Abstract
We describe the development of a spotted array for the delineation of the most common 14 disease causing Salmonella serovars in the United States. Our array consists of 414 70mers targeting core genes of S. enterica, subspecies I specific genes, fimbrial genes, pathogenicity islands, Gifsy elements and other variable genes. Using this array we were able to identify a unique gene presence/absence profile for each of the targeted serovar which was used as the serovar differentiating criteria. Based on this profile, we developed a Matlab programme that compares the profile of an unknown sample to all 14 reference serovar profiles and give out the closest serovar match. Since we have included probes targeting most of the virulence genes and variable genes in Salmonella, in addition to using for serovar detection this array could also be used for studying the virulence gene content and also for evaluating the genetic relation between different isolates of Salmonella.
Keywords: Salmonella, microarray, oligonucleotide
1. Introduction
Salmonellosis is an important public health problem in the United States causing significant economic loss and substantial morbidity. Although most Salmonella infections cause mild to moderate self-limited disease, serious infections leading to death can occur. As per the estimates of Centers for Disease Control and Prevention (CDC; Atlanta, GA) ∼1,400,000 cases of salmonellosis occur annually [15]. Salmonella survive well in a variety of food, particularly of animal origin (e.g., beef, poultry, eggs, and dairy products) and also on fruits and vegetables [24]. Expenditure associated with salmonellosis, including the costs of medical care and lost productivity, may approach several billion dollars annually [9]. Hence it is important to develop methods that can differentiate pathogenic Salmonella.
The genus Salmonella is divided into two species, Salmonella enterica and Salmonella bongori. Salmonella enterica comprises of six subspecies and of these, Salmonella enterica subsp enterica 1 having more than 1,500 serovars, is responsible for almost all Salmonella infections [2]. This classification for the genus Salmonella has evolved from the initial one serotype-one species concept proposed by Kauffmann on the basis of the serologic identification of O (somatic) and H (flagellar) antigens [3]. These surface features tend to be variable because of the strong selection pressure from host and comparative genomics of Salmonella serovars have revealed that there could be significant intra-serovar variations among different isolates [6, 20]. Several phenotypic, genotypic and molecular techniques like biotyping, phage typing, ribotyping IS200 typing, PCR, pulsed-field electrophoresis and nucleic acid hybridization have been developed for Salmonella differentiation [7, 8, 11, 12]. Although serotyping continues to be the most commonly used method of delineation for Salmonella spp. [12], the downside is that it is incapable of revealing the genetic constitution and intra-serovar variations. Genome sequencing of different Salmonella serovars has confirmed that up to 97% of the genome sequence is identical between different serovars of Salmonella enterica [5, 14, 17]. However, comparative studies using microarrays have revealed the conserved/core and variable gene components in Salmonella serovars [1, 4, 19]. These studies show that variability in the Salmonella genome is mainly associated with fimbrial clusters, pathogenecity islands and phage elements. Further comparative genome hybridizations using Salmonella enterica serovar Typhimurium (S. Typhimurium) LT2 genome as reference reveal that closely related serovars are not always genotypically close and these variations are characterized at single-gene resolution [19]. Since serotyping does not detect the intra-serovar variations, a single “multi target” method that would delineate the most common Salmonella serovars will be a valuable tool for diagnostic and epidemiological studies. In this study, drawing on the knowledge of the core genes and genetic elements that cause serovar variability, we developed an oligonucleotide spotted array for the differentiation of serovars of Salmonella enterica. The probe specifity was validated using three sequenced strains, namely Salmonella enterica subspecies I, serovar Typhimurium (S. Typhimurium, Accession numbers AE006468 and AE006471), Salmonella enterica serovar Typhi (S. Typhi, Accession numbers AL513382, AL513383 and AL513384) and Salmonella enterica serovar Choleraesuis (S. Choleraesuis, Accession number NC_006905). Our results show the feasibility of microarray based serovar differentiation of most prevalent serovars in the United States.
2. Materials and Methods
2.1. Bacterial strains and growth conditions
All the reference strains and field isolates of Salmonella serovars were maintained in our laboratory and the details are given in Table 1. The serovars of Salmonella for this study were selected from the most frequently reported Salmonella serovars as listed by the Center for Disease Control and Prevention (CDC) in 2004 (http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm). All these strains/isolates were streaked onto Luria agar plates and a single colony was selected and grown in Luria broth at 37°C for 12 hrs.
