Summary
Members of the interleukin-1 (IL-1) family of cytokines play major roles in host defense and immune system regulation in infectious and inflammatory diseases. IL-1 cytokines trigger a biological response in effector cells by assembling a heterotrimeric signaling complex with two IL-1 receptor chains, a high-affinity primary receptor and a low-affinity co-receptor. To gain insights into the signaling mechanism of the novel IL-1-like cytokine IL-33, we first solved its solution structure and then performed a detailed biochemical and structural characterization of the interaction between IL-33, its primary receptor ST2 and the co-receptor IL-1RAcP. Using NMR data, we obtained a model of the IL-33/ST2 complex in solution that is validated by small-angle X-ray scattering (SAXS) data and is similar to the IL-1β/IL-1R1 complex. We extended our SAXS analysis to the IL-33/ST2/IL-1RAcP and IL-1β/IL-1R1/IL-1RAcP complexes and propose a general model of the molecular architecture of IL-1 ternary signaling complexes.
Introduction
The interleukin-1 (IL-1) family of immune regulatory cytokines in humans comprises a group of eleven secreted factors that include IL-1α, IL-1β, IL-18, and the most recent addition, IL-33 (for review, see Arend et al., 2008; Barksby et al., 2007; Dinarello, 2009). Several members of this cytokine family exhibit an unconventional secretion mechanism with their initial synthesis as leaderless precursors in the cytosol, and then activation by a caspase-1-mediated proteolytic cleavage of their N-terminal prodomains (IL-1β, IL-18, and IL-1F7; Keller et al., 2008; Sharma et al., 2008). Unexpectedly, proIL-33 does not need to be matured by caspase-1 for secretion and bioactivity (Lamkanfi and Dixit, 2009). The mature forms of these cytokines have a single β-trefoil domain that is shared with fibroblast growth factors (FGFs) (Ponting and Russell, 2000), and bind to signaling receptors of the IL-1 receptor family on their target cells. This class of receptors typically possess three extracellular immunoglobulin (Ig)-like repeats, and belong to the greater Toll-IL-1 receptor (TIR) superfamily by virtue of their intracellular signaling domain, a distinctive TIR module (Boraschi and Tagliabue, 2006; O’Neill, 2008). IL-1 cytokine binding drives the assembly of a heterodimer of IL-1 receptor chains, forming a ternary signaling complex with paired TIR domains. These recruit MyD88 and other cytosolic adaptor proteins leading ultimately to the activation of NF-κB and triggering of the proinflammatory response (Boraschi and Tagliabue, 2006; O’Neill, 2008).
A critical trio of proinflammatory cytokines, IL-1α, IL-1β and IL-18, can cause severe pathologic effects if their sustained production or signaling is not carefully regulated (Arend et al., 2008; Barksby et al., 2007; Dinarello, 2009). To this effect, a series of natural antagonists block the ordered assembly of selected IL-1 signaling complexes, revealing insights into the mechanisms of IL-1 receptor binding (Feldmann, 2008). IL-1 receptor antagonist (IL-1Ra) competes with IL-1α and IL-1β for binding to their primary receptor, the IL-1 receptor type 1 (IL-1R1). This IL-1Ra/IL-1R1 complex does not form a productive ternary complex with the co-receptor, the IL-1 receptor accessory protein (IL-1RAcP) (Arend et al., 2008; Barksby et al., 2007; Evans et al., 1995). By contrast, the IL-1 receptor type 2 (IL-1R2), which lacks an intracellular TIR domain, acts as a decoy receptor by competing with IL-1R1 for IL-1α and IL-1β binding. The IL-18 binding protein (IL-18BP) in turn comprises a secreted, single-Ig-domain sink for IL-18 (Arend et al., 2008; Barksby et al., 2007).
IL-33 is the ligand of the ST2 orphan IL-1 receptor (also known as T1, FIT-1 or DER4) (Dinarello, 2005; Schmitz et al., 2005). The signaling form of ST2 (or ST2L, the “long” transmembrane variant) is principally expressed in T-helper cell type 2 (Th2) and mast cells (Lohning et al., 1998; Trajkovic et al., 2004). Accordingly, IL-33 has been shown to be involved in Th2-mediated immune responses, such as asthma, allergy-induced inflammation and parasitic infections, and potently stimulates mast cells, basophils and eosinophils (Kakkar and Lee, 2008; Saenz et al., 2008; Schmitz et al., 2005). Gene loci for both ST2 and IL-33 have been flagged by genome-wide association scans using SNP arrays as linked to asthma, a Th2 immune disorder (Gudbjartsson et al., 2009). Intriguingly, the co-receptor for IL-33, IL-1RAcP, is promiscuously shared with the IL-1α/β signaling complex (Ali et al., 2007; Chackerian et al., 2007; Palmer et al., 2008). The structures of IL-1 family members IL-1α (Graves et al., 1990), IL-1β (Priestle et al., 1989), IL-1Ra (Vigers et al., 1994), IL-18 (Kato et al., 2003) and IL-1F5 (Dunn et al., 2003) share a core β-trefoil fold comprised of 12 β-strands arranged in a distinctive threefold symmetric pattern. How this cytokine fold engages its specific cellular receptors is only partly disclosed by the respective crystal structures of agonist IL-1β and antagonist IL-1Ra bound to the ectodomain (ECD) of IL-1R1 (Schreuder et al., 1997; Vigers et al., 1997). Both complexes show a common Ig-repeat architecture for the IL-1R1 ECD (domains D1 and D2 tightly interacting and linked by a disulfide bond, separated from D3 by an extended eight–residue linker) and differ only in the nature of the contacts with the bound cytokine. The loss of a critical D3 interaction by IL-1Ra (with respect to bound IL-1β) is the likely structural basis for antagonism (Schreuder et al., 1997). By contrast, another antagonist relies exclusively on the maintenance of this D3-cytokine contact for biological activity, as demonstrated by the structure of a poxvirus homolog of IL-18BP, a single D3-like Ig domain, bound to human IL-18 (Krumm et al., 2008). The relative independence of the D1-D2 Ig bloc from the D3 segment was evident in a complex of IL-1R1 with a short antagonistic peptide where D3 rotates by almost 180° to trap the peptide against the concave surface of D1-D2 (Vigers et al., 2000). This suggests that IL-1R1 and the extended family of IL-1 receptors can show significant conformational variability in the unbound state and adopt multiple bound structures.
The pose captured by the archetypal complex of IL-1β with IL-1R1 (Vigers et al., 1997) lacks the additional signaling chain required for bioactivity. The architecture of the full IL-1 ternary receptor complex remains elusive, though the expectation has been that a pseudosymmetric arrangement would provide the best model (Stroud and Wells, 2004; Wiesmann and de Vos, 2000). However, though the distantly related FGF β-trefoil cytokines bind to their Ig-repeat FGFR ectodomains in a similar fashion to IL-1β, the active FGFR assembly is a back-to-back receptor pair with outwardly mounted FGF ligands tied together by a heparan sulfate chain (Mohammadi et al., 2005). Heparin is not a requirement for IL-1 cytokine binding, and furthermore, the suggested 1:1:1 stoichiometry of primary receptor to co-receptor to IL-1 ligand is distinct from the FGF system (Boraschi and Tagliabue, 2006). With FGFs as an unreliable guide, one attempt to sort through IL-1 alternatives invoked the in silico docking of a molecular model of the co-receptor IL-1RAcP onto the existing IL-1β/IL-1R1 structure (Casadio et al., 2001). This resulted in two distinct configurations: in the ‘front’ model, the IL-1RAcP chain approaches the IL-1R1-bound IL-1β from the solvent-exposed face, while in the ‘back’ model, IL-1RAcP is placed on the opposite side of the engaged IL-1R1. A key experiment designed to distinguish between these different models utilized antibodies raised against an exposed loop of IL-1β; these antibodies do not impede IL-1RAcP ternary interaction, suggesting that the ‘front’ model is incorrect. This prediction clearly sets the IL-1 signaling architecture on a different course from other paradigmatic receptor complexes that are nucleated around a centrally placed cytokine ligand (Stroud and Wells, 2004; Wang et al., 2009; Wiesmann and de Vos, 2000). To resolve this issue of the molecular arrangement of IL-1 family heterotrimeric signaling complexes, in the present study we demonstrate the IL-1-like β-trefoil fold of the IL-33 cytokine by NMR spectroscopy, and characterize its sequential assembly with ST2 and IL-1RAcP receptor chains by biochemical methods, NMR and X-ray solution studies. To show that we define a consistent model for the greater IL-1 system, we employ a parallel structural study of IL-1β, IL-1R1 and IL-1RAcP molecules to draw a unified ternary architecture for IL-1 cytokine signaling.
