Abstract
Scientific and technological advances in our understanding of the nature and consequences of human genetic variation are now allowing genetic determinants of susceptibility to common multifactorial diseases to be defined, as well as our individual response to therapy. I review how genome-wide association studies are robustly identifying new disease susceptibility loci, providing insights into disease pathogenesis and potential targets for drug therapy. Some of the remarkable advances being made using current genetic approaches in Crohn's disease, coronary artery disease and atrial fibrillation are described, together with examples from malaria, HIV/AIDS, asthma, prostate cancer and venous thrombosis which illustrate important principles underpinning this field of research. The limitations of current approaches are also noted, highlighting how much of the genetic risk remains unexplained and resolving specific functional variants difficult. There is a need to more clearly understand the significance of rare variants and structural genomic variation in common disease, as well as epigenetic mechanisms. Specific examples from pharmacogenomics are described including warfarin dosage and prediction of abacavir hypersensitivity that illustrate how in some cases such knowledge is already impacting on clinical practice, while in others prospective evaluation of clinical utility and cost-effectiveness is required to define opportunities for personalized medicine. There is also a need for a broader debate about the ethical implications of current advances in genetics for medicine and society.
Introduction
The translation of recent advances in our understanding of the genetic basis of common multifactorial diseases into clinical practice remains limited. However, the extraordinary pace of change in human genetics means that this field of research is now starting to challenge how we understand and manage disease, with opportunities for new insights into pathogenesis, drug development and the tailoring of clinical care for the individual patient. This review provides an introduction to the nature of human genetic variation and its functional consequences for disease. Recent insights into the role of genetic diversity in a number of important common diseases serve to illustrate both the advances achieved to date and the challenges that lie ahead.
Approaches to defining genetic determinants of common disease
Linkage and association
Considerable success was achieved using linkage analysis and positional cloning (for a definition of these and other genetic terms, see Glossary in Appendix 1) to identify rare variants with high penetrance responsible for diseases showing a mendelian pattern of inheritance such as cystic fibrosis and haemochromatosis.1,2 In contrast, progress in defining genetic susceptibility loci in common multifactorial diseases remained frustratingly slow until the advent of genome-wide association studies in 2005.3 Prior to this time, the application of a linkage-based approach to common ‘complex’ traits was recognized to be of limited value as multiple genetic loci were likely to be involved in conjunction with environmental factors; moreover, in contrast to mendelian disorders, the underlying genetic variants were of low penetrance, relatively high allele frequency and typically associated with a modest magnitude of effect.3–6 Despite this, there were some notable successes involving linkage studies such as in Crohn's disease with the demonstration of the important contribution of nucleotide-binding oligomerization domain containing 2 (NOD2) variants as will be described shortly.
Studies seeking evidence of genetic association in common diseases using a candidate gene approach were in general underpowered to detect variants associated with a modest magnitude of effect and were not able to dissect the complex underlying genomic architecture of coinherited variants. This was incompletely understood until recently and typically studies analysed only a limited number of single-nucleotide polymorphisms (SNPs) as genetic markers, making resolution of specific variants difficult. These factors, together with the complexities of gene–gene and gene–environment interactions, meant that the results of candidate gene association studies were often difficult to interpret and replicate.7 There were, however, some notable successes achieved through a candidate gene approach with gene selection based on biological plausibility, for example important genetic determinants of susceptibility to human immunodeficiency virus 1 (HIV-1) and malaria were resolved, together with resistance to new variant Creutzfeldt–Jakob prion disease and norovirus diarrhoea.8–11 An important component to many of these discoveries were initial insights based on microbial challenge experiments or observation of extreme clinical phenotypes.11
Genetic variation and application of genome-wide association studies
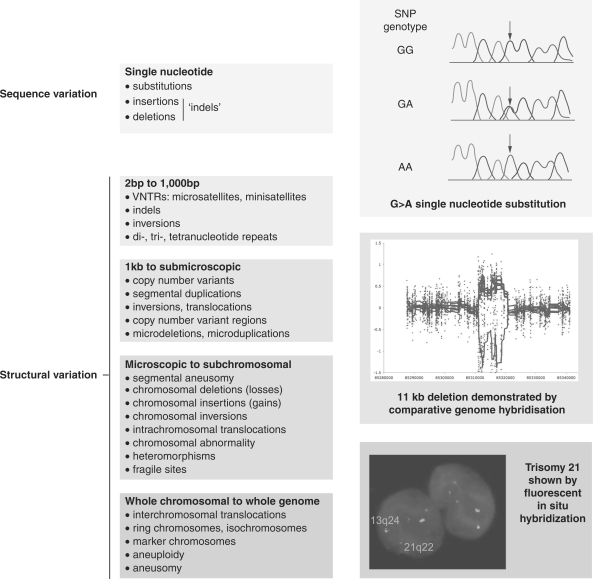
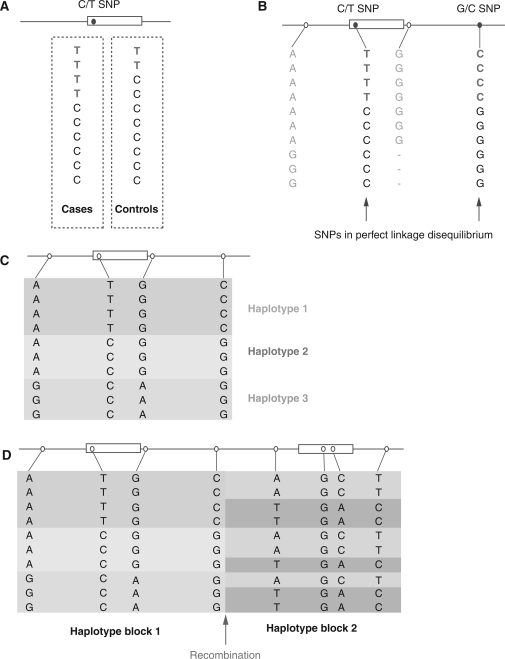
The sequencing of the human genome provided a route map to catalogue and interpret genetic variation among individuals.12,13 The scale of diversity is vast, ranging from microscopically visible variation at the level of individual chromosomes to variation at the deoxyribonucleic acid (DNA) sequence level with nucleotide substitutions, insertions and deletions, including >10 million SNPs (dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/) (Figure 1).14 Submicroscopic structural variation, notably copy number variation, is increasingly recognized and plays a critical role in a number of different diseases.15–17 A number of major collaborative studies, such as the International HapMap Project, aim to understand the genomic architecture of human genetic variation based on very extensive genotyping of common SNPs among different ethnically diverse human populations (http://www.hapmap.org).18,19 This work, together with other studies, enabled a much clearer understanding of how sequence variants are coinherited, with important advances in understanding linkage disequilibrium and haplotypic structure (Figure 2).20,21
Figure 1.