Table 1. Source and strain name of Salmonell Serovars.
| serial no | Number | Name | Biotype | Host | Isolation |
|---|---|---|---|---|---|
| 1 | 3086 | Salmonella enterica serovar Dublin | SARB 12 | SARB | |
| 2 | 2895 | Salmonella enterica serovar Dublin | Unknown | bovine | AHDC |
| 3 | 2905 | Salmonella enterica serovar Dublin | Unknown | bovine | AHDC |
| 4 | 2900 | Salmonella enterica serovar Dublin | Unknown | bovine | AHDC |
| 5 | 3168 | Salmonella enterica serovar Agona | ccc39 | SARB | |
| 6 | 3357 | Salmonella enterica serovar Agona | Unknown | bovine | AHDC |
| 7 | 4294 | Salmonella enterica serovar Agona | S5-667 | human | NYSDOH |
| 8 | 4295 | Salmonella enterica serovar Agona | S5-647 | human | NYSDOH |
| 9 | 3104 | Salmonella bongori | SARC 11 | SARC | |
| 10 | 3082 | Salmonella enterica serovar Choleraesuis | SARB 4 | SARB | |
| 11 | 1777 | Salmonella enterica serovar Choleraesuis* | CN207* | Pig | AHDC |
| 12 | 1778 | Salmonella enterica serovar Choleraesuis | CN214 | Pig | AHDC |
| 13 | 3090 | Salmonella enterica serovar Enteritidis | SARB 16 | SARB | |
| 14 | 4283 | Salmonella enterica serovar Enteritidis | S5-377 | human | NYSDOH |
| 15 | 3124 | Salmonella enterica serovar Heildelberg | SARB 23 | SARB | |
| 16 | 4286 | Salmonella enterica serovar Heildelberg | S5-448 | human | NYSDOH |
| 17 | 3126 | Salmonella enterica serovar Infantis | SARB 26 | SARB | |
| 18 | 4285 | Salmonella enterica serovar Infantis | S5-506 | human | NYSDOH |
| 19 | 4293 | Salmonella enterica serovar Infantis | S5-372 | human | NYSDOH |
| 20 | 3110 | Salmonella enterica serovar Javiana | SGGC 4073 | SARB | |
| 21 | 4310 | Salmonella enterica serovar Javiana | S5-665 | human | NYSDOH |
| 22 | 4311 | Salmonella enterica serovar Javiana | S5-395 | human | NYSDOH |
| 23 | 4281 | Salmonella enterica serovar Muenchen | S5-479 | human | NYSDOH |
| 24 | 4282 | Salmonella enterica serovar Muenchen | S5-636 | human | NYSDOH |
| 25 | 3348 | Salmonella enterica serovar Muenster | Unknown | bovine | AHDC |
| 26 | 3404 | Salmonella enterica serovar Muenster | Unknown | bovine | AHDC |
| 27 | 3136 | Salmonella enterica serovar Newport | SARB 36 | SARB | |
| 28 | 3438 | Salmonella enterica serovar Newport | Unknown | bovine | AHDC |
| 29 | 3439 | Salmonella enterica serovar Newport | Unknown | bovine | AHDC |
| 30 | 3440 | Salmonella enterica serovar Newport | Unknown | bovine | AHDC |
| 31 | 4283 | Salmonella enterica serovar Newport | S5-413 | human | NYSDOH |
| 32 | 3098 | Salmonella enterica serovar St. paul | SARB 55 | SARB | |
| 33 | 4291 | Salmonella enterica serovar St. paul | S5-485 | human | NYSDOH |
| 34 | 3148 | Salmonella enterica serovar Thompson | SARB 62 | SARB | |
| 35 | 4298 | Salmonella enterica serovar Thompson | S5-472 | human | NYSDOH |
| 36 | 3158 | Salmonella enterica serovar Typhimurium* | LT2* | SARB | |
| 37 | 3347 | Salmonella enterica serovar Typhimurium | Unknown | bovine | AHDC |
| 38 | 3414 | Salmonella enterica serovar Typhimurium | Unknown | bovine | AHDC |
| 39 | 3429 | Salmonella enterica serovar Typhimurium | Unknown | bovine | AHDC |
| 40 | 4306 | Salmonella enterica serovar Typhimurium | S5-381 | human | NYSDOH |
| 41 | 4307 | Salmonella enterica serovar Typhimurium | S5-370 | human | NYSDOH |
| 42 | 3150 | Salmonella enterica serovar Typhimurium | SARB 65 | SARB | |
| 43 | 3109 | Salmonella enterica serovar Typhi* | CT18* | ||
| 44 | 3102 | Salmonella enterica serovar Typhi | SARB 63 | SARB |
SARB: Salmonell reference B collection; AHDC: Animal Health Diagnostic center, New York; NYSDOH; New york State Department of Health, Albany, NY
Indicates sequenced strains
2.2. Construction of sub species I specific, core genes of Salmonella enterica and variable gene probes
Comparative genome analysis of different serovars of Salmonella, E. coli K12 and E. coli O157 has provided a list of core genes of Salmonella enterica, subspecies I specific genes. Fimbriae, prophage like elements, pathogenic islands and other genes such as LPS encoding genes have also been reported to encode for diversity within serovars of Salmonella. Therefore, gene probes were constructed on core genes of S. enterica, subspecies I specific genes, fimbrial, pathogenicity islands, Gifsy-1 and 2 elements and other variable genes. The DNA sequences of these genes for S. Typhimurium LT2 and S. Typhi genomes were downloaded from the NCBI database, and the probes (70 mers) were designed using Arrayoligoselector (http://arrayoligosel.sourceforge.net/) as described previously [16]. The gene name, gene accession number, source, probe sequence, are listed in Appendix 1.