Results
Solution structure of human IL-33
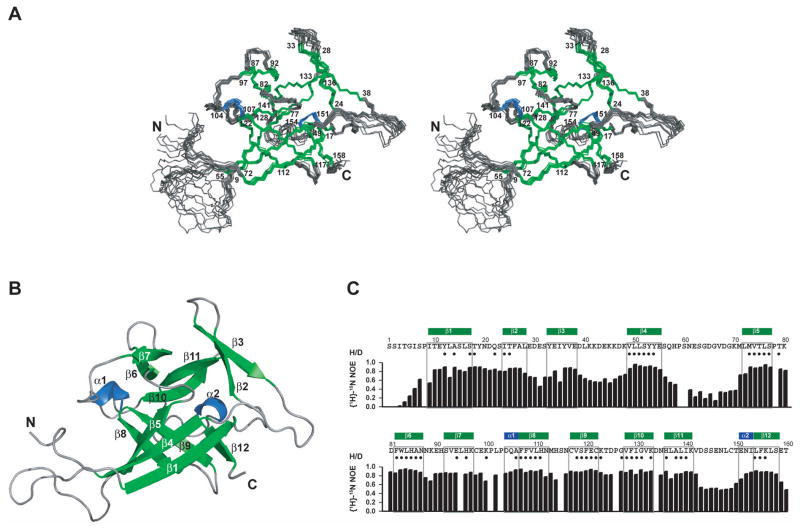
The IL-1-like fold of mature IL-33 was recognized by a sensitive computational screen utilizing sequence profiles based on the structural alignment of IL-1 and FGF β-trefoil cytokines (at a low identity level, 16% over 139 residues aligned with human IL-1β). To confirm the fold prediction, we have solved the solution structure of recombinant human IL-33 by multidimensional heteronuclear NMR spectroscopy (Figure 1A and 1B, Table 1). IL-33 indeed adopts a β-trefoil fold similar to that described for IL-1α, IL-1β, IL-1Ra and IL-18 (Figure 1B). In brief, the 12 β-strands of the β-trefoil (numbered β1-β 12) are arrayed in three pseudorepeats of four β-strand units, of which the first and last β-strands are antiparallel staves in a six-stranded β-barrel, while the second and third β-strands of each repeat form a β-hairpin sitting atop the β-barrel. Two short helices (α1 and α2) in IL-33 precede strands β8 and β12, and are located at equivalent positions to analogous helices in IL-1β and IL-18, respectively. Superposition by SSM (Krissinel and Henrick, 2004) of the lowest energy model of IL-33 with other IL-1 cytokine folds shows that the β-trefoil core is well conserved while differences apportion to the loops. In particular, the β4-β5 linker in IL-33 has a 10-amino acid loop insertion relative to other IL-1 cytokines (Figure S1); this loop is flexible and unstructured in solution as evidenced by low {1H}-15N heteronuclear NOE values (Figure 1C) and the lack of any long-range NOEs. Other regions of the protein with increased backbone dynamics (heteronuclear NOE values < 0.6) are the N-terminus (residues 1–9), the β3-β4 loop (residues 40–46), the β 11-β12 loop (residues 143–150) and the C-terminal tail (residues 159 and 160).
Figure 1. Solution structure and backbone dynamics of IL-33.
(A) Stereo view of the IL-33 ensemble. The ensemble of the 10 lowest-energy NMR structures is shown. Helices are shown in blue, β-strands in green. Secondary structure elements are labeled by residue numbers (numbering is based on the proposed caspase-1 cleavage site).
(B) Ribbon representation of IL-33, secondary structure elements are labeled.
(C) Additional NMR data on IL-33. The secondary structure is shown above the sequence. Solvent-protected amide protons (slow H/D exchange in NMR measurements) are indicated by filled circles. Heteronuclear {1H}-15N NOE data (black bars) report regions of IL-33 that are rigid and regions of increased backbone dynamics.
Table 1.
Structural statistics for the solution structure of IL-33
| Number of structural restraints | 〈SA〉* | 〈SA〉water refined* |
|---|---|---|
| NOE-derived distance restraints | ||
| Total (unambiguous/ambiguous) | 4693 (4661/32) | |
| intraresidual | 1714 (1712/2) | |
| sequential | 909 (898/11) | |
| medium range | 423 (419/4) | |
| long range | 1647 (1632/15) | |
| Dihedral restraints | 245 | |
| φ | 123 | |
| ψ | 122 | |
| Hydrogen bonds † | 76 | |
| Violations | ||
| R.m.s.d from experimental restraints | ||
| NOE distance restraints (Å) | 0.0055 ± 0.0006 | 0.0115 ± 0.0018 |
| Dihedral restraints (°) | 0.15 ± 0.02 | 0.43 ± 0. 05 |
| NOE distance violations | ||
| Number > 0.1 Å | 1.9 ± 0.9 | 13.6 ± 3.2 |
| Number > 0.3 Å | 0 | 0.7 ± 1.0 |
| Dihedral violations | ||
| Number > 1° | 1.3 ± 0.9 | 13.7 ± 2.8 |
| Number > 2° | 0 | 3.2 ± 1.3 |
| R.m.s.d. from idealized geometry | ||
| Bonds (Å) | 0.00114 ± 0.00003 | 0.00353 ± 0.00009 |
| Angles (°) | 0.278 ± 0.002 | 0.486 ± 0.010 |
| Impr (°) | 0.149 ± 0.007 | 1.518 ± 0.096 |
| Coordinate precision ¶ | ||
| Backbone (all residues) | 1.30 ± 0.27 | 1.36 ± 0.23 |
| All heavy atoms (all residues) | 1.50 ± 0.20 | 1.54 ± 0.18 |
| Backbone (structured regions) | 0.36 ± 0.05 | 0.46 ± 0.06 |
| All heavy atoms (structured regions) | 0.74 ± 0.06 | 0.81 ± 0.06 |
| Stereochemistry # | ||
| Ramachandran plot (%) (all residues) | ||
| Percentage in most favored regions | 75.2 ± 3.0 | 77.2 ± 1.6 |
| Percentage in additionally allowed regions | 23.7 ± 3.2 | 21.5 ± 1.7 |
| Percentage in generously allowed regions | 0.8 ± 0.8 | 0.8 ± 0.7 |
| Percentage in disallowed regions | 0.3 ± 0.6 | 0.6 ± 0.3 |
| Ramachandran plot (%) (structured regions) | ||
| Percentage in most favored regions | 82.6 ± 2.5 | 83.8 ± 2.0 |
| Percentage in additionally allowed regions | 17.4 ± 2.5 | 16.1 ± 2.2 |
| Percentage in generously allowed regions | 0 | 0.2 ± 0.4 |
| Energies (kcal mol−1) ‡ | ||
| ENOE | 7.5 ± 1.7 | 32. 2 ± 10 |
| ECDIH | 0.4 ± 0.1 | 2.9 ± 0.7 |
| Ebond | 3.3 ± 0.2 | 31.5 ± 1.6 |
| Eangles | 54.1 ± 0.8 | 165.0 ± 7.0 |
| Eimpr | 4.4 ± 0.4 | 458.5 ± 60 |
〈SA〉 is an ensemble of the ten lowest energy solution structures of IL-33 (out of 100 calculated). The CNS Erepel function was used to simulate van der Waals interactions with an energy constant of 25.0 kcal mol−1Å−4 using ‘PROLSQ’ van der Waals radii. 1 kcal=4.18 kJ. For 〈SA〉 water refined, the ensemble of 〈SA〉 structures was refined in a shell of water.
Hydrogen bonds were derived from slowly exchanging amide protons and were applied as two distance restraints with bounds of 1.8–2.3 Å (H–O) and 2.8–3.3 Å (N–O).
Coordinate precision is given as the Cartesian coordinate r.m.s.d. of the ten lowest-energy structures in the NMR ensemble with respect to their mean structure for the full length IL-33 and the structured regions of IL-33 (residues 10–39, 47–56, 71–142 and 151–158).
Structural quality was analyzed using PROCHECK (Laskowski et al., 1996) for full length IL-33 and for the structured regions of IL-33 (residues 10–39, 47–56, 71–142 and 151–158).
NOESY derived distance restraints were used with a soft square-well potential using an energy constant of 50 kcal mol−1 Å−2. Dihedral angle restraints were applied using an energy constant of 200 kcal mol−1 rad−2. The force constants were 1000 kcal mol−1 Å−2 for bond length and 500 kcal mol−1 rad−2 for angles and improper dihedrals.