Classes of genetic variation. Classification of genetic variation based on size according to Scherer and colleagues with illustrations of a single-nucleotide substitution, a copy number variant and presence of an additional chromosome. Reproduced and adapted by permission from Macmillan Publishers Ltd: Nat Genet14 copyright (2007) and Nat Rev Genet118 (2004); and http://www.sanger.ac.uk/humgen/cnv/.
Figure 2.
Linkage disequilibrium and haplotypes. (A) For a C>T SNP, the minor T allele shows association with cases of disease, being present twice as often as in controls. (B) However, when further variants are genotyped, a second G>C SNP shows perfect linkage disequilibrium with the initial disease associated SNP: everyone who has the T allele of the first SNP always has the C allele of the second SNP and it is not possible to say whether the disease association is due to the first or second SNP. (C) For four SNPs there are 16 possible combinations in which they could theoretically occur together in a population. However, variants occur together more often than expected by chance and the observed coinherited blocks of variants are described as haplotypes, of which three are present in this theoretical population sample. (D) Haplotype blocks break down at sites of recombination. Understanding the haplotype block structure of the genome across populations is highly informative for population genetics studies.20
This has provided a remarkable resource of sequence level diversity in the form of common SNP markers and showed how considerable economies of scale could be achieved based on genotyping informative SNPs (sometimes described as ‘tag SNPs’) which define particular haplotypes. This, together with major advances in the technologies needed for accurate, affordable and high throughput genotyping, has enabled the successful application of genome-wide association studies in a number of common diseases, ranging from early successes with age-related macular degeneration22 to type 1 diabetes23 and Crohn's disease.24 There have also been very significant advances in our ability to statistically analyse and interpret such data, exemplified by the Wellcome Trust Case Control Consortium (WTCCC) study of seven common diseases.25 This study highlighted the utility in a UK population of common controls for multiple diseases as well as the ability of genome-wide genotyping data to resolve potential population stratification. The WTCCC also showed the value of statistical approaches to infer (by a process described as imputation) additional genotypes based on the genomic architecture from the HapMap studies to allow finer resolution of observed associations. Growing numbers of genome-wide association studies have underlined the importance of sample size to ensure adequately powered studies able to detect small effect sizes; of replication in independent samples given the issues relating to statistical correction for multiple testing when hundreds of thousands of SNP markers are analysed for evidence of association; and the persisting ‘winners curse’ in association studies in which initial studies will tend to overestimate the magnitude of association. Clear, minimally heterogeneous phenotypes and substantially increased sample sizes involving tens of thousands of cases together with meta-analysis are currently advocated to allow detection of smaller odds ratios (typically <1.5).
Challenges to be addressed
A note of caution is, however, appropriate as despite the successes of genome-wide association studies, only a minority of the attributable genetic risk is typically being explained for a given disease. This may be improved by larger studies with increasing numbers of common SNP markers but there may be other explanations. For example, we are not yet able to effectively assess the contribution of low penetrance, less common alleles.26 There is also increasing interest in rare variants and the role of structural genomic variants which have until recently been very hard to detect and analyse. Technological advances in high throughput sequencing and microarray-based comparative genome hybridization are rapidly advancing this field, the latter allowing detection of increasingly small structural variants such that the extent of copy number variation across the human genome is now being appreciated and associated with susceptibility to a number of common diseases ranging from Parkinson's disease to HIV-1 infection.27–30 Such technologies have significant potential clinical utility, for example in relation to causes of learning disability,31,32 although issues relating to predictive value for specific phenotypes and clinical outcomes need to be resolved before such testing could be considered evidence based. Finally, it is clear that gene–gene and gene–environment interactions play an important and potentially confounding role in current approaches, which also do not account for epigenetic effects.