2.3. Microarray printing
The probes (70 mer) were synthesized (Illumina Inc, CA) and suspended in 50% dimethyl sulfoxide and spotted onto Ultra-GAPS glass slides (Corning Inc., Corning, N.Y.) with a spot size of 100μm, in triplicates arranged next to each other using a custom build microarray spotter following the original design of Pat Brown lab at Stanford University (http://cmgm.stanford.edu/pbrown/mguide/index.html). Autoblank (buffer without oligos) was used as a negative control.
2. 4. DNA preparation and labeling
Genomic DNA from Salmonella was prepared according to the manufacturer's instructions using DNeasy kit (Qiagen). 2μg of genomic DNA was digested with Sau3AI (New England Biolabs) and purified using Qiaquick PCR purification kit (Qiagen). The purified fragments were labeled as described previously [16] but by replacing random hexamers with 3′ phosphorothioated random hexamers. The labeled probes were purified using Qiaquick PCR purification kit (Qiagen) and were vacuum dried. To the vacuum dried probes, 1μl of 10 mg/mL Salmon sperm DNA, 1 μl of 4mg/mL yeast tRNA, 16.0 μl of resuspension buffer (25% deionized formamide, 5× SSC, 0.1% SDS) were added, reconstituted and was boiled for 3 minutes and the denatured probes were kept at 37°C for 10 minutes prior to hybridization.
2.5. Microarray experiments and data analysis
Immediately before use, the slides were incubated in prehybrization solution (5× SSC, 0.2% sarkosyl, 25% formamide, 1% BSA) at 42°C for 1 h, followed by rinsing with Milli-Q water and were dried by blowing with nitrogen gas. Denatured probe DNA was applied slowly to the slide through the edges of LifterSlip coverslip (Erie Scientific) and the slide was immediately placed in a hybridization chamber (Corning), and hybridized overnight by submerging in a 42°C water bath. Slides were washed for 5 minutes once with wash buffer 1 (2× SSC, 0.1% SDS at 42°C) twice with (1× SSC, at 37°C) and rinsed with MilliQ water for 2 minutes, dried by blowing with nitrogen gas and scanned on a GenePix 4000A scanner (Axon Instruments, Union city, CA). Each slide had triplicate spots of each feature and every experiment was repeated twice. Fluorescence data from the scanned images was extracted by genepix pro 6.0 software. For each feature, the background subtracted median foreground intensity value was averaged for every triplicate spot and was log-transformed. Then mean and standard deviation (SD) of the sub species 1 and Salmonella core gene specific features were calculated. Features with 1.5 SD below mean log values were considered absent and features more than 1.0 SD below mean log values were considered present and the range in between this was considered uncertain. This cut off criteria was selected after testing several cut off thresholds on the reference strains. The above probe classification was carried out using Avadis (Avadis software, Strand genomics) and Microbial Diagnostic Array Workstation (http://www.arraydb.org/). Hierarchical clustering of the strains/isolates based on these values was performed using Manhattan median method as the distance matrix (Avadis software, Strand genomics).