Biochemical interaction of IL-33 with ST2 and IL-1RAcP
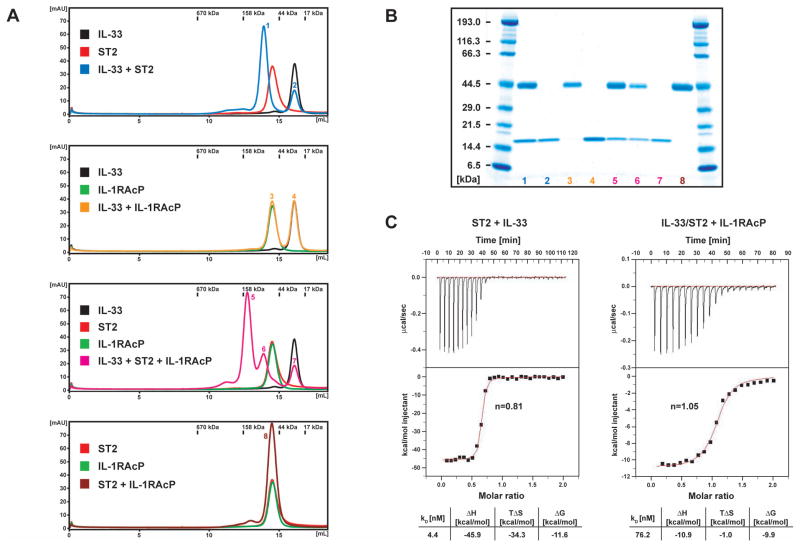
The primary, high-affinity receptor for IL-33 is the IL-1 receptor family member ST2. The IL-33/ST2 complex has been shown to use the same co-receptor as the IL-1β/IL-1R1 system, namely IL-1RAcP (Ali et al., 2007; Chackerian et al., 2007; Palmer et al., 2008). To characterize these interactions in solution, the ectodomains of ST2 and IL-1RAcP were produced by insect cells using the baculovirus expression system. Both receptor chains were secreted into the medium and purified to homogeneity in milligram quantities (Figure S2). To test whether the proteins were folded, we performed an initial binding analysis by size exclusion chromatography. The mixture of IL-33 and ST2 showed a decreased retention volume compared to ST2 alone, indicating complex formation between IL-33 and ST2 (Figure 2A and 2B). There was an additional shift towards higher molecular weight when all three proteins were combined, indicating ternary complex formation. However, addition of the IL-1RAcP co-receptor to either IL-33 or ST2 did not result in a change of the elution profile of the individual proteins. Lack of binding of IL-1RAcP to IL-33 with even low affinity was further confirmed by an NMR experiment, in which IL-1RAcP was added to IL-33 without any observable spectral changes (data not shown). We conclude that binding of IL-1RAcP co-receptor requires the initial complex formation between IL-33 and ST2 and furthermore does not engage either of the proteins alone. Two reasons could explain this observation: the binding site for the co-receptor is a composite surface with contributions from both IL-33 and ST2, and/or binding of the co-receptor requires a conformational shift in the ST2 primary receptor. Significant structural variation in IL-33 is unlikely, since IL-1β, IL-1Ra and IL-18 experience only minor changes upon binding to IL-1R1 and IL-18BP, respectively (Krumm et al., 2008; Schreuder et al., 1997; Vigers et al., 1997).
Figure 2. Binding of IL-33 to ST2 and IL-1RAcP.
(A) Size exclusion chromatography analysis
Left: Individual proteins were mixed and analyzed on a Superdex 200 10/300 GL column. The color of the individual traces matches the labeled boxes and numbers on peaks in the profiles correspond to lanes in the gel shown in (B). Protein molecular weights of a gel filtration standard are indicated above the chromatograms.
(B) SDS-Page of the individual peaks of the size exclusion chromatography analysis shown in (A).
(C) ITC binding curves. ST2 was titrated with IL-33 (left) and the IL-33/ST2 complex was titrated with IL-1RAcP (right). The binding curves were fit with a 1:1 binding model, resulting in the deduced stoichiometry n and the thermodynamic parameters listed in the table.
To gain a quantitative measure of the binding events during the formation of the ternary complex, we performed isothermal titration calorimetry (ITC) experiments. ST2 binds IL-33 with a dissociation constant of 4 nM and a stoichiometry of 1:1 (Figure 2C). Analysis of the thermodynamic contributions to complex formation reveals that the binding interaction is largely driven by enthalpy (ΔH ~ −46 kcal/mol). In agreement, the mapped binding interface between IL-33 and ST2 is mainly polar (see below). The large enthalpy compensates the considerable, unfavorable negative entropic term of the binding reaction (TΔS ~ −34 kcal/mol). This can be rationalized by a conformational change that the ST2 receptor has to undergo when binding to the cytokine. Although there are no structural data on unbound forms of any of the IL-1 receptors, it is highly likely that the linker between the D1-D2 Ig bloc and D3 is flexible and allows multiple ectodomain conformations (as discussed above). The negative entropy of the binding reaction can therefore be explained by the constraining of the conformational variability of ST2 on forming the cytokine complex.
The IL-33/ST2 complex in turn binds IL-1RAcP with a KD of 76 nM and a stoichiometry of 1:1:1 (Figure 2C). The interaction has a negative binding enthalpy as well (ΔH ~ −11 kcal/mol), although it is significantly smaller than that released during the binding of ST2 to IL-33. The reduction of entropy is small (TΔS ~ −1 kcal/mol), suggesting that any conformational fixing of the co-receptor upon assembly is compensated in large part by the release of water molecules from a hydrophobic area that is part of the binding interface. The interaction between the IL-33/ST2 complex and IL-1RAcP is therefore likely characterized by a combination of polar and non-polar contacts.
Chemical shift mapping of the interaction of IL-33 with ST2
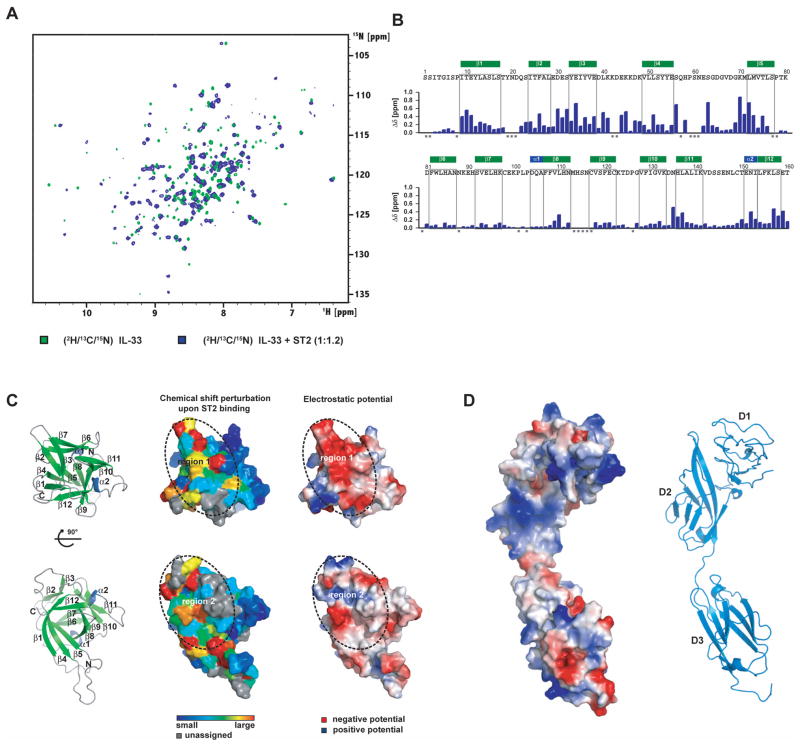
Addition of ST2 to IL-33 results in a large number of chemical shift perturbations suggesting an extensive interface between IL-33 and ST2 (Figure 3A). During the titration, resonances of the unbound IL-33 disappear gradually and new signals emerge at different locations in the spectrum. The binding is therefore in the slow exchange regime on the NMR time scale, as expected for a high-affinity interaction. The large degree of chemical shift changes and the type of interaction did not allow us to easily assign IL-33 in the complex by following the resonances of IL-33 during the titration with ST2. We therefore performed a completely new backbone assignment of IL-33 in complex with ST2 using fully 2H/13C/15N-labeled IL-33. This enabled assignment of ~ 90% of the backbone resonances of IL-33 in complex with ST2, and determination of chemical shift perturbations on a per-residue basis (Figure 3B). Residues with the largest changes form two clusters in the IL-33 fold, which are connected by residues that experience moderate changes (Figure 3B and 3C). The first cluster comprises the solvent exposed side of the β2-β3 hairpin, with E34 showing a very large chemical shift change, together with residues in the β10-β11 turn and residues in the β3-β4 loop, which is highly charged (Figure 3C). Two cross peaks (for K42 and D43) are too broad to assign, presumably due to exchange. A second cluster is formed by residues at the bottom of the β-barrel, notably residues at the N-termini of β1 and β5, in the β8-β9 turn (where the signals broaden due to binding), and the C-terminus of β12. Both spatial clusters in IL-33 overlap with the IL-1R1-binding surface of IL-1β, conveying a similarly positioned binding site for ST2. Absent an available structure for the human ST2 ectodomain, a comparative molecular model was built with MODELLER (Sali and Blundell, 1993) based on the IL-1R1 template taken from the IL-1β/IL-1R1 complex (PDB entry 1ITB). While the two receptor chains have a sequence identity of only 23% (over 308 aligned residues), key disulfide bonds are preserved along with the D1-D2 repeat segment structure; in addition, model quality assessment by VERIFY3D (quality score of 91.18 (Luthy et al., 1992)) is consistent with a good ST2 ectodomain model. The concave interaction surface of repeats D1-D2 is positively charged, complementing the negatively charged first residue cluster identified in IL-33 (Figure 3D). By contrast, the cytokine-facing surface of D3 is partly hydrophobic and partly electronegative, and represents therefore a complementary binding site for the second cluster region on IL-33.
