SUMMARY
RNAi-related pathways regulate diverse processes, from developmental timing to transposon silencing. Here, we show that in C. elegans the Argonaute CSR-1, the RNA-dependent RNA polymerase EGO-1, the Dicer-related helicase DRH-3, and the Tudor-domain protein EKL-1 localize to chromosomes and are required for proper chromosome segregation. In the absence of these factors chromosomes fail to align at the metaphase plate and kinetochores do not orient to opposing spindle poles. Surprisingly, the CSR-1 interacting small RNAs (22G-RNAs) are antisense to thousands of germline-expressed protein-coding genes. Nematodes assemble holocentric chromosomes in which continuous kinetochores must span the expressed domains of the genome. We show that CSR-1 interacts with chromatin at target loci, but does not down-regulate target mRNA or protein levels. Instead, our findings support a model in which CSR-1 complexes target protein-coding domains to promote their proper organization within the holocentric chromosomes of C. elegans.
INTRODUCTION
In many organisms, centromeric regions are flanked by repetitive sequences that assemble into densely packed heterochromatin (reviewed in Carroll and Straight, 2006; Vos et al., 2006). These pericentromeric heterochromatin domains are thought to play a role in stabilizing kinetochores, the proteinaceous structures to which spindle attachments are made (reviewed in Cheeseman and Desai, 2008; Welburn and Cheeseman, 2008). In plants (Arabidopsis thaliana) (Kasschau et al., 2007), fission yeast (Schizosaccharomyces pombe) (Reinhart and Bartel, 2002; Buhler et al., 2008), and fruit flies (Drosophila melanogaster) (Brennecke et al., 2007), deep-sequencing studies have identified abundant endogenous small RNAs derived from repetitive regions, including the pericentromeric heterochromatin.
In S. pombe, transcripts generated from the repetitive pericentromeric regions become substrates for an RNA-dependent RNA polymerase (RdRP). After processing by the ribonuclease Dicer, small RNAs derived from these transcripts are loaded into an Argonaute (AGO) complex (the RNA-Induced Transcriptional Silencing complex; RITS). The RITS complex targets pericentromeric heterochromatin and is thought to function in a feedback loop to reinforce chromatin marks that stabilize centromeres during mitosis (reviewed in Buhler and Moazed, 2007).
Not all organisms exhibit repetitive heterochromatin domains associated with centromeric regions. A striking example of this is the organization of the holocentric chromosomes of nematodes (reviewed in Dernburg, 2001). Holocentric, or holokinetic, chromosomes were first described over 100 years ago, in a series of elegant cytological studies by Theodor and Marcella Boveri. In these classic studies, the large presomatic germline chromosomes of the parasitic nematode, Parascaris, were shown to make multiple spindle attachments along their length (reviewed in Pimpinelli and Goday, 1989; Satzinger, 2008). Remarkably, in the somatic cells of the early embryo, the large germline chromosomes were observed to undergo fragmentation resulting in the elimination of heterochromatin and the production of over 40 small, euchromatic chromosomes that comprise the somatic genome. Despite the elimination of heterochromatin, these newly formed chromosomes continued to exhibit holocentric features including continuous kinetochores and multiple spindle attachments along their lengths (Goday et al., 1992).
Although C. elegans chromosomes do not exhibit chromosomal fragmentation, they are similar to the somatic chromosomes of Parascaris in that they are largely euchromatic and exhibit a well-defined holokinetic structure (Albertson and Thomson, 1982). Despite superficial differences, the kinetochores of holocentric and monocentric chromosomes are assembled from a set of highly conserved proteins (reviewed in Maddox et al., 2004), including the histone variant CENP-A/HCP-3. However, unlike monocentric chromosomes, CENP-A/HCP-3 is incorporated into nucleosomes along the entire poleward face of condensed holocentric chromosomes (Buchwitz et al., 1999; Nagaki et al., 2005). The underlying sequences required for the assembly of holokinetic centromeres, and the potential involvement of Argonaute/small-RNA pathways in their assembly and function has not yet been explored.
In C. elegans, previous studies have shown that depletion of drh-3, a Dicer-related helicase, or csr-1, an AGO, result in similar anaphase bridging and chromosome segregation defects (Duchaine et al., 2006; Yigit et al., 2006; Nakamura et al., 2007). Both factors are also required for RNAi (Duchaine et al., 2006; Yigit et al., 2006), and in vitro studies suggest that DRH-3 is required for the synthesis of small RNAs by RdRPs, while CSR-1 has been shown to cleave complementary RNA targets when loaded with triphosphorylated small RNAs (Aoki et al., 2007).