For finding the closest serovar match for an unknown sample we developed a Matlab programme that is implemented as follows. Let Pz denote the set of negative probes in the input data sample and each reference has its own set of known negative probes. Let P1, P2, …, P14 (for each of the 14 references) denote these sets and let the number of probes in each of these sets be denoted N1, N2, …, N14. Then algorithm finds the Pz probes in each of P1, P2, … P14. Let us denote these numbers Pz1, Pz2, …, Pz14. Then the ratios Pz1/N1, Pz2/N2, …, Pz14/N14 are calculated. The maximum value of this ratio points to the reference to which the input sample is closest. Fig.1 explains this diagrammatically. This programme is included in appendix 4 as a zip file. The instructions for running this programme, sample input data and reference data set are included in this zip file.
Fig. 1.
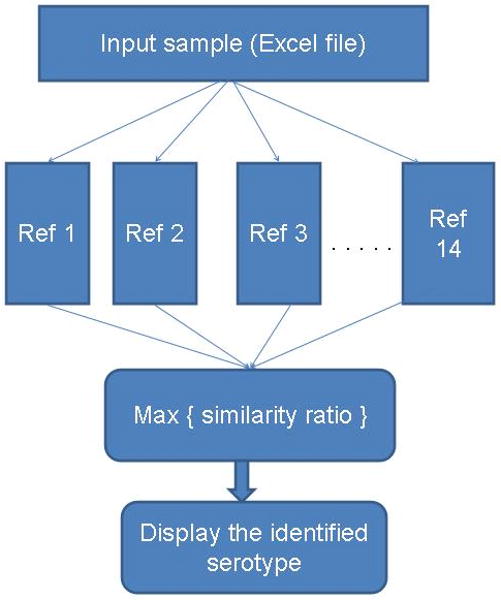
Implementation of the Matlab Programme for serovar assignment. The programme reads the input data from an excel file then it compares the values from input file with that of each of the reference serovar profile that is stored in the programme. It them calculate the maximum similarity ratio of the sample to the references and display the closest serovar match. The instruction for running the programme is given in appendix 4.
3. Results
3.1. Establishment probe classification and specificity
Recent studies using microarrays have identified the core/invariable and variable component of Salmonella genome [1, 4, 19]. Based on these findings we have developed a microarray for the differentiation of the most common disease causing Salmonella serovars in USA. Our array consists of 414 probes targeting core genes of S. enterica, subspecies I specific genes, fimbrial, pathogenicity islands, Gifsy-1 and 2 elements and other variable genes (Appendix 1). A set of 14 probes were designed on core genes of Salmonella which are also specific to Salmonella subspecies I and served to delineate the sample from E. coli and all other types of Salmonella. Another 71 probes were designed on core genes of Salmonella and served as the positive control for genus Salmonella. The rest of the probes were designed using variable genes in S. Typhimurium LT2 and S. Typhi CT18 genome. The stringency of microarray cutoff was verified using the sequenced strains (S. Typhimurium LT2 and S. Typhi CT18, S. Choleraesuis Sc-B67). The microarray results after making the cut off were compared against the BLAST results of all the probes against the sequenced strains (Appendix 2). There were 17 probes classified as negative for S. Typhimurium LT2. Of these, 10 were designed for genes specific for S. Typhi, hence expected to be negative in S. Typhimurium. The remainder is only 7 false negatives (1.7% of total probe set). When array results of S. Typhi CT18 were compared against BLAST results, there were 22 false negative probes. However 21 of these probes had 1-5 base mismatches between the probe and the corresponding gene. Hence, it's natural that these probes bind less efficiently with S. Typhi DNA. If the probes with mismatches are excluded, there was only one false negative (0.24% of total probe set) for S. Typhi. For S. Choleraesuis, there were 3 false negatives (0.72% of total probes) and 2 false positives (0.48% of total probes). There were only a limited number of probes in the uncertain (twilight zone) category and most of these had mismatches with the S. Typhi and S. Choleraesuis genome. Hence the probe classification and specificity which we establish here can be viewed with a high degree of confidence.
The tcf, sta, and ste fimbrial operons and Vi polysaccharide genes are absent in S. Typhimurium but present in S. Typhi. The probes targeting these genes were all negative for S. Typhimurium LT2. In the case of S. Typhi CT18, all these S. Typhi specific probes were positive. All the reference and clinical strains were 98-100% positive for the subspecies I specific gene probes while all these probes were negative for S. bongori. The core gene probes (71 probes in total) were designed for genes that are present in at least five out of six Salmonella subspecies and with respect to these probes all reference and clinical strains were 98-100% positive. This established the validity of the classification scheme and the specificity of the probes. For delineation of different serovars, the gene presence/absence profile obtained based on this parameter was examined for each SARB strain and clinical strains for every serovar and probes which are negative across all strains. For each serovar, such a signature was obtained (Appendix 3) and we used this gene signature as the inter serovar differentiating criteria. This profile of each of the 14 serovars studied in this work was mosaic in nature with respect to the gene presence.