Figure 3. NMR analysis of ST2 binding to IL-33.
(A) Chemical shift perturbation after addition of ST2 to 2H/13C/15N-labeled IL-33. Chemical shifts were monitored in 1H, 15N TROSY correlation spectra at 800 MHz 1H Larmor frequency. Slow exchange on the NMR chemical shift timescale between free and bound states indicates tight binding with nanomolar affinity.
(B) Bar diagram of chemical shift perturbations. Δδ is the weighted chemical shift change of IL-33 backbone amides upon addition of ST2 (see Supplemental Experimental Procedures). Gray squares indicate prolines and unassigned residues.
(C) Ribbon representation of the two binding regions, surface coloring according to chemical shift change and electrostatic surfaces of IL-33. Left panel: ribbon representation of IL-33 in the same orientation as in the other panels. The views in the upper and lower panel are correlated by a 90° rotation around the x-axis. Middle panel: Surface representation of IL-33 with residues colored according to the chemical shift perturbation upon binding of ST2. Prolines and unassigned residues are shown in gray. The two regions of major chemical shift changes are outlined by a dotted line and labeled. Right panel: Surface representation of IL-33 colored by electrostatic potential. White, blue and red corresponds to neutral, positive and negative electrostatic potential, respectively.
(D) Electrostatic surface and ribbon representation of the ST2 model Surface representation of the ST2 model colored by electrostatic potential. A ribbon diagram is shown in the same orientation with the individual Ig domains labeled.
The long β4-β5 loop, which is flexible in the free IL-33 (see above), is close to cluster 2 and contains several residues displaying chemical shift changes upon ST2 binding. Heteronuclear {1H}-15N NOE analysis of the complex reveals that this region is less flexible when IL-33 is bound to ST2, but still has NOE values lower than residues in regular secondary structure (data not shown). Binding analysis shows that the β4-β5 loop is dispensable for receptor interaction, as a mutated form of IL-33 lacking this loop (residues H58 to V67 deleted) is still capable of binding to ST2 with a similar affinity. This suggests that most of the chemical shift changes in this region are due to secondary binding effects.
In order to substantiate the predicted interface between IL-33 and ST2 and to search for possible ‘hot spot’ residues of this interaction, we introduced mutations based both on the amino acid clusters defined by the chemical shift perturbation data, and on residue conservation between IL-33, IL-1β and IL-18. In total, 28 mutations were made and all proteins were expressed in E. coli and purified (Figure S3) for binding trials. In order to screen quickly the large number of IL-33 variants for ST2 binding, we used a binding assay based on biolayer interferometry (Octet). Multiple independent screens revealed that only one of our mutants, E34K, had a pronounced effect on the interaction between IL-33 and ST2 (Figure S4). Surface plasmon resonance (Biacore) analysis provided a more precise comparison for the wild-type versus charge-reversal E34K IL-33 proteins in binding ST2 (Figure S5), showing a reduction in affinity for the mutant by seven fold from 0.45 nM to 3.11 nM. The absence of additional ‘hot spots’ implies that the binding energy for the interaction between IL-33 and ST2 is a sum of many small contributions spanning the large contact site. By contrast, for both the IL-1β/IL-1R1 (Boraschi et al., 1995; Evans et al., 1995) and IL-18/IL-18Rα (Kato et al., 2003; Kim et al., 2002; Krumm et al., 2008) interactions, similar individual mutations had a larger effect on the binding affinity.
Small-angle X-ray scattering analysis of the IL-33/ST2 complex
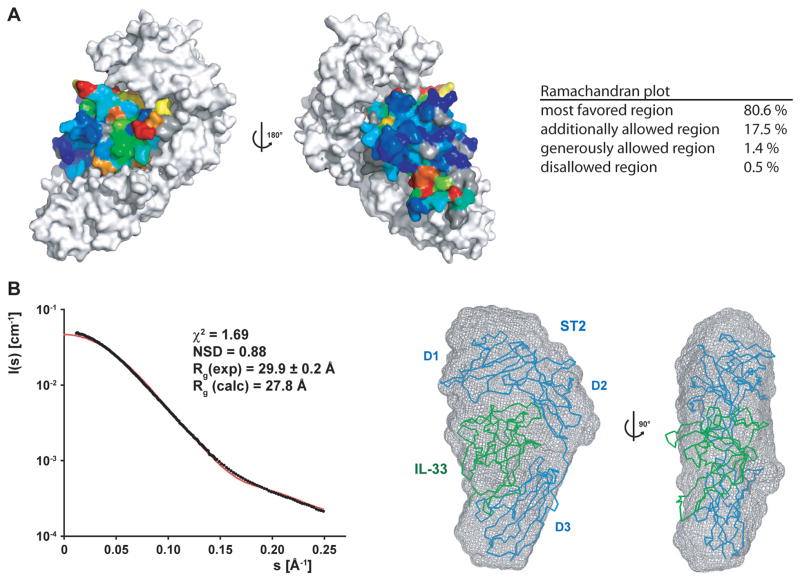
The chemical shift perturbation data suggest that the complex of IL-33 and ST2 is architecturally very similar to the IL-1β/IL-1R1 complex. With this assumption, we employed the program HADDOCK (Dominguez et al., 2003) to construct a model of the IL-33/ST2 complex. Restraints for interaction residues of IL-33 were defined based on the chemical shift perturbation data; residues in ST2 were chosen from the superposition of the IL-33 structure and ST2 model with the respective cytokine/receptor chains in the IL-1β/IL-1R1 complex. This latter structure also served as a starting pose for the IL-33/ST2 rigid-body docking step in HADDOCK. In the second docking step, loop sequences in IL-33 and ST2 were allowed to be fully flexible, resulting in an energy-minimized model of the IL-33/ST2 complex without steric clashes and with good stereochemistry (Figure 4A). To obtain independent structural information on the IL-33/ST2 complex, we performed small-angle X-ray scattering (SAXS) analysis, which provides information about the size and overall shape of macromolecules in solution (Putnam et al., 2007; Svergun and Koch, 2002). The calculated scattering curve of the IL-33/ST2 model fits the experimental data well, with a low χ2 value of 1.69 (Figure 4B), and a value for the radius of gyration (Rg) of the IL-33/ST2 complex derived from the experimental scattering curve as 29.9 Å–which is close to the back-calculated Rg of our IL-33/ST2 model (27.8 Å) using CRYSOL (Svergun et al., 1995). The shape of the complex in solution was also reconstructed from the experimental scattering curves using GASBOR (Svergun et al., 2001). 15 independent models were calculated and averaged using DAMAVER (Volkov and Svergun, 2003). The envelope of the reconstructed shape was superposed on the IL-33/ST2 model with SUBCOMB (Kozin and Svergun, 2001), with a very good fit showing a low normalized spatial discrepancy (NSD) of 0.88 (Figure 4B). Together, this provides strong evidence that our model of the heterodimeric complex is very close to the structure adopted in solution.
Figure 4. Model and SAXS validation of the IL-33/ST2 complex.
(A) A model of the IL-33/ST2 complex was generated based on chemical shift perturbation data on IL-33 and homology to the IL-1β/IL-1R1 complex using the molecular docking software HADDOCK. The surface of the ST2 receptor is shown in white, whereas the residues of IL-33 are colored according to the degree of chemical shift change (same coloring scheme as in Figure 3C). The views are correlated by a 180° rotation around the y-axis. The Ramachandran plot statistics are shown on the right.