Recent genome-wide associations have been scientifically compelling and have highlighted a number of new biological pathways in disease processes, as highlighted by studies in Crohn's disease described below, but it is unclear the extent to which we are ready to use such information in a clinical arena. The prediction of risk is complex and at present it has been advocated that our estimation of risk remains imprecise for possession of particular genetic variants, and indeed is likely to change in magnitude as more data are accumulated.33 Moreover, how we combine possession of individual variants into a ‘total genetic risk’ score in a clinically validated manner remains in its infancy, and the use and interpretation of genetic testing for individual patients, particularly in a ‘direct-to-consumer’ setting, remains highly controversial.34,35 Careful prospective clinical evaluation considering sensitivity, specificity, predictive value of testing, relationship with outcome and cost-effectiveness need to be combined with wider public and patient education to establish clear guidelines and ethical processes for regulated implantation of genetic testing in common diseases.36,37
Coronary artery disease and atrial fibrillation
Among the many associations found in the WTCCC study, the association of coronary artery disease with SNP markers on chromosome 9p21.3 highlights both the excitement and the work remaining to be done inherent in current genome-wide association studies. This was the most significant association seen across the genome for this disease and has been independently replicated in different populations.25,38–40 Meta-analysis of >12 000 cases and ∼29 000 controls shows that possession of each copy of the risk allele is associated with an odds ratio of 1.24 (95% confidence interval 1.20–1.29).41 The magnitude of risk is relatively modest but highly significant; the biological mechanism is unknown although nearby are CDKN2A and CDKN2B, genes encoding cyclin-dependent kinase inhibitors involved in cancer cell biology with a postulated role in atherosclerosis through modulation of transforming growth factor beta. Intriguingly, there is now evidence of a large noncoding ribonucleic acid (RNA) in the risk locus, ANRIL, specific transcripts of which are differentially expressed in the presence of the risk allele which is thought to relate to regulatory variants in a conserved enhancer element.42 This correlates with altered expression of CDKN2B consistent with a regulatory role for ANRIL and is postulated to modulate disease by altering cellular proliferation.
Further work is required to resolve specific regulatory variants and the functional mechanisms involved. However, the disease association may prove highly informative in terms of both novel insights into pathogenesis and in the clinic. A prospective study, for example, shows a role in risk assessment for predicting the presence of angiographic coronary artery disease but not severity, which is independent of family history and other known risk factors,43 although a recent study of cardiovascular disease in the Women's Genome Health Study found no benefit in risk prediction.44 It is also striking that within 10 kb on a neighbouring linkage disequilibrium block is a SNP marker showing strong association with type 2 diabetes,45–47 although the specific variants and their functional consequences relating to coronary artery disease and type 2 diabetes at 9p21 remain unknown.
Success has also been achieved using genome-wide association studies in atrial fibrillation with association at chromosome 4q25.48 Again, extensive replication has confirmed this association among individuals of North European descent with a meta-analysis showing an odds ratio of 1.9 (1.6–2.26) for the most strongly associated SNP with atrial fibrillation.49 In this case, the associated SNP is in a gene desert ∼50 000 bases away from the nearest gene, but that gene is particularly intriguing as it is paired-like homeodomain 2 (PITX2) which encodes a transcription factor important to cardiac development, notably the sleeve of cardiac tissue extending out over the pulmonary vein known to be important in the aetiology of atrial fibrillation and the target of therapeutic ablation therapy for this condition.50 Further work is needed to resolve the specific variants responsible for this association, but the result illustrates how an unbiased genetics approach has implicated a specific regulatory protein which may unlock the underlying mechanism of disease.
Insights from Crohn's disease
Research in Crohn's disease highlights many important issues related to the genetics of common disease. The pathological basis of this disease is thought to involve an environmental trigger such as an infectious agent(s) leading to disease in genetically susceptible individuals. An early striking result was achieved through genome-wide linkage analysis which identified a major genetic risk locus51 and subsequently specific coding SNPs within the NOD2 gene.52–54 NOD2 encodes a protein critical to the recognition of bacteria and subsequent proinflammatory response. Individuals inheriting one risk allele have an odds ratio of 2.4 (2–2.9) for disease compared to those without a copy, this increases to 17.1 (10.7–27.2) for carriage of at least two risk alleles.55
The advent of genome-wide association studies has dramatically increased the number of genetic susceptibility loci in Crohn's disease to over 30.24,56 It was notable, however, that initial scans did not highlight the known role of NOD2 as the panel of SNP markers did not include the known coding variants associated with disease risk. Significant association was seen for a genotyped SNP in modest linkage disequilibrium with those variants, but the observed effect size was considerably lower (odds ratios of 1.3 and 1.9 for heterozygotes and homozygotes, respectively), showing how SNP coverage on the genotyping platform used can be very important.25 Overall, the effect sizes seen with the many highly significant new susceptibility loci in Crohn's disease are modest, usually of the order of 1.5-fold. This is a typical finding in genome-wide association scans of common multifactorial diseases and highlights how, even with several thousand cases, such studies are relatively underpowered.24
Genomic signposts
It is important to remember that genome-wide association studies are using SNPs as markers to define association. The finding of association does not imply causation to that particular variant but rather the first insights into the likely genomic location of functionally important variants. This could be compared to planting a signpost in a highly variable landscape to indicate the region of interest and helping to guide us in the right direction on the path to resolving causative variants. The ‘fine mapping’ of disease associations represents a major hurdle still to be overcome, both at the level of understanding exactly what variation is present to more clearly resolve the genetic association, and in establishing the functional significance of specific variants.57 Approaches currently employed in fine mapping loci identified by genome-wide association studies include denser genotyping of specific regions and local resequencing, although to uncover rare and structural genomic variants relatively large numbers of individuals may need to be analysed using sophisticated approaches. Coinheritance or linkage disequilibrium between variants adds considerably to the difficulties of fine mapping as it can be very hard to dissect apart the effects of individual variants. It is worth noting that functional effects may be operating at a considerable distance, in Crohn's disease, for example, strong hits of association have been found within ‘gene deserts’ in which associated SNP markers are far from any known gene.
Relationship to structural genomic variation
A further insight into genome-wide association studies in Crohn's disease is the recent striking result that a large deletion, involving 20 kb of DNA upstream of the immunity-related GTPase family, M (IRGM) gene, is responsible for the observed association with SNP markers at that gene.25 The presence of this deletion, which is associated with increased disease risk, significantly altered levels of gene expression.58 Similar structural variation, notably copy number variation, is likely to underpin many observed disease associations found using DNA sequence variants.