Here we have analyzed the role of DRH-3 and CSR-1 in chromosome segregation and have identified endogenous small RNAs that interact with CSR-1. The CSR-1-interacting small RNAs are members of a class of endogenous small RNAs that are neither microRNAs nor piRNAs (Ambros et al., 2003; Ambros and Lee, 2004; Ruby et al., 2006; Pak and Fire, 2007; Guang et al., 2008; Gu et al., cosubmitted). These abundant small-RNAs (termed 22G-RNAs) are primarily 22 nucleotides in length, with a 5′ triphosphate and a strong bias for a 5′ Guanosine (Ambros et al., 2003; Ruby et al., 2006; Gu et al., cosubmitted). Together with Gu et al. (cosubmitted), we demonstrate that the CSR-1-interacting small RNAs comprise one of two major 22G-RNA pathways. The second 22G-RNA system is dependent on the worm-specific AGOs (WAGOs) and functions to silence transposons, pseudogenes and cryptic loci, as well as certain protein-coding genes (Gu et al., cosubmitted).
We provide evidence that EGO-1, an RNA-dependent RNA-polymerase (RdRP) (Smardon et al., 2000), and EKL-1, a tudor-domain protein (Rocheleau et al., 2008), function along with DRH-3 and CSR-1 to promote chromosome segregation. Together, these factors are required for the biogenesis of CSR-1-interacting 22G-RNAs, which, surprisingly, are antisense to thousands of germline-expressed genes. CSR-1 interacts with chromatin at its target loci, but does not appear to silence mRNA or protein expression. We hypothesize that the role of CSR-1 in chromosome segregation in C. elegans is analogous to that of Ago1 in the S. pombe chromosome segregation pathway. However, instead of targeting repetitive pericentromeric heterochromatin, CSR-1 targets protein-coding euchromatic domains to promote their proper organization within the holocentric chromosomes of C. elegans.
RESULTS
A set of RNAi-related factors required for chromosome segregation
To identify additional genes that function with drh-3 and csr-1 to promote chromosome segregation, we examined the mutant phenotypes of genes previously implicated in RNAi-related pathways for evidence of chromosome segregation defects. We found that one of four RdRP genes, ego-1 (Smardon et al., 2000), and the tudor-domain-containing gene, ekl-1 (Rocheleau et al., 2008), exhibited defects in fertility and chromosome segregation, similar to those described previously for drh-3 and csr-1 (Duchaine et al., 2006; Yigit et al., 2006; Nakamura et al., 2007) (see below). EKL-1 had been implicated in several silencing pathways by RNAi-based screens (Kim et al., 2005; Robert et al., 2005; Rocheleau et al., 2008). We found that a null allele of ekl-1(tm1599) was deficient for both germline and somatic RNAi, and in addition caused a fully penetrant sterile phenotype (Figure S1 and data not shown).
Mutation or RNAi depletion of drh-3, csr-1, ego-1, and ekl-1 resulted in a similar spectrum of meiotic and mitotic defects. The germlines of each mutant are underproliferated, with nuclei of abnormal shape and size (Maine et al., 2005; Vought et al., 2005; Duchaine et al., 2006; She et al., 2009) (data not shown and see Figure 3E). Chromosomal abnormalities were evident in DAPI-stained oocytes, which occasionally possessed more than six DAPI-staining bodies (Figure 1A) (Nakamura et al., 2007, She et al., 2009). One measure of chromosome segregation defects in the hermaphrodite germline is the proportion of XO male progeny, which arise via spontaneous loss of the X chromosome at a frequency of 0.1–0.2% in wild type populations (Meneely et al., 2002). We found that a partially rescued transgenic csr-1(tm892) strain (Figure S1) generated approximately 6% male progeny (a high incidence of males, or him, phenotype) (Figure 1B). A similar him phenotype was also observed in strains homozygous for hypomorphic alleles of drh-3 (Gu et al., cosubmitted). These observations suggest that the loss of csr-1 or drh-3 can lead to defects in chromosome segregation during either mitotic or meiotic divisions in the germline. Despite the evidence described above for chromosomal abnormalities in the germline, we failed to directly observe mitotic or meiotic chromosome mis-segregation (n=greater than 100 germlines examined, data not shown). In most cases, the dividing nuclei exhibited either wild type segregation or already contained an abnormal DNA complement. The relative paucity of abnormalities observed in csr-1(tm892) germlines could reflect a perdurance of maternally loaded CSR-1.