(B) SAXS analysis of the IL-33/ST2 complex. Left: The experimental (black dots) and back-calculated scattering curves (red line) of the IL-33/ST2 complex are shown, the scattering intensities are displayed as a function of the momentum transfer s. The fitting statistics are shown above the two curves. Right: The IL-33/ST2 model was placed into the ab initio SAXS envelope; two orientations related by a 90° rotation are shown. IL-33 is colored in green and ST2 in blue.
Binding of IL-1RAcP to the IL-33/ST2 complex induces few additional chemical shift changes in IL-33
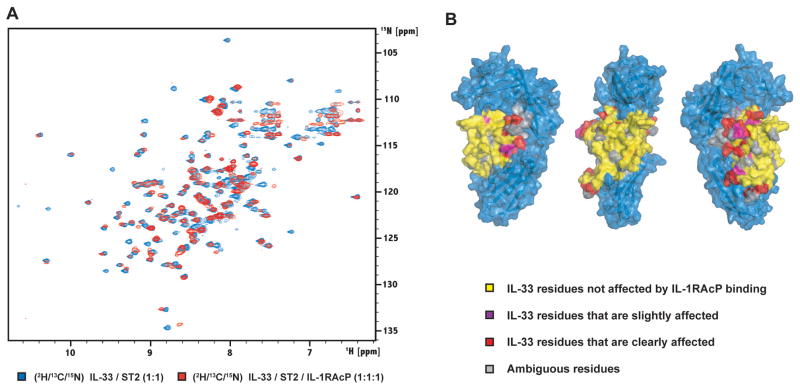
Having assigned the backbone resonances of IL-33 in the IL-33/ST2 heterodimer, we asked next if the addition of IL-1RAcP to create the ternary complex induces further resonance changes in IL-33, which could be explained by a direct interaction between the cytokine and the co-receptor. To this effect, IL-1RAcP was added in three steps to a purified complex of fully 2H/13C/15N-labeled IL-33 and ST2, until a 1:1:1 molar ratio was reached (Figure 5A). Similar to the interaction between IL-33 and ST2, we observed binding in the slow exchange regime on the NMR time scale. Due to the increasing molecular weight of the complex and the faster transverse relaxation, the peaks experience additional broadening which results in a decrease of intensity. Although most of the IL-33 resonances don’t change their position in the spectrum significantly, we observe some chemical shift perturbation for a number of peaks. Because it was not feasible to perform a new backbone assignment on the ternary complex, we performed a careful comparison of the TROSY spectra of the heterodimeric and ternary complexes and grouped residues into four different classes based on their assigned position in the heterodimeric complex: (1) no change, peak is at the same position in the spectrum of the ternary complex, (2) the peak position changed between half a line width and one line width, (3) the peak position changed by • one line width, and (4) residues for which the peak position information is missing. With this analysis, we found that addition of IL-1RAcP to the IL-33/ST2 complex only causes additional chemical shift perturbations of residues that are proximal to the interface between IL-33 and ST2 (Figure 5B). Strikingly, there were no changes observed for residues facing the solvent-exposed side of IL-33, showing that the co-receptor is not binding to this surface area.
Figure 5. Binding of the co-receptor monitored by NMR.
(A) Chemical shift perturbation after addition of IL-1RAcP to 2H/13C/15N-labeled IL-33 in complex with ST2. Chemical shifts were monitored in 1H, 15N TROSY correlation spectra at 900 MHz 1H Larmor frequency. Slow exchange on the NMR chemical shift timescale between free and bound states indicates tight binding with sub-micromolar affinity.
(B) IL-33 residues are colored according to the different classes used to analyze chemical shift perturbations upon addition of the IL-1RAcP co-receptor to the IL-33/ST2 complex (see text). The surface of ST2 is shown in blue.
As additional experiments that would probe the spatial positioning of the co-receptor chain in the ternary complex, two IL-33 variants were produced to further test the binding of both ST2 and IL-1RAcP. The first variant was an IL-33 C-terminal fusion (with a short Gly-Ser linker) to mannose-binding protein (MBP), and the second an insertion of green-fluorescent protein (GFP) into the β11-β12 loop of IL-33. Both IL-33 variants retain binding to both ST2 and IL-1RAcP, in spite of the large masses that ‘balloon’ out to the ‘right’ (MBP) and ‘left’ (GFP) sides of the disk-like IL-33/ST2 complex (Figure S6). A recent finding that pro-IL-33 is active and can bind to both of its receptors (Lamkanfi and Dixit, 2009) is consistent with our results, since the 111 residue prodomain would be fused to the N-terminal residue of IL-33, spatially close to the C-terminal MBP fusion.
Structural characterization of IL-1/IL-1 receptor holocomplexes by small-angle X-ray scattering
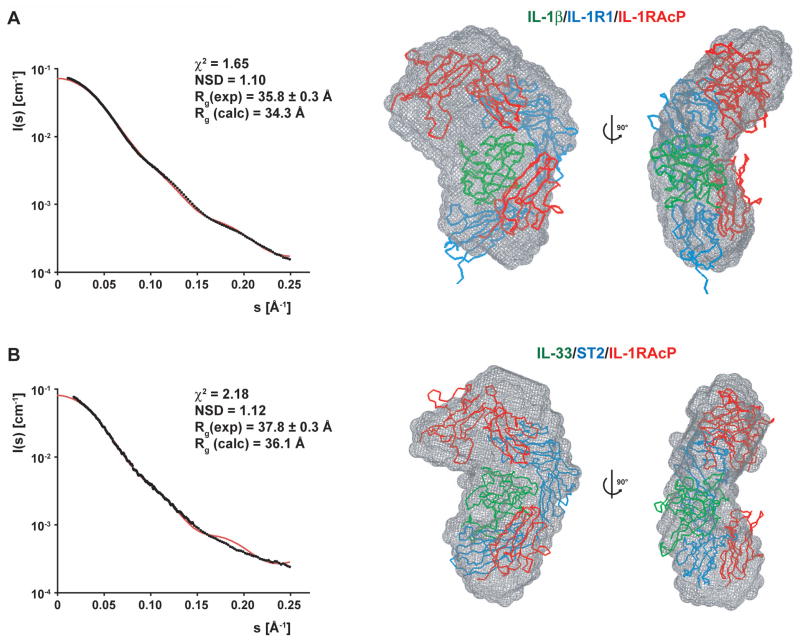
As there are no structural data available for the heterotrimeric complexes IL-1-like cytokines form with their respective receptors and co-receptors, we performed additional SAXS experiments on the homologous IL-1β/IL-1R1/IL-1RAcP and IL-33/ST2/IL-1RAcP complexes. Since IL-1RAcP is the common co-receptor chain for both IL-1β and IL-33 cytokines, a similar spatial engagement is expected for both ternary complexes. Analysis of the scattering curves of the IL-1β- and IL-33-containing heterotrimers resulted in very similar Rg values of 35.8 Å and 37.3 Å for the IL-1β/IL-1R1/IL-1RAcP and IL-33/ST2/IL-1RAcP complexes, respectively. Furthermore, the ab initio reconstruction-derived SAXS envelopes look globally similar with a NSD of 0.64 (Figure S7). Thus, the IL-1 cytokine and its two receptor chains seem to adopt an equivalent and conserved arrangement with a curved kidney-like shape.
To reveal the position of the individual proteins in the trimeric complexes, we applied the unbiased approach of preparing possible models of the holocomplexes and testing which of them best reflects the experimental scattering curve. As building block components of the respective complexes, we took the crystal structure of the IL-1β/IL-1R1 complex and the calculated model of the IL-33/ST2 complex, together with the common subunit, a homology model of the co-receptor (also built on the IL-1R1 template). The IL-1RAcP ectodomain features the family-distinctive set of three Ig repeats. D1 and D2 are very likely interacting closely with each other as in IL-1R1 and ST2 because of the conserved register of Cys residues; likewise, a flexible linker of 7 amino acids links domain D3 to the D1-D2 segment. Given the reasonable assumption that the linker can adopt a wide range of conformations, we independently positioned the Ig repeat segments D1-D2 and D3 within a distance that can be bridged by the linker (~ 25 Å). Although this procedure is introducing an additional degree of uncertainty to our model building approach, it allows us to account for the expected plasticity of the linker.
