H. Chemical shifts in denatured proteins: Resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J Biomol NMR. 2001;19:153–165. doi: 10.1023/a:1008307323283. [DOI] [PubMed] [Google Scholar]
- 160.Atkinson RA, Saudek V. The direct determination of protein structure by NMR without assignment. FEBS Lett. 2002;510:1–4. doi: 10.1016/s0014-5793(01)03208-2. [DOI] [PubMed] [Google Scholar]
- 161.Grishaev A, Llinas M. CLOUDS, a protocol for deriving a molecular proton density via NMR. Proc Natl Acad Sci U S A. 2002;99:6707–12. doi: 10.1073/pnas.082114199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee MS, Cao B. Nuclear magnetic resonance chemical shift: comparison of estimated secondary structures in peptides by nuclear magnetic resonance and circular dichroism. Protein Eng. 1996;9:15–25. doi: 10.1093/protein/9.1.15. [DOI] [PubMed] [Google Scholar]
- 163.Chou PY. Prediction of protein structural class from amino acid composition. In: Fasman GD, editor. Prediction of protein structure and principles of protein conformation. Plenum Press; New York: 1989. pp. 549–586. [Google Scholar]
- 164.Havlin RH, Laws DD, Bitter HML, Sanders LK, Sun HH, Grimley JS, Wemmer DE, Pines A, Oldfield E. An experimental and theoretical investigation of the chemical shielding tensors of C-13(alpha) of alanine, valine, and leucine residues in solid peptides and in proteins in solution. J Am Chem Soc. 2001;123:10362–10369. doi: 10.1021/ja0115060. [DOI] [PubMed] [Google Scholar]
- 165.Pervushin K, Riek R, Wider G, Wuthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J Am Chem Soc. 1998;120:6394–6400. [Google Scholar]
- 166.McConnell HM. Theory of nuclear magnetic shielding in molecules I Long-range dipolar shielding of protons. J Chem Phys. 1957;27:226–229. [Google Scholar]
- 167.Buckingham AD, Schaefer T, Schneider WG. Solvent effects in nuclear magnetic resonance spectra. J Chem Phys. 1960;32:1227–1233. [Google Scholar]
- 168.Haigh CW, Mallion RB. Ring current theories in nuclear magnetic resonance. Prog Nucl Magn Reson Spectros. 1980;13:303–344. [Google Scholar]
- 169.Wang CC, Chen JH, Yin SH, Chuang WJ. Predicting the redox state and secondary structure of cysteine residues in proteins using NMR chemical shifts. Proteins. 2006;63:219–26. doi: 10.1002/prot.20875. [DOI] [PubMed] [Google Scholar]
- 170.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR. 2002;24:149–54. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 171.Osapay K, Case DA. Analysis of Proton Chemical Shifts in Regular Secondary Structure of Proteins. J Biomol NMR. 1994;4:215–230. doi: 10.1007/BF00175249. [DOI] [PubMed] [Google Scholar]
- 172.Xu XP, Case DA. Automated prediction of 15N, 13Calpha, 13Cbeta and 13C′ chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21:321–33. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- 173.Moon S, Case DA. A new model for chemical shifts of amide hydrogens in proteins. J Biomol NMR. 2007;38:139–50. doi: 10.1007/s10858-007-9156-8. [DOI] [PubMed] [Google Scholar]
- 174.Williamson MP, Asakura T. Calculation of Chemical Shifts of Protons On Alpha Carbons in Proteins. J Magn Reson. 1991;94:557–562. [Google Scholar]
- 175.Williamson MP, Asakura T, Nakamura E, Demura M. A Method For the Calculation of Protein Alpha-Ch Chemical Shifts. J Biomol NMR. 1992;2:83–98. doi: 10.1007/BF02192802. [DOI] [PubMed] [Google Scholar]
- 176.Williamson MP, Asakura T. The Application of H-1-NMR Chemical Shift Calculations to Diastereotopic Groups in Proteins. FEBD Lett. 1992;302:185–188. doi: 10.1016/0014-5793(92)80436-k. [DOI] [PubMed] [Google Scholar]
- 177.Williamson MP, Asakura T. Empirical Comparisons of Models For Chemical-Shift Calculation in Proteins. J Magn Reson Series B. 1993;101:63–71. [Google Scholar]
- 178.Williamson MP, Kikuchi J, Asakura T. Application of H-1 NMR Chemical Shifts to Measure the Quality of Protein Structures. J Mol Biol. 1995;247:541–546. doi: 10.1006/jmbi.1995.0160. [DOI] [PubMed] [Google Scholar]