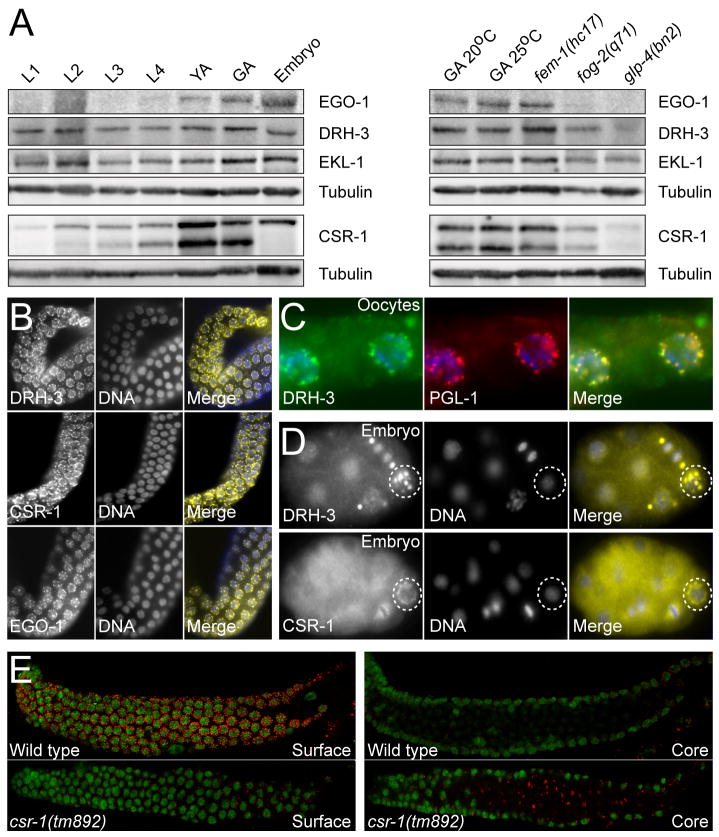
Figure 3. CSR-1, DRH-3, EKL-1 and EGO-1 are expressed in the germline.
(A) Western blots of developmentally staged protein lysates (left) or various germline mutant lysates (right) probed for EGO-1, DRH-3, EKL-1, CSR-1 (multiple isoforms), and tubulin (as a loading control). L1, L2, L3 and L4 are larval stages; YA, Young Adults; GA, Gravid Adults; Embryos, mixed stage embryos. GA 25°C, Gravid Adults grown at 25°C; fem-1(hc17), no sperm at 25°C; fog-2(q71) enriched to 95% males by filtration (20°C); and glp-4(bn2), no germline at 25°C.
(B) Wild type perinuclear germline localization of DRH-3, CSR-1, and EGO-1 (left, yellow) (DNA, center, blue).
(C) DRH-3 (left, green) colocalizes with the P Granule component, PGL-1 (center, red; DNA, blue).
(D) DRH-3 and CSR-1 (left, yellow) remain localized to P Granules in the embryonic P cell lineage (dashed circles; DNA, center, blue).
(E) Single confocal sections of PGL-1 (red) in wild type and csr-1(tm892) mutant germlines through the germline surface and core. P Granules become detached from the nuclear periphery in csr-1(tm892) (DNA, green; distal is to the left).
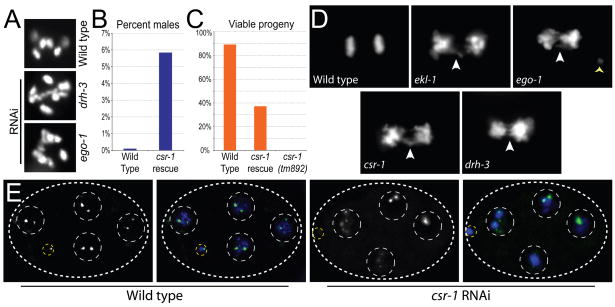
Figure 1. csr-1, ego-1, ekl-1 and drh-3 mutants display chromosome segregation defects in mitosis and meiosis.
(A) Diakinetic oocyte chromosomes in wild type and drh-3 or ego-1 RNAi-depleted animals. Six discrete DAPI figures are observed in wild type, while greater than six figures are present in mutant oocytes.
(B) Incidence of males in wild type (N2) and 3× Flag csr-1 rescue.
(C) Viable progeny per brood in wild type (N2), 3× Flag csr-1 rescue, and csr-1(tm892).
(D) DAPI-stained wild type (N2) and RNAi-depleted embryos undergoing the first mitotic division. Anaphase bridging is evident (white arrowhead). An aberrant piece of DNA is visible in ego-1 (yellow arrowhead).