All models in which the co-receptor binds over the solvent-exposed surface of the cytokine did not correspond to the experimental scattering curves, resulting in very high χ2 values (Figure S8A and S8B). This agrees well with the observation that residues of the solvent-exposed surface of IL-33 did not experience any chemical shift perturbations when we added IL-1RAcP to the IL-33/ST2 heterocomplex. We can thus rule out any arrangement of the trimeric complexes that involves binding of the co-receptor from the ‘front’. Even higher χ2 values were observed for models wrapping IL-1RAcP on the reverse side of the dimeric complexes in a diagonal fashion, as previously proposed in the ‘back’ model of Casadio et al. (Casadio et al., 2001) (Figure S8C and S8D). A lower χ2 value is observed for a model that has the co-receptor mounted completely on the convex spine of the primary receptor in parallel (Figure S8E), supporting the notion that the trimeric complex has a much thinner profile than the ‘front’ or the ‘back’ models. However, the χ2 value of this arrangement is still above a value that gives confidence that the model is optimally representing the particle in solution. Therefore, we also investigated models that place the co-receptor in parallel next to the primary receptor on either side (akin to the FGFR homodimer stance; (Mohammadi et al., 2005), which resulted in slightly worse statistics (Figure S9A and S9B). We could improve the agreement between the back-calculated scattering curves and the experimental curves by moving the IL-1RAcP chain upwards when the co-receptor is placed on the ‘right’ side of the dimeric complex seen from the front. This resulted in low χ2 values of 1.65 and 2.19 for the IL-1β/IL-1R1/IL-1RAcP and IL-33/ST2/IL-1RAcP complexes, respectively (Figure 6A and 6B). Furthermore, the complexes fit well into the reconstructed shapes with NSDs of 1.10 and 1.12. In these models, the D3 module of IL-1RAcP is packed close to the D3 domains of ST2 or IL-1R1, bringing the two C-termini of the signaling receptor chains into proximity. The D1-D2 segment of the co-receptor is placed close to the top-mounted D1 domain of the primary receptor in such a fashion that the D2 domain of IL-1RAcP interacts with the D1 module and the D1-D2 interface of ST2 and IL-1R1. In our models, the flexible linker between the separated D1-D2 and D3 IL-1RAcP domains bridges 24.9 and 22.3 Å distances respectively in the IL-1β and IL-33 ternary complexes.
Figure 6. Characterization of the IL-1β/IL-1R1/IL-1RAcP and IL-33/ST2/IL-1RAcP complexes by SAXS analysis.
(A) SAXS data from the IL-1β/IL-1R1/IL-1RAcP complex. Left: The experimental scattering curve is shown with black dots, the scattering profile of the IL-1β/IL-1R1/IL-1RAcP model is shown as a red line. The fitting statistics are shown above the two curves. Right: The IL-1β/IL-1R1/IL-1RAcP model was placed into the ab initio SAXS envelope; two orientations related by a 90° rotation are shown. IL-1β is colored in green, IL-1R1 in blue, and IL-1RAcP in red.
(B) SAXS data from the IL-33/ST2/IL-1RAcP complex. Left: The experimental scattering curve is shown with black dots, the scattering profile of the IL-33/ST2/IL-1RAcP model is shown as a red line. The fitting statistics are shown above the two curves. Right: The IL-33/ST2/IL-1RAcP model was placed into the ab initio SAXS envelope; two orientations related by a 90° rotation are shown. IL-33 is colored in green, ST2 in blue and IL-1RAcP in red.
Discussion
The modular architecture of IL-1 receptors places them at the evolutionary crossroads of two ancient structural superfamilies, with a triple-Ig-repeat ectodomain that recognizes β-trefoil-class cytokines, an interaction conserved between IL-1 and FGF cytokine/receptor families (Ponting and Russell, 2000; Vigers et al., 1997), and a cytosolic TIR domain shared with Toll-like receptors (TLRs) that links the respective molecules to a common NF-κB signaling pathway (O’Neill, 2008). However, FGFRs differently harness cytokine binding to dimerization of a cytosolic tyrosine kinase domain, while pathogen recognition drives the assembly of distinctive Leu-rich repeat TLR ectodomains to trigger intracellular TIR signaling (Jin and Lee, 2008; Mohammadi et al., 2005). Modular diversity aside, these structurally interlinked families of receptors share a common functional imperative: cytokine (or pathogen-derived molecule, in the case of TLRs) binding drives the oligomerization of receptor chains into a ternary complex that critically bundles together transmembrane helices and cytosolic signaling domains. The attendant cytokine recognition and assembly mechanisms of IL-1 receptors were only partly disclosed by the structure of the IL-1β/IL-1R1 heterodimer complex (Vigers et al., 1997), which further left open the inviting prospect of a face-to-face (or pseudosymmetric) docking of the unseen IL-1RAcP co-receptor chain. This ‘front’ binding model has been questioned by Casadio et al. (Casadio et al., 2001), who suggested that the IL-1RAcP co-receptor adopts an alternative and unorthodox orientation in the ternary complex. The present work strives to answer this outstanding question by applying NMR-based structural, biochemical and SAXS methods to show how IL-33 interacts with both its primary receptor ST2 and co-receptor IL-1RAcP; we further extend our studies to the homologous IL-1β cytokine/receptor complex.
The solution structure of human IL-33 shows that this distant member of the IL-1 family adopts a β-trefoil fold similar to that of the other IL-1 cytokines, but with a unique insert of approximately 10 residues in the β4-β5 loop. This insert is flexible in solution and is not required for folding or solubility. IL-33 binds its primary receptor ST2 in solution with high affinity, a 1:1 stoichiometry and an extensive buried surface area of contact. As deduced from chemical shift binding data, the interaction site in IL-33 comprises two separated contact regions: region 1 includes mainly strand β3 and the β1-β2 and β3-β4 loops. E34 in β3 provides a large contribution to the binding energy, as a charge-reversal mutation decreases the affinity by approximately 7-fold; region 2 is otherwise located at the base of the β-barrel. Drawing from the crystal structure of the IL-1β/IL-1R1 complex, IL-1β is likewise shown to have two detached binding sites for IL-1R1: the one referred to as site A nestles against the D1-D2 Ig-repeat segment whereas the other site B contacts D3 (Vigers et al., 1997). The interaction mode between cytokines and their cognate receptors tends to be strongly conserved in families despite great sequence divergence (Stroud and Wells, 2004), so we expect that the IL-1β/IL-1R1 binding mode is preserved in the interaction of IL-33 with its primary receptor, where region 1 of IL-33 would bind the ST2 D1-D2 segment, and region 2 would engage D3. Our model of the IL-33/ST2 complex based on the chemical shift perturbation data and homology to the IL-1β/IL-1R1 complex supports these predictions. Furthermore, the solution scattering curve of the IL-33/ST2 complex validates our model, which fits also very well into the SAXS-derived shape reconstruction. This result represents the first structural demonstration of the architecture of IL-1-like cytokines bound to their primary receptor in solution.
Using purified recombinant proteins, we have shown that IL-1RAcP binds to the complex of IL-33 and ST2 with a 1:1:1 stoichiometry, forming a stable heterotrimeric assembly. Casadio et al. showed that an antibody directed against residues in the β6-β7 turn and strand β7 of IL-1β (residues 73–81) did not inhibit the biological effect of IL-1β in cell-based assays, thus disfavoring their computationally-derived ‘front’ model for the interaction of IL-1RAcP with the IL-1β/IL-1R1 complex (Casadio et al., 2001). A similar conclusion can be drawn from a study showing that mutations in the residue 74–80 area did not affect the biological activity of IL-1β (Simon et al., 1993). By contrast, an antibody directed against the β5-β6 turn and strand β6 of IL-1β (residues 61–70) inhibited IL-1β-directed signaling without interfering with IL-1R1 binding (Casadio et al., 2001; D’Ettorre et al., 1997). In a study that attempted to map the binding site for IL-1β/IL-1R1 on IL-1RAcP using antibodies raised against various Ig repeat-derived co-receptor peptides, Yoon et al. reported data suggesting that repeats D2 and D3 of IL-1RAcP are primarily involved in the interaction with IL-1β/IL-1R1 (Yoon and Dinarello, 1998). In addition, the juxtamembrane module D3 of the co-receptor appeared important for proper dimerization of the intracellular chains of IL-1R1 and IL-1RAcP, which is required for activation. Interestingly, the IL-1β antagonist, IL-1Ra, engages the D1-D2 segment of IL-1R1 almost exclusively (with a binding surface corresponding to our site A), whereas few contacts are maintained with D3, which appears rotated away by 20° in the IL-1Ra/IL-1R1 complex compared to the IL-1β/IL-1R1 structure (Schreuder et al., 1997). Furthermore, the IL-1Ra/IL-1R1 heterodimer does not bind the co-receptor chain, suggesting that the area of greatest structural divergence with the agonist IL-1β complex–the vicinity of the cytokine-disengaged D3 domain–might be involved in the loss of recognition of IL-1RAcP.