Insights into disease pathogenesis
The association with IRGM suggested that autophagy may play an important role in Crohn's disease pathogenesis, an unexpected result added further credence by the association with ATG16 autophagy related 16-like 1 (ATG16L1).59,60 An additional novel insight into disease pathogenesis is the strong association with variants involving genes in the interleukin 23 receptor (IL23R) signaling pathway.60,61 IL23 plays a key role in innate and T-cell-mediated intestinal inflammation62 and the significance of the association with IL23R SNPs is substantially increased by the finding of disease association with variants in genes that are part of the IL23 signalling pathway namely interleukin 12B (IL12B), signal transducer and activator of transcription 3 (STAT3) and Janus kinase 2 (JAK2).24
Lessons from infectious diseases including HIV and malaria
Functional consequences of genetic variants in malaria
Malaria has exerted a major selective pressure on genetic diversity in human populations and provides elegant examples of how variants may exert their functional effects.10 It bears repetition to briefly consider again two classic examples from this field, namely sickle cell and Duffy blood group antigen, to highlight how functional consequences may operate at a structural level (changing the structure and function of the encoded protein) or by variation in noncoding DNA modulating levels of gene expression. In the former case, the selective advantage gained by individuals with sickle cell trait (having one copy of the allele encoding haemoglobin S) in areas endemic for Plasmodium falciparum has driven a monogenic disorder to high allele frequencies in particular populations notably in sub-Saharan Africa.63 Hemoglobin S results from an A to T nucleotide substitution in the coding sequence of the hemoglobin beta (HBB) gene which leads to an amino acid substitution from glutamic acid to valine and a radically altered phenotype in terms of malarial disease risk, in particular protection from severe manifestations of the disease.64
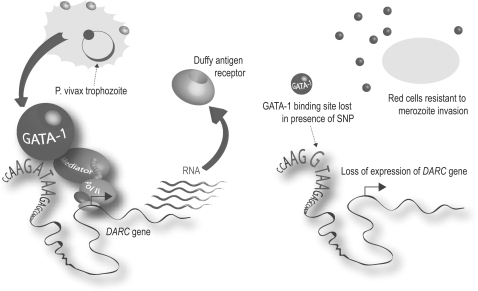
Malaria provides a further example of the consequences of diversity at the level of gene regulation through a SNP modulating expression of the duffy antigen receptor for chemokines gene (DARC) (also known as the FY gene). Individuals who do not have the Duffy blood group antigen on the surface of their red blood cells are completely protected from P. vivax malaria.65 This failure of expression is the result of a T to C nucleotide substitution in the regulatory sequence (the promoter region) found upstream of the DARC gene which disrupts the binding of the transcription factor GATA-1, a protein which binds DNA and helps regulate gene expression (Figure 3).66
Figure 3.
Duffy blood group antigen and P. vivax malaria. Schematic representation of how an A to G SNP disrupts a GATA-1 binding site resulting in loss of expression of the DARC gene and hence the cell surface expression of the receptor that renders red blood cells resistant to invasion by the malarial parasite. The effect of the SNP is highly specific to red blood cells where the transcription factor GATA-1 is found. Selective advantage is thought to have driven the variant to a very high allele frequency in Africa such that it is almost universally found in most West and Central African populations. Reproduced from57 by permission of Oxford University Press.
Consideration of these two examples serves as an introduction to the functional consequences of genetic diversity.57 Early work focused on variation in coding DNA, but more recently it is clear that variation in noncoding DNA is also important.67 The consequences are wide ranging for the magnitude and timing of gene expression at the transcription level, as well as how RNA is alternatively spliced, its stability and processing.68
HIV-1 and the chemokine (C-C motif) receptor 5 ΔΔ32 deletion
Like malaria, research into human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) has advanced our understanding of the nature and implications of genetic variation.9 Host genetic factors have now been found to operate at a number of different levels in relation to HIV-1, including how the virus gains entry into cells, barriers to infection within cells and modulation of cell-mediated and -innate immunity.
Sequencing the CC chemokine receptor CCR5 gene (the major host coreceptor protein for HIV-1) among individuals who were HIV-1 seronegative and whose T cells were resistant to viral infection revealed a 32 bp deletion which leads to a frameshift and dramatic truncation of the protein such that the receptor fails to be expressed on the cell surface.69 When present in a homozygous state, the deletion completely protects against viral entry to cells.69–71 When a single copy is inherited, the consequences are dependent on other variation involving the second allele.72–74 This highlights both the important role of structural variation in common disease and of the CCR5 coreceptor which is now a drug target through reversible small molecule antagonists such as maraviroc.75
Copy number variation and HIV-1
Research into genetic susceptibility to HIV-1 also highlighted how copy number variation may modulate susceptibility to common disease with the discovery that possession of lower numbers of copies of the chemokine (C-C motif) ligand 3-like 1 (CCL3L1) gene, when compared to the median for a given population, were highly significant in terms of susceptibility to HIV-1 and rate of disease progression.28 CCL3L1 is a natural ligand of CCR5 that inhibits infection of cells by HIV-1 strains which use this coreceptor. Increased expression seen with higher copy number is thought to result in more binding to CCR5 and reduced accessibility to HIV-1.