- 179.Iwadate M, Asakura T, Williamson MP. C-alpha and C-beta carbon-13 chemical shifts in proteins from an empirical database. J Biomol NMR. 1999;13:199–211. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- 180.Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR. 2003;26:215–40. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 181.Neal S, Berjanskii M, Zhang H, Wishart DS. Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn Reson Chem. 2006;44(Spec No):S158–67. doi: 10.1002/mrc.1832. [DOI] [PubMed] [Google Scholar]
- 182.Wishart DS, Watson MS, Boyko RF, Sykes BD. Automated H-1 and C-13 chemical shift prediction using the BioMagResBank. J Biomol NMR. 1997;10:329–336. doi: 10.1023/a:1018373822088. [DOI] [PubMed] [Google Scholar]
- 183.Meiler J. PROSHIFT: protein chemical shift prediction using artificial neural networks. J Biomol NMR. 2003;26:25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- 184.Berjanskii M, Wishart DS. NMR: prediction of protein flexibility. Nat Protoc. 2006;1:683–8. doi: 10.1038/nprot.2006.108. [DOI] [PubMed] [Google Scholar]
- 185.Berjanskii MV, Wishart DS. The RCI server: rapid and accurate calculation of protein flexibility using chemical shifts. Nucl Acid Res. 2007 doi: 10.1093/nar/gkm328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Arun K, Langmead CJ. Structure-Based Chemical Shift Prediction using Random Forests Non-Linear Regression. Proc. of the The 4th Asia-Pacific Bioinformatics Conference, (APBC); 2006. p. 4. [Google Scholar]
- 187.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 188.Jabs A, Weiss MS, Hilgenfeld R. Non-proline cis peptide bonds in proteins. J Mol Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 189.Weiss MS, Metzner HJ, Hilgenfeld R. Two non-proline cis peptide bonds may be important for factor XIII function. FEBS Lett. 1998;423:291–6. doi: 10.1016/s0014-5793(98)00098-2. [DOI] [PubMed] [Google Scholar]
- 190.Dorman DE, Torchia DA, Bovey FA. Carbon-13 and proton nuclear magnetic resonance observations of the conformation of poly(L-proline) in aqueous salt solutions. Macromolecules. 1973;6:80–2. doi: 10.1021/ma60031a013. [DOI] [PubMed] [Google Scholar]
- 191.Siemion IZ, Wieland T, Pook KH. Influence of the distance of the proline carbonyl from the beta and gamma carbon on the 13C chemical shifts. Angew Chem Int Ed Engl. 1975;14:702–3. doi: 10.1002/anie.197507021. [DOI] [PubMed] [Google Scholar]
- 192.Richarz R, Wuthrich K. Carbon-13 NMR chemical shifts of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-LAla-OH. Biopolymers. 1978;17:2133–2141. [Google Scholar]
- 193.Bundi A, Wuthrich K. 1H -NMR parameters of the common amino acid a\residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-LAla-OH. Biopolymers. 1979;18:285–297. [Google Scholar]
- 194.Glushka J, Lee M, Coffin S, Cowburn D. 15N chemical shifts of backbone amides in bovine pancreatic trypsin inhibitor and apamin. J Am Chem Soc. 1989;111:7716–7722. [Google Scholar]
- 195.Glushka J, Lee M, Coffin S, Cowburn D. 15N chemical shifts of backbone amides in bovine pancreatic trypsin inhibitor and apamin. (correction) J Am Chem Soc. 1990;112:2843. [Google Scholar]
- 196.Thanabal V, Omecinsky DO, Reily MD, Cody WL. The 13C chemical shifts of amino acids in aqueous solution containing organic solvents: application to the secondary structure characterization of peptides in aqueous trifluoroethanol solution. J Biomol NMR. 1994;4:47–59. doi: 10.1007/BF00178335. [DOI] [PubMed] [Google Scholar]
- 197.Merutka G, Dyson HJ, Wright PE. Random Coil H-1 Chemical Shifts Obtained As a Function of Temperature and Trifluoroethanol Concentration For the Peptide Series Ggxgg. J Biomol NMR. 1995;5:14–24. doi: 10.1007/BF00227466. [DOI] [PubMed] [Google Scholar]
- 198.Bienkiewicz EA, Lumb KJ. Random-coil chemical shifts of phosphorylated amino acids. J Biomol NMR. 1999;15:203–206. doi: 10.1023/a:1008375029746. [DOI] [PubMed] [Google Scholar]
- 199.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. H-1, C-13 and N-15 Random Coil NMR Chemical Shifts of the Common Amino Acids.1. Investigations of Nearest-Neighbor Effects (Vol 5, Pg 67, 1995) J Biomol NMR. 1995;5:332–332. doi: 10.1007/BF00211764. [DOI] [PubMed] [Google Scholar]