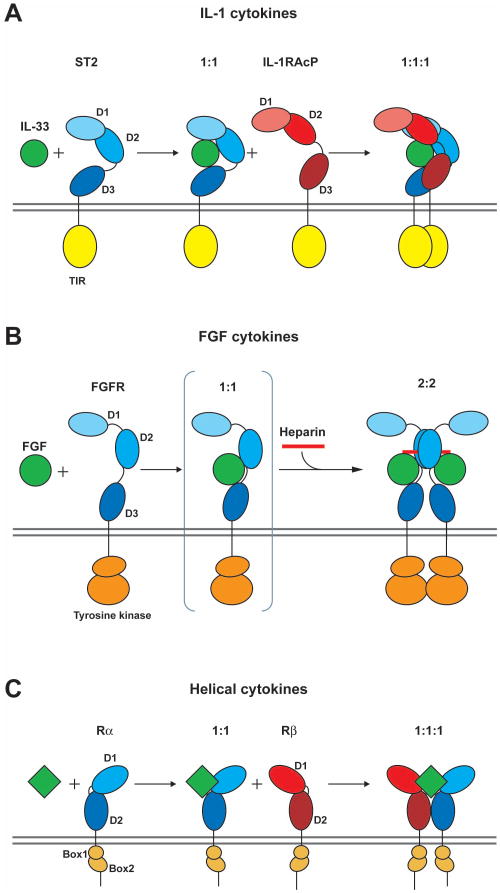
Our NMR-based study of the interaction of the IL-33/ST2 complex with IL-1RAcP and the SAXS analysis of the IL-1β and IL-33 holocomplexes clearly rule out any ternary arrangement that places the co-receptor on the open, cytokine-exposed ‘front’ face of the heterodimeric complexes. However, our SAXS data are also clearly incompatible with ‘back’ models where the co-receptor wraps across the convex spine of the primary receptor, and instead we provide evidence that both IL-1β and IL-33 holocomplexes feature a similar and conserved binding mode of IL-1RAcP to the ‘right’ side of both complexes. The proposed models reveal a novel architecture of IL-1-type β-trefoil cytokine signaling complexes. Receptor-ligand complexes formed by the FGF β-trefoil cytokines minimally comprise two FGF molecules and two identical FGFR chains in a 2:2 stoichiometry, with symmetrically placed heterocomplexes bridged by a heparin strand (Figure 7B) (Mohammadi et al., 2005). Only FGFR Ig repeats D2 and D3 are needed to bind cytokine, and furthermore, the receptor homodimer contact is mediated mainly by D2. In contrast, IL-1 cytokines bind their primary receptor in a 1:1 stoichiometry and therefore need to recruit a second and distinct co-receptor chain to form the bioactive 1:1:1 heterotrimeric complex. This staged assembly resembles the strategy of helical cytokine complexes, with important architectural differences: four-helix bundle cytokines bind in the heart of the complex between two different receptor chains (Figure 7C) (Wang et al., 2009). Surprisingly, the assembly of IL-1 receptors is not centered around the ligand in a pseudosymmetric fashion and is thus counter to most other cytokine systems. In our present model, the IL-1RAcP co-receptor interacts mainly with Ig repeats D1 and D3 of the primary receptor (ST2 in the case of IL-33, IL-1R1 for IL-1β) and makes minor contacts with the bound cytokine, principally along the seam of its binding interface with the primary receptor (Figure 7A). Since we cannot measure any interaction in solution between IL-1RAcP and either cytokine or primary receptor alone, the docking surface for co-receptor on the engaged IL-33/ST2 or IL-1β/IL-1R1 heterodimers must only be present in the bound configuration. Taken together, our models of the IL-33 and IL-1β holocomplexes derived from a combination of NMR structural and binding analysis, and low-resolution SAXS data, are consistent with the majority of experimental data available so far and advance a novel mode of cytokine-receptor ternary association. Our structural model of the IL-33 receptor signaling complex has broad mechanistic implications for the larger IL-1 cytokine/receptor family, as the close architectural resemblance to the IL-1β complex attests, but in particular, it should facilitate the design and targeting of therapeutics that specifically (positively or negatively) regulate the IL-33 signal.
Figure 7. Cartoons comparing the formation of active signaling complexes for IL-1, FGF, and helical cytokines.
(A) The IL-33 system is shown as a representative for the IL-1 cytokine family. IL-33 (green) is binding to the ST2 receptor (blue): complex formation enables the recruitment of the co-receptor IL-1RAcP (red). In the ternary 1:1:1 complex, the two cytoplasmic TIR domains (yellow) are proximal resulting in activation of the signaling cascade.
(B) In contrast, FGF cytokines (green) bind to FGF receptor (blue) forming a 2:2 complex in the presence of heparin (red). The proximity of the intracellular tyrosine kinase domains (orange) leads to activation.
(C) In the helical cytokine system, the ternary signaling complex also consists of the cytokine and a pair of receptors (primary Rα and secondary Rβ). This can be a homodimeric pair of receptor chains in the case of growth hormones, or a heterodimeric pair of receptor chains as seen for the short chain helical cytokines like IL-2, IL-4/13 or GMCSF. Activation is achieved by clustering of intracellular chains bearing JAK family kinases bound to conserved, juxtamembrane Box1–2 motifs.
Experimental Procedures
Protein preparation
Human IL-33 and IL-1β were expressed in E. coli as an N-terminal His-tag fusion protein and as a fusion protein with an N-terminal His-tagged maltose-binding protein, respectively. For uniform 15N and 15N/13C labeling, cells were grown in M9 minimal medium supplemented with 15NH4Cl without or with 13C6-glucose. For the preparation of 2H/13C/15N-labeled sample, the cells were grown in minimal medium prepared with 100% D2O and supplemented with 15NH4Cl and 2H/13C-rich glucose. His-tagged fusion proteins were affinity-purified, followed by protease cleavage. After removing the tags, final purification was by gel filtration.
The extracellular domains of ST2, IL-1RAcP, and IL-1R1 were secreted from insect cells as His-tag fusion proteins. Supernatants were subjected to affinity purification, the eluted fusion proteins protease cleaved and further purified by size exclusion chromatography. For SAXS measurements, the insect cell-secreted receptors were deglycosylated using a mixture of four different enzymes: F1, F2, F3 and Endo H (all QA-Bio).
NMR spectroscopy
NMR spectra of IL-33 were analyzed using the CCPN analysis software (Vranken et al., 2005). 1H, 13C and 15N chemical shifts and sets of distance and dihedral restraints were determined by standard methods. Slowly exchanging amide protons were identified from 1H, 15N correlation experiments after redissolving lyophilized protein in D2O. Heteronuclear {1H}–15N NOE was measured on a 15N-labeled IL-33 sample at 35°C and 800 MHz 1H Larmor frequency and analyzed with the CCPN analysis software.
The IL-33/ST2 complex for analysis by NMR spectroscopy was formed by titrating the 2H/13C/15N-labeled IL-33 with ST2 protein until a molar stoichiometry of 1:1.2 was reached. For backbone assignments of the complex, TROSY versions of backbone experiments were employed. The ternary complex comprised of IL-33, ST2 and IL-1RAcP was prepared by adding IL-1RAcP to purified IL-33/ST2 complex, in which IL-33 was 2H/13C/15N-labeled. Changes were observed by TROSY 1H, 15N correlation spectra.
Structure calculation
The experimentally determined distance and dihedral restraints for IL-33 (see Table 1) were applied in a simulated annealing protocol using ARIA2.2 (Rieping et al., 2007) and CNS (Brunger et al., 1998). The final ensemble of NMR structures was refined in a shell of water molecules (Linge et al., 2003). Structural quality was analyzed using PROCHECK (Laskowski et al., 1996). Ribbon and surface representations were prepared with PyMOL (DeLano Scientific).
Isothermal titration calorimetry
Proteins were dialyzed in 50 mM sodium phosphate pH 6.5, 150 mM NaCl and afterwards measured using a VTC calorimeter (Microcal) at 30°C. For the analysis of the interaction between IL-33 and ST2, the sample cell was loaded with 2.5 μM ST2 and IL-33 was added over 20 injections of 5 μL of a 50 μM solution. Interaction of the IL-33/ST2 complex with IL-1RAcP was measured using 5 μM of the purified complex in the sample cell and injecting 20 × 5 μL of 75 μM IL-1RAcP.
Octet and Biacore analysis
Protein-protein interactions were measured by biolayer interferometry using the Octet QK system (ForteBio). Streptavidin biosensors were loaded with 10 μg/mL biotinylated ST2, followed by an association of IL-33 at 20 nM concentration and a dissociation phase in the absence of IL-33.
For all Biacore experiments, the running and dilution buffer was HBS-EP. A CM5 chip was coated with ~7000 RU of anti-human IgG Fc antibody per flow cell using amine coupling chemistry. An ST2-Fc fusion protein (R&D Systems) was then immobilized on the surface of one of the flow cells and a control-Fc fusion protein in the other flow cell. IL-33 was injected at different concentrations and association/dissociation phases were obtained by subtracting the signal of the control flow cell.