Genetic determinants of viral load
HIV-1 infection also provides an elegant example of how genome-wide association scans can be applied to other phenotypes, in this case to the amount of virus in plasma during the asymptomatic phase prior to AIDS (the viral set point). This has been found to vary significantly between patients with variants in the major histocompatibility complex (MHC) on chromosome 6 explaining about 15% of the observed variability in viral load.76 The observed associations are intriguing, as the most strongly associated SNP marker is within HLA complex P5 (HCP5), a gene itself thought to have arisen from an endogenous retroviral element. Expression of this gene may potentially interfere with the virus but more likely the association is due to linkage disequilibrium with HLA-B*5701. The second strongest association is with a SNP near to HLA-C, which is also associated with expression of HLA-C.
Human leukocyte antigen, killer-cell immunoglobulin-like receptors and HIV-1
Genetic variation in the MHC, notably human leukocyte antigen (HLA) class I alleles such as HLA-B57 and HLA-B35, are important determinants of disease progression. For example, individuals homozygous for HLA-B*35 progress to AIDS in half the median time of those without a copy.77 The situation is, however, complex as diversity in other genomic loci, specifically the highly polymorphic KIR genes encoding killer immunoglobulin-like receptors, is important with the combination of high expressing KIR allotypes and specific HLA-B alleles most informative for AIDS progression and levels of HIV-1 RNA.78
Pharmacogenomics: abacavir hypersensitivity
HLA-B*5701 and HIV-1 also provide one of the few clear examples of how knowledge of genetic variation can directly impact on clinical care in terms of drug use, in this case to avoid drug hypersensitivity. There is now robust evidence from a prospective, randomized, multicenter, double-blind study of nearly 2000 adult patients infected with HIV-1 that prospective screening for this HLA allele can eliminate immunologically proven hypersensitivity reactions to abacavir, a reverse transcriptase inhibitor used to treat HIV-1.79 This is thought to involve the endogenous pathway for antigen presentation and is highly specific to HLA-B*5701 and not closely related alleles, with the drug or a metabolite of it interacting directly with the antigen-binding cleft or modifying it to allow self-antigens to bind.80 There is also recent evidence from a genome-wide association study linking possession of HLA-B*5701 with risk of drug-induced liver injury due to flucloxacillin.81
Gene expression, asthma and regulatory variants
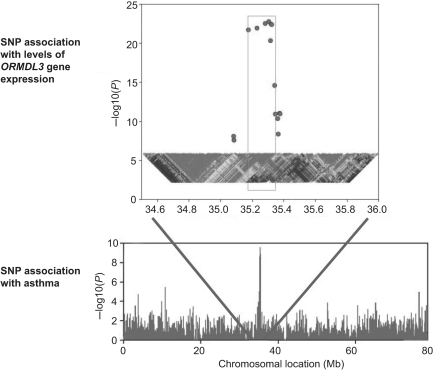
Gene expression varies between individuals and has been successfully mapped as a quantitative trait in model organisms and more recently in humans.82,83 Genome-wide mapping of disease association and of global gene expression offers a complimentary and synergistic approach, which was elegantly demonstrated by a recent study of asthma. Here, a strong signal of disease association was found at chromosome 17q21 which replicated in different European populations; analysis of gene expression showed that the same SNPs were also those most strongly associated with expression of a neighboring gene, ORM1-like 3 (ORMDL3) (Figure 4).84 The SNPs accounted for ∼30% of the variance in expression of this gene which encodes a widely expressed transporter protein.
Figure 4.
Genetic association with disease and gene expression. A genome-wide association study of asthma revealed a strong association with disease at chromosome 17q21. The same SNPs also showed evidence of association with levels of expression of the nearby gene ORMDL3. Adapted and reprinted by permission from Macmillan Publishers Ltd: Nature,84 copyright (2007).
Efforts to map variants associated with gene expression have highlighted the role of regulatory variants in particular classes of genes such as encoding cellular chaperones, cell cycle, RNA processing and immune function. Improved understanding of gene regulation through characterization of regulatory elements across the genome by projects such as ENCODE (ENCyclopedia Of DNA Elements) (http://www.genome.gov/10005107) will provide a route map to identify functional variants. Effects are likely to be highly context specific, for example to specific cell or tissue types, developmental stages or particular cellular conditions.67
Genome-wide association studies and prostate cancer
Many new susceptibility loci have been defined among common cancers occurring in European populations (breast, lung, colorectal, prostate and melanoma) through the application of genome-wide association studies over the last 3 years.85 These germline variants are associated with modest increased risks (typically odds ratios <1.3) although as they are typically relatively common, the population attributable risk is higher. Moreover, for the individual, possession of multiple risk alleles may confer a relatively large risk. Genome-wide association studies using common SNP markers have highlighted the polygenic nature of the identified genetic risk for these cancers, but taking into account the rare high penetrance variants (such as involving BRCA1 and BRCA2 in breast and ovarian cancer) which have been identified, still only explain a minority of the familial risk. Moreover, the utility for clinical testing in cancer remains to be resolved, as the reported associations do not generally help in predicting the aggressiveness of the disease process which may be of greatest benefit. Analysis of somatic mutations may be more informative for this purpose notably gene fusion events such as between TMPRSS2 (transmembrane protease serine 2) and EGR (ETS related gene) which commonly occurs in prostate cancer and results in overexpression of oncogenic transcription factors.86,87 The main benefit of the new genetic knowledge gained through association studies, as seen with other multifactorial diseases such as Crohn's disease, is likely to be in providing novel insights into disease pathogenesis and potential new targets for intervention.