(E) Fluorescence in situ hybridization with probes for chromosome V 5S rDNA in wild type and csr-1 RNAi-depleted embryos (DNA, blue; FISH signal, green). Left panels in each set show FISH signal alone. White dotted lines indicate embryo (large oval) and nuclei (circles). Yellow dotted lines indicate polar bodies. Images are projections of Z-stacks through the entire embryo after deconvolution.
In addition to the him phenotype, dead embryos were also prevalent in both the the csr-1(tm892) rescued strain and the hypomorphic drh-3 strains (Gu et al., cosubmitted). For instance, the csr-1(tm892) rescued strain only generated approximately 38% viable progeny (Figure 1C). The dead embryos produced by this strain arrested at various points in embryogenesis, up to approximately the 100-cell stage, and accumulated nuclei with abnormal DNA content (data not shown). To better examine chromosome morphology and segregation defects in the absence of csr-1, ekl-1, ego-1, and drh-3, we used DAPI, as well as histone-GFP and tubulin-GFP (Figure 1D, Movies S1–S4) in RNAi depleted embryos. Chromosomes appeared to condense during prophase with wild type timing and morphology. However, as the cell cycle progressed, the following defects were evident during essentially every cell division, beginning with the first cell division of the embryo. At metaphase, chromosomes failed to align into well-organized plates perpendicular to the long axis of the spindle. At anaphase, chromosomal bridging was evident in the spindle midzone (Figure 1D, Movies S1–S4) and at cytokinesis the lagging chromosomes were bisected by the cleavage furrow. As embryogenesis progressed, abnormally shaped nuclei, with greater or less than wild type chromosomal complements, accumulated until cell division arrested at about the 50-cell stage (visible in Figures S2, S3 and S5).
To examine the chromosome segregation abnormalities resulting from loss of these RNAi factors at the molecular level, we utilized Fluorescence In Situ Hybridization (FISH) with 5S rDNA probes to chromosome V. Of 32 wild type embryos, only two showed aberrant FISH signals in one or more nuclei (van Wolfswinkel et al., cosubmitted). In contrast, more than half (10/19) of the csr-1 depleted embryos displayed abnormal numbers of FISH positive chromosomes along with a range of additional abnormalities including aberrantly sized and shaped nuclei (Figure 1E and S2).
DRH-3, EKL-1, EGO-1 and CSR-1 promote the proper organization and alignment of metaphase chromosomes
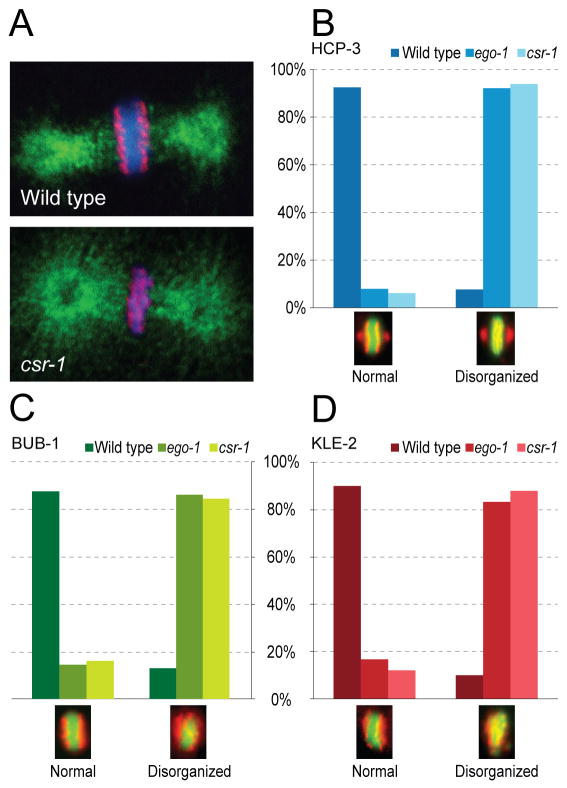
We next examined three related aspects of chromosome structure that are essential for faithful chromosome segregation: kinetochore formation, condensin loading, and cohesin loading. During mitotic divisions in wild type C. elegans embryos, HCP-3 localizes to the poleward faces of metaphase chromosomes (Buchwitz et al., 1999; Oegema et al., 2001). In csr-1, drh-3, ekl-1 and ego-1 RNAi-depleted embryos, HCP-3 was loaded onto chromosomes, but was dramatically disorganized. Instead of poleward localization on both sides of the metaphase plate, HCP-3 was distributed over the metaphase chromosomes in an interrupted pattern that extended throughout the spindle midzone (Figure 2A–B). This pattern could reflect a defect in chromosome alignment and/or compaction, or could indicate that, even though HCP-3 is loaded, it is not targeted to the appropriate regions of the chromosome. Another conserved inner centromeric protein, HCP-4/CENP-C (Moore and Roth, 2001), displayed the same disorganized localization (data not shown). Finally, to assess whether the kinetochores were fully assembled in csr-1, drh-3, ekl-1 and ego-1 RNAi-depleted embryos, we examined the outer kinetochore proteins KLP-7/MCAK (a kinesin) and the conserved spindle checkpoint protein, BUB-1 (Oegema et al., 2001). Both were loaded onto mitotic chromosomes in the RNAi-depleted embryos but were disrupted in a manner similar to HCP-3 (Figure 2C, S3 and data not shown).