The majority of virulence genes of Salmonella are clustered in regions distributed over the chromosome called Salmonella pathogenicity islands (SPI). SPI 1, 4 and 5 homologue regions were present in almost all the reference serovars of Salmonella. Variation was detected across the serovars in SPI-3 genes. SP1-3 genes are divided into three parts, part I, II and III [14]. Accordingly, SPI-3 part I corresponds to genes present in S. Typhimurium, S. Paratyphi and S. Typhi genes (probes Sal429- Sal435). Part II (Probes Sal436-Sal447) represents the genes present in subspecies I and S. bongori and Part III (probes Sal448-Sal461) represents genes present in subspecies I. 30-40% of part I probes were positive for S. Dublin, S. Enteritidis and S. Newport while the other serovars were almost 100% positive. Except for one gene (STM3768), all other SPI-3 Part II genes was present in all the serovars of Salmonella. In the case of S. Dublin, S. Enteritidis, S. Heidelberg, S. SaintPaul and S. Typhimurium 95-100% of probes were positive for SPI-3 part III genes. However, a genetic variation of 10-30% was detected in other serovars of Salmonella enterica and also S. bongori. A similar trend was seen for propahges (Gifsy1, Gisfsy2, Fels1, Fels2, Phage5), fimbrial operons (pef, bcf, sti, stf, saf, stb, stc, std, ipf, stj, sth, csf, and sef) and other variable genes (rfb locus, allantoin/glyoxylate cluster, dgo, and other hypothetical genes) where for some serovars majority of probes targeting these genes are positive while some other serovars have only smaller number of positive probes. Genome level sequence comparisons have indicated that the housekeeping genes of Salmonella are 97-99% similar while great degree of variation exists in fimbrial operons, pathogenicity islands, phage elements and other variable genes [6]. The previous microarray based genome hybridization studies have also arrived at the same conclusion [1, 4, 19]. When compared to these studies we have used only a subset of genes focusing on variable regions from the whole genome of S. Typhimurium and S. Typhi. However, by comparing the presence/ absence profile across all the probes for 14 serovars, we were able to identify a unique gene signature which can be used for serovar delineation. A visual comparison of this pattern is given in Fig.2 and the detailed classification is given in Appnedix 3.
Fig. 2.
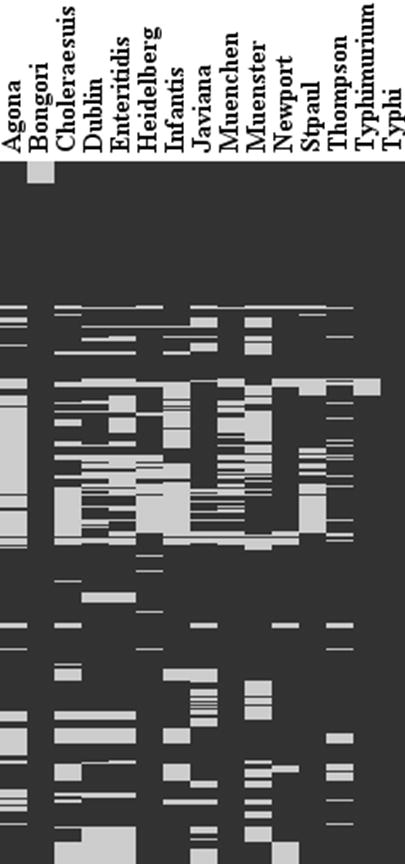
Heat map of the identified gene presence/absence signature by the array. Black regions represent regions that are present. Grey represent the regions that are absent in each serovar.
3.2. Strain clustering
To explore the possibility of using our array not only for serovar detection but also assessing the phylogenetic relationship among the strains based on the class score of the genes, hierarchical clustering of the strains/isolates was performed using Manhattan median method as the distance matrix [10]. In the cluster tree, S. bongori formed a separate branch while all other serovars were clustered in another complex branch (Fig.3). Most of the clinical strains clustered with their reference strains. Although the tree generated was in general agreement with the serovar assignment of the isolates, exceptions to this were observed in the case of S. Newport, S. Thompson, S. Muenchen and S. Heidelberg. It has been reported that S. Newport and S. Muenchen are polyphyletic according to MLEE data [22] and in CGH data [19]. Also intra-serovar differences were evident in many instances. As in the case of the clinical isolates of S. Newport, bovine and human strains showed differences from the reference strains in the Gifsy-2 and Gifsy-1 elements. The bovine and human isolates of S. Typhimurium also showed a similar pattern of gene profile with reference strain but the differences were evident in the individual genes such as STM266, STM2617, STM2238, hilD, STM2868, YciM, STM4259-4262, gop, allC, allD, fdrA and yeb. Despite clustering with their reference in the dendrogram, other clinical isolates of various serovars also showed intraserovar variations.