Prostate cancer has seen the largest number of new susceptibility loci identified of any cancer to date from genome-wide association scans.85,86 Perhaps most striking among these are the associations with chromosome 8q24, a genomic region initially identified from linkage and association88 together with admixture mapping89 and subsequently genome-wide association studies.90–93 Remarkably, within a 1.2 Mb ‘gene desert’ at 8q24, three independent susceptibility loci for prostate cancer are present containing multiple risk alleles. For one of these loci, the most significantly associated SNP is also associated with colorectal cancer94; the region also contains a distinct breast cancer susceptibility locus.95 The basis for these associations remains unresolved although the proto-oncogene MYC is one of the genes flanking the region. Indeed, recent evidence shows that the variant associated with colorectal and prostate cancer is located in a conserved enhancer element which interacts with MYC and shows allele-specific binding of a transcription factor activated in colorectal cancer.96,97
Other disease-associated loci include chromosome 10q11.2 in the region of a postulated tumour suppressor gene MSMB encoding β microseminoprotein;90,91 and chromosome 17q12, where intriguing evidence of association was found with increased risk of prostate cancer and protection from type 2 diabetes for a variant in HNF1B (TCF2) encoding the transcription factor HNF1 homeobox B.98 This gene had been previously implicated in maturity onset diabetes of the young (MODY)99 and the association is consistent with epidemiological evidence of an inverse risk between prostate cancer and type 2 diabetes.100,101 Different variants involving the transcriptional repressor JAZF1 (JAZF zinc finger 1) at chromosome 7p15 have also been found to show contrasting associations with these two conditions, of protection from prostate cancer91 and susceptibility to type 2 diabetes102 suggesting possible commonality in disease pathogenesis and new avenues for investigation.103
Venous thrombosis and warfarin dose
Venous thromboembolism is a commonly encountered clinical problem, in which genetic testing for inherited causes of thrombophilia when clinically indicated is an important component of patient care. The genetic basis for venous thrombosis includes multiple rare variants affecting inhibitors of the pro-coagulant system such as deficiency of antithrombin, protein C and protein S, but together these account for only a small minority of cases of familial thrombophilia. Other variants common in the general population and accounting for a significant genetic risk have been defined including factor V Leiden which confers activated protein C resistance.104 Here, a specific SNP in exon 10 of coagulation factor V (F5), the gene encoding coagulation factor V, results in an amino acid substitution from arginine to glutamine which makes the protein less susceptible to degradation. The effect in terms of disease is large with a 5- or 50-fold increased risk of venous thrombosis associated with having a single or two copies of the variant, respectively.105 Other common variants have also been identified, notably in the coagulation factor II (F2) gene encoding prothrombin.106 The indications for thrombophilia screening remain controversial, but evidence relating to clinical outcomes, absolute and relative risks is being established to aid clinical decision making.107–110
Pharmacogenomics of warfarin therapy
Venous thromboembolism is an important indication for warfarin therapy, but as a treatment, warfarin is associated with a high risk of adverse events. This is related to the narrow therapeutic range of warfarin and the marked inter-individual variation in response. This variability is modulated by many factors including patient age and size, together with concomitant drug use, diet and the underlying disease process. Genetic factors also play an important role and illustrate the developing field of pharmacogenomics. Cytochrome P450, family 2, subfamily C, polypeptide 9 (CYP2C9) encodes a cytochrome P450 family enzyme involved in elimination of the more active form of warfarin, and varies in activity more than 20-fold between individuals. Reduced enzyme activity and warfarin dosage was associated with two common alleles of CYP2C9.111 A recent prospective randomized study of warfarin dosing incorporating knowledge of CYP2C9 genotype showed a significant reduction in time to stable anticoagulation, a reduction of minor bleeding episodes and the number of monitoring blood tests required.112 Genetic variation in vitamin K epoxide reductase complex, subunit 1 (VKORC1), which encodes vitamin K epoxide reductase complex 1, has also been associated with levels of gene expression and warfarin dosage.113
This genetic evidence led the US Food and Drug Administration in August 2007 to approve a labelling change for warfarin on package insert such that ‘lower initiation doses should be considered for patients with certain genetic variations in CYP2C9 and VKORC1 enzymes’.114 Combining genetic data for these two genes with information on age, sex and drug interactions explains 59% of the variance in warfarin dose.115 Dosage algorithms incorporating genetic data are available (http://www.warfarindosing.org), and while initial work suggests use of pharmacogenomic information may be cost-effective only in patients at high risk of haemorrhage116 the falling costs of genetic testing and growing feasibility of rapid ‘inhouse’ tests mean that further studies to assess this and clinical utility across different patient groups will be important, together with a clearer understanding of molecular mechanisms and interactions.117 Consideration of clinical outcomes, whether in terms of warfarin use or specific disease traits, is an essential part of evaluation of the clinical utility of genetic testing for evidence-based practice but substantial challenges remain in how to acquire such data.
Conclusions
There has been massive research investment in the field of human genetics over the last 20 years and remarkable advances achieved (Table 1 ). There remains a gap, however, in terms of this growing genetic knowledge and translation into clinical practice, and more broadly with how as a society we choose to use and manage such information. As knowledge of the basic science and associated technologies has advanced, so has our awareness of the daunting task ahead; the scale and complexity of genetic variation in human populations is vast and still incompletely understood, and is manifested in terms of common disease risk through multiple genetic, epigenetic and environmental interactions. It remains a major hurdle to resolve current genetic associations to specific functionally important genetic variants and understand their mechanism of action. We are only now beginning to address the role of rare and structural variants and much of the genetic risk in common disease remains to be explained. We need to more robustly understand the clinical utility of genetic testing, to appreciate how this will impact on screening, diagnosis and management, and to more clearly establish the cost-effectiveness of these approaches. As genetic testing for the individual becomes increasingly available through the private sector, understanding of risk and the caveats that need to be placed on its interpretation in the context of common disease are critically important if we are to use and manage such information. ‘Personalised medicine’ may not yet be a reality, but the potential of recent genetic advances are clear. How we use and regulate this new information for the benefit of patients requires a more active debate in which clinicians and other health care professionals should play a greater role.