Small-angle X-ray scattering experiments
The small-angle X-ray scattering (SAXS) data were collected at the beamline 4-2 of the Stanford Synchrotron Radiation Light Source (SSRL) (Smolsky et al., 2007). Sample-to-detector distances of 0.5 m and 2.5 m were used to cover the Q range 0.01 – 0.3 Å−1 (Q is the momentum transfer 4 sin(θ/2)/λ with θ and λ being the scattering angle and X-ray wavelength, respectively). Details of the SAXS data analysis are included in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Jeff Pelton at the Central California NMR Center (University of California, Berkeley) for measurement time and instrumental help and the protein expression group at Genentech for expressing ST2, IL-1RAcP and IL-1R1 proteins. SAXS experiments were carried out at the Stanford Synchrotron Radiation Light Source. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program. A.L., B.P., C.W., J.F.B, and W.J.F. are employees of Genentech, Inc.
Footnotes
Protein Data Bank accession number
The coordinates and the NMR data of IL-33 have been deposited at the Protein Data Bank and the BioMagResBank under accession codes 2kll and 16317, respectively.
Detailed Experimental Procedures, Supplemental Figures S1–S9.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali S, Huber M, Kollewe C, Bischoff SC, Falk W, Martin MU. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc Natl Acad Sci USA. 2007;104:18660–18665. doi: 10.1073/pnas.0705939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- Barksby HE, Lea SR, Preshaw PM, Taylor JJ. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol. 2007;149:217–225. doi: 10.1111/j.1365-2249.2007.03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraschi D, Bossu P, Ruggiero P, Tagliabue A, Bertini R, Macchia G, Gasbarro C, Pellegrini L, Melillo G, Ulisse E, et al. Mapping of receptor binding sites on IL-1 beta by reconstruction of IL-1ra-like domains. J Immunol. 1995;155:4719–4725. [PubMed] [Google Scholar]
- Boraschi D, Tagliabue A. The interleukin-1 receptor family. Vitam Horm. 2006;74:229–254. doi: 10.1016/S0083-6729(06)74009-2. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr, D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Casadio R, Frigimelica E, Bossu P, Neumann D, Martin MU, Tagliabue A, Boraschi D. Model of interaction of the IL-1 receptor accessory protein IL-1RAcP with the IL-1beta/IL-1R(I) complex. FEBS Lett. 2001;499:65–68. doi: 10.1016/s0014-5793(01)02515-7. [DOI] [PubMed] [Google Scholar]
- Chackerian AA, Oldham ER, Murphy EE, Schmitz J, Pflanz S, Kastelein RA. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179:2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- D’Ettorre C, De Chiara G, Casadei R, Boraschi D, Tagliabue A. Functional epitope mapping of human interleukin-1 beta by surface plasmon resonance. Eur Cytokine Network. 1997;8:161–171. [PubMed] [Google Scholar]
- Dinarello CA. An IL-1 family member requires caspase-1 processing and signals through the ST2 receptor. Immunity. 2005;23:461–462. doi: 10.1016/j.immuni.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Dunn EF, Gay NJ, Bristow AF, Gearing DP, O’Neill LA, Pei XY. High-resolution structure of murine interleukin 1 homologue IL-1F5 reveals unique loop conformations for receptor binding specificity. Biochemistry. 2003;42:10938–10944. doi: 10.1021/bi0341197. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Bray J, Childs JD, Vigers GP, Brandhuber BJ, Skalicky JJ, Thompson RC, Eisenberg SP. Mapping receptor binding sites in interleukin (IL)-1 receptor antagonist and IL-1 beta by site-directed mutagenesis. Identification of a single site in IL-1ra and two sites in IL-1 beta. J Biol Chem. 1995;270:11477–11483. doi: 10.1074/jbc.270.19.11477. [DOI] [PubMed] [Google Scholar]
- Feldmann M. Many cytokines are very useful therapeutic targets in disease. J Clin Invest. 2008;118:3533–3536. doi: 10.1172/JCI37346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves BJ, Hatada MH, Hendrickson WA, Miller JK, Madison VS, Satow Y. Structure of interleukin 1 alpha at 2.7-A resolution. Biochemistry. 1990;29:2679–2684. doi: 10.1021/bi00463a009. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342 – 347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008;7:827–840. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Z, Jee J, Shikano H, Mishima M, Ohki I, Ohnishi H, Li A, Hashimoto K, Matsukuma E, Omoya K, et al. The structure and binding mode of interleukin-18. Nat Struct Biol. 2003;10:966–971. doi: 10.1038/nsb993. [DOI] [PubMed] [Google Scholar]
- Keller M, Ruegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- Kim SH, Azam T, Novick D, Yoon DY, Reznikov LL, Bufler P, Rubinstein M, Dinarello CA. Identification of amino acid residues critical for biological activity in human interleukin-18. J Biol Chem. 2002;277:10998–11003. doi: 10.1074/jbc.M108311200. [DOI] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr, D: Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krumm B, Meng X, Li Y, Xiang Y, Deng J. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proc Natl Acad Sci USA. 2008;105:20711–20715. doi: 10.1073/pnas.0809086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. IL-33 Raises Alarm. Immunity. 2009;31:5–7. doi: 10.1016/j.immuni.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Lohning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez-Ramos JC, Levinson D, Radbruch A, Kamradt T. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA. 1998;95:6930–6935. doi: 10.1073/pnas.95.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- Palmer G, Lipsky BP, Smithgall MD, Meininger D, Siu S, Talabot-Ayer D, Gabay C, Smith DE. The IL-1 receptor accessory protein (AcP) is required for IL-33 signaling and soluble AcP enhances the ability of soluble ST2 to inhibit IL-33. Cytokine. 2008;42:358–364. doi: 10.1016/j.cyto.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Russell RB. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all beta-trefoil proteins. J Mol Biol. 2000;302:1041–1047. doi: 10.1006/jmbi.2000.4087. [DOI] [PubMed] [Google Scholar]
- Priestle JP, Schar HP, Grutter MG. Crystallographic refinement of interleukin 1 beta at 2.0 A resolution. Proc Natl Acad Sci USA. 1989;86:9667–9671. doi: 10.1073/pnas.86.24.9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Schreuder H, Tardif C, Trump-Kallmeyer S, Soffientini A, Sarubbi E, Akeson A, Bowlin T, Yanofsky S, Barrett RW. A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist. Nature. 1997;386:194–200. doi: 10.1038/386194a0. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kulk N, Nold MF, Graf R, Kim SH, Reinhardt D, Dinarello CA, Bufler P. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines. J Immunol. 2008;180:5477–5482. doi: 10.4049/jimmunol.180.8.5477. [DOI] [PubMed] [Google Scholar]
- Simon PL, Kumar V, Lillquist JS, Bhatnagar P, Einstein R, Lee J, Porter T, Green D, Sathe G, Young PR. Mapping of neutralizing epitopes and the receptor binding site of human interleukin 1 beta. J Biol Chem. 1993;268:9771–9779. [PubMed] [Google Scholar]
- Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle x-ray scattering facility at the Stanford synchrotron radiation laboratory. J Appl Crystallogr. 2007;40:S453–S458. [Google Scholar]
- Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE. 2004;23:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI, Koch MH. Advances in structure analysis using small-angle scattering in solution. Curr Opin Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic V, Sweet MJ, Xu D. T1/ST2--an IL-1 receptor-like modulator of immune responses. Cytokine Growth Factor Rev. 2004;15:87–95. doi: 10.1016/j.cytogfr.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Anderson LJ, Caffes P, Brandhuber BJ. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1beta. Nature. 1997;386:190–194. doi: 10.1038/386190a0. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Caffes P, Evans RJ, Thompson RC, Eisenberg SP, Brandhuber BJ. X-ray structure of interleukin-1 receptor antagonist at 2.0-A resolution. The Journal of biological chemistry. 1994;269:12874–12879. [PubMed] [Google Scholar]
- Vigers GP, Dripps DJ, Edwards CK, 3rd, Brandhuber BJ. X-ray crystal structure of a small antagonist peptide bound to interleukin-1 receptor type 1. J Biol Chem. 2000;275:36927–36933. doi: 10.1074/jbc.M006071200. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, de Vos AM. Variations on ligand-receptor complexes. Nat Struct Biol. 2000;7:440–442. doi: 10.1038/75825. [DOI] [PubMed] [Google Scholar]
- Yoon DY, Dinarello CA. Antibodies to domains II and III of the IL-1 receptor accessory protein inhibit IL-1 beta activity but not binding: regulation of IL-1 responses is via type I receptor, not the accessory protein. J Immunol. 1998;160:3170–3179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.