Fig. 3.
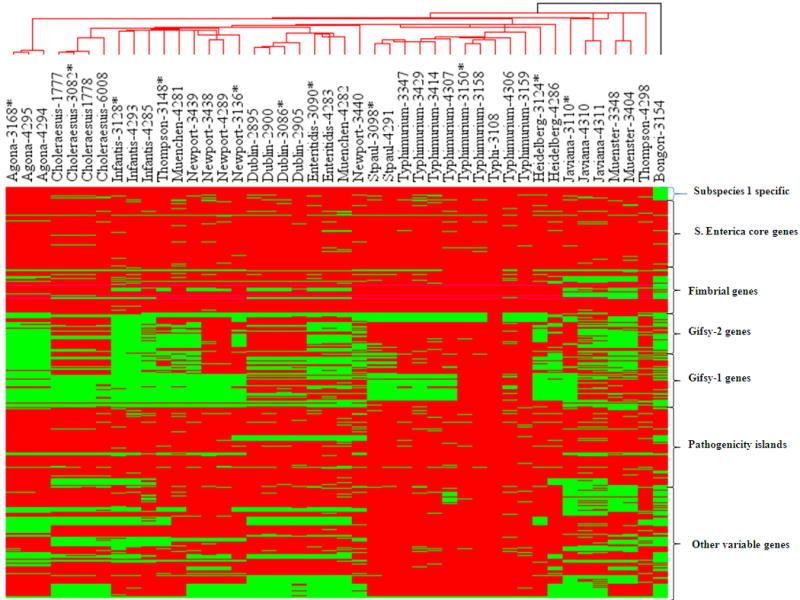
Hierarchical clustering of reference strains (SARB collection) and clinical isolates of Salmonella. Genomic DNA from the strains/isolates were fluorescently labeled with Cy3 and subjected to hybridization with gene probes present on the slides. Gene probes were targeted on core genes of S. enterica, subspecies I specific genes, fimbrial genes, genes from pathogenic Islands, genes of Gifsy-1 and 2 and variable genes to differentiate the serovars of Salmonella. Dendrogram represents the relatedness among the serovars of Salmonella. The close relationship of S. Typhimurium with S. SaintPaul; S. Thompson with S. Infantis; and S. Enteritidis with S. Dublin were observed. SARB strains are denoted by asterix.
3.3 Matlab programme for automatic serovar identification
Although clustering would provide a visual means of data analysis and exploration, this would require knowledge of different clustering methods and distance matrices. As an alternative to this, we developed a custom Matlab programme that takes the data as MS excel spreadsheet and then compare the probe profile of any unknown strain with the profile of 14 reference serovars and then automatically display the closest serovar match. When tested with the data in appendix 3, this programme correctly assigned the clinical samples to the respective reference serovars. Instructions for running this programme, sample input data and reference data set are included in appendix 4 as a zip file.