Table 1.
Summary points
| There have been radical advances over the last 20 years in our understanding of the nature of the human genome, its remarkably complex regulation and the extent of genetic diversity between individuals |
| Genome-wide association studies using hundreds of thousands of polymorphic common genetic markers (single nucleotide polymorphisms or SNPs) across the genome are now allowing many novel disease susceptibility loci to be resolved in common multifactorial diseases |
| The proportion of the genetic risk currently explained by such studies remains disappointingly low however future studies of rarer variants enabled by recent advances in high throughput DNA sequencing, and of structural genomic variants such as copy number variation, are likely to be highly informative |
| Genetic variation may modulate the structure or function of the protein encoded by a gene or how gene expression is regulated at a transcriptional or post transcriptional level, including effects on epigenetic control mechanisms and alternative splicing |
| Defining functionally important variants remains challenging and requires analysis in the relevant cell or tissue type in the specific disease context |
| Important lessons are being learned from diseases such as Crohn's disease and HIV/AIDS where many different genetic variants have been implicated, and new insights into disease pathogenesis and potential drug targets gained, with direct implications for patient care |
| Much work remains to be done to establish the clinical utility of genetic testing in common diseases and there is a need for a broader debate on the ethical implications of current advances in genetics |
| Pharmacogenomics is likely to have the most immediate impact on the clinical practice of the general physician |
Funding
Wellcome Trust (grant 074318 to J.C.K.).
Conflict of interest: None declared.
Acknowledgements
The author is grateful to friends and colleagues at the Wellcome Trust Centre for Human Genetics and John Radcliffe Hospital in Oxford for their helpful discussions relating to aspects of this review.
Appendix 1: Glossary
Allele: two alternative forms of a gene, found at a given chromosomal locus.
Alternative splicing: process controlling how introns are removed and exons joined together to generate diversity in protein products encoded by a given gene.
Chromatin: dynamic protein packaging of DNA notably involving histone proteins which is important to control of gene expression.
Copy number variant: a structural variant involving gain (for example by insertion or duplication) or loss (for example by deletion) of regions of DNA more than 1000 bases in size relative to a reference sequence; when present at a frequency of greater than 1% in a population, described as a copy number polymorphism.
Epigenetic: heritable change not involving a difference in DNA sequence, for example DNA methylation.
Genome-wide association study: a study utilizing several hundred thousand SNP markers across the genome to look for evidence of statistical association, for example between disease cases and controls, with particular SNPs.
Genotyping: a generic term for techniques to determine genotype, for example which nucleotide is present in a biallelic SNP, based on DNA sequencing, array hybridization or a PCR based method.
Haplotype: the combination of genetic markers or alleles found along a region of a chromosome.
Heterozygote: individual possessing one of each of two possible alleles for a given locus (for example with a G>A SNP, having genotype GA).
Homozygous: individual having identical alleles for a given locus (for example with a G>A SNP, having genotype GG or AA).
Linkage: tendency of characters or marker alleles to be coinherited as responsible loci lie close together on a chromosome.
Linkage disequilibrium: a measure of statistical association or correlation between alleles at different loci such that the presence of one allele can be predicted by another.
Microsatellite: polymorphic DNA sequence involving repeated DNA sequences (of up to six base pairs) that are up to 100 base pairs in overall length; also known as short tandem repeats.
Penetrance: proportion of people with a genetic variant who manifest the associated phenotype.
Phenotype: observed characteristic.
Positional cloning: identification of a gene based on chromosomal location rather than function.
Promoter: region of noncoding DNA found typically upstream of a gene that facilitates and regulates transcription.
Recombination: also known as crossing over, the process involves breakage of a DNA strand and joining to a new strand, for example, homologous recombination during meiosis leading to genetic diversity.
Single nucleotide polymorphisms (SNPs): single-nucleotide substitutions present at a frequency of >1% in a population.
Structural genomic variation: describes variation involving DNA segments more than 1000 bases in size which may vary in number, position or orientation; for example copy number variants, translocations and inversions.
Transcription: the process of recruiting RNA polymerase and generating RNA from DNA.
References
- 1.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 2.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 3.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–17. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 5.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 6.Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9:314–18. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–74. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 9.Heeney JL, Dalgleish AG, Weiss RA. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–6. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- 10.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–92. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet. 2006;40:469–86. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 14.Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, Carter NP, et al. Completing the map of human genetic variation. Nature. 2007;447:161–5. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–71. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- 17.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 18.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–29. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 27.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–69. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 29.Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet. 2006;15:R57–66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- 30.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–50. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 31.Knight SJ, Regan R. Idiopathic learning disability and genome imbalance. Cytogenet Genome Res. 2006;115:215–24. doi: 10.1159/000095917. [DOI] [PubMed] [Google Scholar]
- 32.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–16. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraft P, Hunter DJ. Genetic risk prediction—are we there yet? N Engl J Med. 2009;360:1701–03. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 34.Kaye J. The regulation of direct-to-consumer genetic tests. Hum Mol Genet. 2008;17:R180–3. doi: 10.1093/hmg/ddn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirts BH, Parker LS. Changing interpretations, stable genes: responsibilities of patients, professionals, and policy makers in the clinical interpretation of complex genetic information. Genet Med. 2008;10:778–83. doi: 10.1097/GIM.0b013e31818bb38f. [DOI] [PubMed] [Google Scholar]
- 36.Melzer D, Hogarth S, Liddell K, Ling T, Sanderson S, Zimmern RL. Genetic tests for common diseases: new insights, old concerns. BMJ. 2008;336:590–3. doi: 10.1136/bmj.39506.601053.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamberts SW, Uitterlinden AG. Genetic testing in clinical practice. Annu Rev Med. 2009;60:431–42. doi: 10.1146/annurev.med.60.103007.132540. [DOI] [PubMed] [Google Scholar]
- 38.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–93. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 39.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–84. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, et al. Arterioscler Thromb Vasc Biol. 2009. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. doi:10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 43.Anderson JL, Horne BD, Kolek MJ, Muhlestein JB, Mower CP, Park JJ, et al. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156:1155–62. doi: 10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–45. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–36. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 48.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 49.Kaab S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–9. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–9. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 51.Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, et al. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–3. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 52.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–28. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 53.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 54.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 55.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 56.Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat Rev Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 57.Knight JC. Human Genetic Diversity. Functional Consequences for Health and Disease. 1st edn. Oxford: Oxford University Press; 2009. [Google Scholar]
- 58.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 60.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genomewide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–63. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neurath MF. IL-23: a master regulator in Crohn disease. Nat Med. 2007;13:26–8. doi: 10.1038/nm0107-26. [DOI] [PubMed] [Google Scholar]
- 63.Frenette PS, Atweh GF. Sickle cell disease: old discoveries, new concepts, and future promise. J Clin Invest. 2007;117:850–8. doi: 10.1172/JCI30920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams TN, Mwangi TW, Wambua S, Alexander ND, Kortok M, Snow RW, et al. Sickle cell trait and the risk of Plasmodium falciparum malaria and other childhood diseases. J Infect Dis. 2005;192:178–86. doi: 10.1086/430744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 66.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 67.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 69.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 70.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 71.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96:12004–9. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mangano A, Gonzalez E, Dhanda R, Catano G, Bamshad M, Bock A, et al. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J Infect Dis. 2001;183:1574–85. doi: 10.1086/320705. [DOI] [PubMed] [Google Scholar]
- 74.Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, Sakchalathorn P, et al. Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism-2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–84. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman-Blum SS, Fung HB, Bandres JC. Maraviroc: a CCR5-receptor antagonist for the treatment of HIV-1 infection. Clin Ther. 2008;30:1228–50. doi: 10.1016/s0149-2918(08)80048-3. [DOI] [PubMed] [Google Scholar]
- 76.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 78.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 80.Chessman D, Kostenko L, Lethborg T, Purcell AW, Williamson NA, Chen Z, et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity. 2008;28:822–32. doi: 10.1016/j.immuni.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 81.Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Pe'er I, Floratos A, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–9. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 82.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–72. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 83.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 85.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 86.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 89.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 91.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 92.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 93.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome–wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 94.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 97.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 99.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet. 1997;17:384–5. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 100.Kasper JS, Liu Y, Giovannucci E. Diabetes mellitus and risk of prostate cancer in the health professionals follow-up study. Int J Cancer. 2009;124:1398–403. doi: 10.1002/ijc.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kasper JS, Giovannucci E. A meta-analysis of diabetes mellitus and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2056–62. doi: 10.1158/1055-9965.EPI-06-0410. [DOI] [PubMed] [Google Scholar]
- 102.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frayling TM, Colhoun H, Florez JC. A genetic link between type 2 diabetes and prostate cancer. Diabetologia. 2008;51:1757–60. doi: 10.1007/s00125-008-1114-9. [DOI] [PubMed] [Google Scholar]
- 104.Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–7. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 105.Dahlback B. Advances in understanding pathogenic mechanisms of thrombophilic disorders. Blood. 2008;112:19–27. doi: 10.1182/blood-2008-01-077909. [DOI] [PubMed] [Google Scholar]
- 106.Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3'-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–703. [PubMed] [Google Scholar]
- 107.Whitlatch NL, Ortel TL. Thrombophilias: when should we test and how does it help? Semin Respir Crit Care Med. 2008;29:25–39. doi: 10.1055/s-2008-1047560. [DOI] [PubMed] [Google Scholar]
- 108.Marchiori A, Mosena L, Prins MH, Prandoni P. The risk of recurrent venous thromboembolism among heterozygous carriers of factor V Leiden or prothrombin G20210A mutation. A systematic review of prospective studies. Haematologica. 2007;92:1107–14. doi: 10.3324/haematol.10234. [DOI] [PubMed] [Google Scholar]
- 109.Ho WK, Hankey GJ, Quinlan DJ, Eikelboom JW. Risk of recurrent venous thromboembolism in patients with common thrombophilia: a systematic review. Arch Intern Med. 2006;166:729–36. doi: 10.1001/archinte.166.7.729. [DOI] [PubMed] [Google Scholar]
- 110.Wu O, Robertson L, Twaddle S, Lowe G, Clark P, Walker I, et al. Screening for thrombophilia in high-risk situations: a meta-analysis and cost-effectiveness analysis. Br J Haematol. 2005;131:80–90. doi: 10.1111/j.1365-2141.2005.05715.x. [DOI] [PubMed] [Google Scholar]
- 111.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–98. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 112.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 113.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 114.Vladutiu GD. The FDA announces new drug labeling for pharmacogenetic testing: is personalized medicine becoming a reality? Mol Genet Metab. 2008;93:1–4. doi: 10.1016/j.ymgme.2007.10.133. [DOI] [PubMed] [Google Scholar]
- 115.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, Holm L, McGinnis R, Rane A, Deloukas P. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–92. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 117.Lesko LJ. The critical path of warfarin dosing: finding an optimal dosing strategy using pharmacogenetics. Clin Pharmacol Ther. 2008;84:301–3. doi: 10.1038/clpt.2008.133. [DOI] [PubMed] [Google Scholar]
- 118.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–38. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]