4. Discussion
Microarrays although most commonly used for comparative genome hybridizations and gene expression studies, their application for diagnostic purposes is becoming more popular. Examples of such arrays include arrays to detect E. coli pathotypes [16] and viruses [25]. Recently using S. Typhimurium and S. Typhi whole genome arrays, the core and variable gene component in their genome has been elucidated [1, 4, 19]. Drawing on these studies we have developed a new diagnostic array to differentiate the most common Salmonella serovars in United States. Our array consists of 414 probes targeting core genes of S. enterica, subspecies I specific genes, fimbrial, pathogenicity islands, Gifsy-1 and 2 elements and other variable genes. The subspecies I specific genes and core genes of Salmonella were used to differentiate the sample between Salmonella subspecies I and all other bacteria and also to define the statistical cutoff for calling the rest of gene probes present or absent. For delineation of 14 targeted serovars, the gene presence/absence profile obtained based on this parameter was examined for each SARB strain and clinical strains for every serovar and probes which are negative across all strains in a particular serovar was considered negative. Except for the S. Typhi probes (10 probes) all the other 404 probes were designed based on the sequenced genome of S. Typhimurium LT2 (reference genome). As an alternate of way of verifying the probe specificity and validity of the cut off points with several PCR reactions, we compared the microarray results with BLAST output of probes against S. Typhi CT18 and S. Choleraesuis SC-B67 genomes (Appendix 2). According to this data, 99% of all the genes that were classified as negative by array had not hit in the BLAST. Also the majority of the genes that were classified as uncertain had mismatches between probe sequence and the corresponding gene sequence from both S. Typhi CT18 and S. Choleraesuis SC-B6, hence leading to weak binding and weak signals in the array. This highlights the high specificity of the probes because the BLAST mismatches between the probes and S. Typhi CT18 and S. Choleraesuis SC-B67genomes represent the divergence of genes between S. Typhimurium LT2, S. Typhi CT18 and S. Choleraesuis SC-B6. However, the number of probes that were falling into the twilight zone was limited and did not impede the serovar differentiation capacity of the array. Instead of making arbitrary cut off, we based our threshold on a large set of subspecies I and core gene specific probes (20% of the total set) that are generally invariant in Salmonella and thereby obtaining higher statistical confidence. This is corroborated by the fact that in all reference and clinical samples the common gene probes were positive in most instances at the statistical criterion used. When the samples were clustered as a dendrogram, most of the clinical strains (except S. Thompson, S. Newport and S. Muenchen) clustered with the reference strains. As pointed out earlier, all isolates of a serovar may not always cluster together due to polyphyletic nature [19]. Polyphyletic behaviour is a situation where strains belonging to same serovar cluster with a different group in the microarray phylogenetic tree due to the differences in genes that are not used for serotyping. Hence the clustering pattern of these strains could be due to polyphyletic behavior. We observed a close relationship of S. Typhimurium with S. SaintPaul and S. Enteritidis with S. Dublin; this is in agreement with previous findings [19].
Serotyping of Salmonella is a very popular method employed worldwide. However this has many limitations like the long turnaround time high cost of producing antisera and lack of unified standardization among reference laboratories. Formerly different strategies have been used to develop microarrays for Salmonella serovar detection [13, 18, 23, 26]. As pointed out by Tankouo-Sandjong et al, MLST is a fast and cost effective method of molecular typing. However this method relies on a limited number of genes (3-5 genes) and does not reveal any information on the virulence and other variable genes which are far in excess of those included in MLST. On the other hand, the oligonucleotide array reported by Yoshida et al, contained 117 probes targeting O and H antigen genes [26]. Although this could detect several serovars, it was too focused on just two antigen regions as opposed to our array which has wider coverage. The study by Reen et al. employed a whole genome array of S. Typhimurium LT2 to compare S. Dublin, S. Agona and S. Typhimurium [21]. Although this type of whole genome arrays is powerful, for diagnostic applications these are not cost effective and also the data analysis will be difficult. Since most part of Salmonella genome is invariant, we have designed most probes based on the variable component that is less than 4-5 % of total genome of Salmonella. Thus instead of representing the whole genome we were able to limit the probe set to a small population without reducing the representation of diversity genes. Using this method we identified a unique gene profile for each of the 14 most common disease causing serovars in United States. One drawback of this method is that the identified gene profile is a mosaic structure and if one has to use this profile to identify an unknown test strain, it would be time consuming to do the profile matching manually. However, as seen in Fig.3, most clinical strains cluster with the reference strains and hence if the profile of the unknown strain is clustered with reference strains, one should be able to identify the serovar assignment based on the clustering behavior. As an alternative to clustering, we also developed a Matlab programme that performs automatic serovar assignments. Thus in addition to using for serovar detection of any of the 14 common disease causing Salmonella serovars in United States our array could also be used for the virulence and variable gene detection among these serovars.
Supplementary Material
Acknowledgments
This project was supported with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institute of Health, Department of Health and Human Services under contract, N01-AI-30054, Project No. ZC002-03 and the Federal Formula Fund from the Cornell University Agricultural Experiment Station.
References
- 1.Anjum MF, Marooney C, Fookes M, Baker S, Dougan G, Ivens A, Woodward MJ. Identification of core and variable components of the Salmonella enterica subspecies I genome by microarray. Infect Immun. 2005;73:7894–7905. doi: 10.1128/IAI.73.12.7894-7905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bopp CA, Brenner FW, WElls J, Strockbine NA. Escherichia, Shigella, and Salmonella. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. Vol. ASM Press; Washington, D.C.: 1999. [Google Scholar]
- 3.Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38:2465–2467. doi: 10.1128/jcm.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J Bacteriol. 2003;185:553–563. doi: 10.1128/JB.185.2.553-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu CH, Tang P, Chu C, Hu S, Bao Q, Yu J, Chou YY, Wang HS, Lee YS. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;33:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards RA, Olsen GJ, Maloy SR. Comparative genomics of closely related salmonellae. Trends Microbiol. 2002;10:94–99. doi: 10.1016/s0966-842x(01)02293-4. [DOI] [PubMed] [Google Scholar]
- 7.Esteban E, Snipes K, Hird D, Kasten R, Kinde H. Use of ribotyping for characterization of Salmonella serotypes. J Clin Microbiol. 1993;31:233–237. doi: 10.1128/jcm.31.2.233-237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezquerra E, Burnens A, Jones C, Stanley J. Genotypic typing and phylogenetic analysis of Salmonella paratyphi B and S. java with IS200. J Gen Microbiol. 1993;139:2409–2414. doi: 10.1099/00221287-139-10-2409. [DOI] [PubMed] [Google Scholar]
- 9.Frenzen PD, Riggs LT, Buzby JC, Breuer T, Roberts T, Voetsch D, Reddy S. Salmonella Cost Estimate Updated Using FoodNet Data. Foodreview. 1999;22:10–15. [Google Scholar]
- 10.Kimmel AR, OLiver B. Methods in Enzymology: DNA Microarrays- Part B: Databases and Statistics. Vol. 411. Elsevier Science & Technology Books; 2006. pp. 197–198. [Google Scholar]
- 11.Lagatolla C, Dolzani L, Tonin E, Lavenia A, Di Michele M, Tommasini T, Monti-Bragadin C. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J Clin Microbiol. 1996;34:2440–2443. doi: 10.1128/jcm.34.10.2440-2443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukinmaa S, Nakari UM, Eklund M, Siitonen A. Application of molecular genetic methods in diagnostics and epidemiology of food-borne bacterial pathogens. Apmis. 2004;112:908–929. doi: 10.1111/j.1600-0463.2004.apm11211-1213.x. [DOI] [PubMed] [Google Scholar]
- 13.Malorny B, Bunge C, Guerra B, Prietz S, Helmuth R. Molecular characterisation of Salmonella strains by an oligonucleotide multiprobe microarray. Mol Cell Probes. 2007;21:56–65. doi: 10.1016/j.mcp.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 14.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 15.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palaniappan RU, Zhang Y, Chiu D, Torres A, Debroy C, Whittam TS, Chang YF. Differentiation of Escherichia coli pathotypes by oligonucleotide spotted array. J Clin Microbiol. 2006;44:1495–1501. doi: 10.1128/JCM.44.4.1495-1501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 18.Pelludat C, Prager R, Tschape H, Rabsch W, Schuchhardt J, Hardt WD. Pilot study to evaluate microarray hybridization as a tool for Salmonella enterica serovar Typhimurium strain differentiation. J Clin Microbiol. 2005;43:4092–4106. doi: 10.1128/JCM.43.8.4092-4106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porwollik S, Boyd EF, Choy C, Cheng P, Florea L, Proctor E, McClelland M. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J Bacteriol. 2004;186:5883–5898. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porwollik S, Wong RM, Helm RA, Edwards KK, Calcutt M, Eisenstark A, McClelland M. DNA amplification and rearrangements in archival Salmonella enterica serovar Typhimurium LT2 cultures. J Bacteriol. 2004;186:1678–1682. doi: 10.1128/JB.186.6.1678-1682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reen FJ, Boyd EF, Porwollik S, Murphy BP, Gilroy D, Fanning S, McClelland M. Genomic comparisons of Salmonella enterica serovar Dublin, Agona, and Typhimurium strains recently isolated from milk filters and bovine samples from Ireland, using a Salmonella microarray. Appl Environ Microbiol. 2005;71:1616–1625. doi: 10.1128/AEM.71.3.1616-1625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selander RK, Caugant DA, Ochman H, Musser JM, Gilmour MN, Whittam TS. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tankouo-Sandjong B, Sessitsch A, Liebana E, Kornschober C, Allerberger F, Hachler H, Bodrossy L. MLST-v, multilocus sequence typing based on virulence genes, for molecular typing of Salmonella enterica subsp. enterica serovars. J Microbiol Methods. 2007;69:23–36. doi: 10.1016/j.mimet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38 3:S127–134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida C, Franklin K, Konczy P, McQuiston JR, Fields PI, Nash JH, Taboada EN, Rahn K. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J Microbiol Methods. 2007;70:261–271. doi: 10.1016/j.mimet.2007.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