Figure 2. csr-1, ego-1, ekl-1 and drh-3 RNAi-depleted embryos display defects in chromsome organization.
(A) Single confocal sections showing kinetochore organization in the first cell division of wild type (N2) and csr-1 RNAi-depleted embryos (HCP-3, red; tubulin, green; DNA, blue).
(B) HCP-3/inner kinetochore disorganization frequency in wild type (N2), vs. ego-1 and csr-1 RNAi-depleted embryos (example metaphase images, HCP-3, red; DNA, green).
(C) BUB-1/outer kinetochore disorganization frequency in wild type (N2), vs. ego-1 and csr-1 RNAi-depleted embryos (example metaphase images, BUB-1, red; DNA, green).
(D) KLE-2/condensin disorganization frequencty in wild type (N2), vs. ego-1 and csr-1 RNAi-depleted embryos (example metaphase images, KLE-2, red; DNA, green).
Because the observed chromosome segregation defects could result from problems in chromosome condensation or cohesion, we examined the localization of proteins involved in these processes. Both the Condensin I/Condensin IDC protein CAPG-1 and the Condensin II protein KLE-2 (Csankovszki et al., 2009), as well as the cohesins SCC-1 and SCC-3 (Mito et al., 2003; Kirsten Hagstrom, unpublished) were loaded onto mitotic chromosomes in csr-1, drh-3, ekl-1 and ego-1 RNAi-depleted embryos, but displayed highly disorganized localization patterns, in a manner similar to HCP-3 (Figure 2D, S3 and data not shown).
Expression studies reveal localization to P Granules and mitotic chromosomes
To explore the role of these RNAi components in chromosome segregation, we examined the expression and localization patterns of DRH-3, EGO-1, EKL-1, and CSR-1. Western blot analyses revealed that DRH-3, EKL-1, and two isoforms of CSR-1 are present at all developmental stages, and that EGO-1 and CSR-1 are most enriched in young adults, gravid adults, and embryos (Figure 3A) (Vought et al., 2005). DRH-3 and EKL-1 were detected in glp-4(bn2) adults which fail to develop a germline and are thus greatly enriched in post-mitotic cells (Beanan and Strome, 1992). This finding is consistent with the role of DRH-3 and EKL-1 in the biogenesis of a broader set of somatically-expressed 22G-RNAs (Gu et al., cosubmitted). The larger CSR-1 isoform was expressed throughout larval development, and was also present at low levels in post-mitotic populations lacking a germline. Quantitative real-time RT-PCR analysis of both csr-1 transcripts indicated that their expression recapitulates the protein expression pattern (Figure S4).
DRH-3, EGO-1, and CSR-1 colocalize in the germline with PGL-1, a previously characterized component of the germline nuage structures called P granules (Figure 3B–C and data not shown) (Kawasaki et al., 1998). EKL-1 was not detected in P granules (data not shown). While many developmentally important factors transiently localize to P granules, DRH-3 and CSR-1 maintained their P granule localization in germ cells throughout the life cycle (Figure 3D and data not shown). As was previously shown for ego-1 mutants (Vought et al., 2005), mutations in ekl-1, csr-1, and drh-3 also caused a striking disruption in the perinuclear localization of P granules (Figure 3E and data not shown), indicating that these factors function more intimately in promoting or maintaining P granule structure and association with the nuclear periphery.
As oocytes matured, EGO-1 was lost from the P granules (data not shown), while DRH-3 (Figure 3C) and CSR-1 (Figure 4A) maintained P granule association. In mature oocytes, CSR-1 (Figure 4A) and EGO-1 (data not shown) both became enriched in nuclei, where CSR-1 was enriched on the diakinetic chromosomes.
Figure 4. CSR-1, DRH-3, EKL-1 and EGO-1 localize to chromosomes.
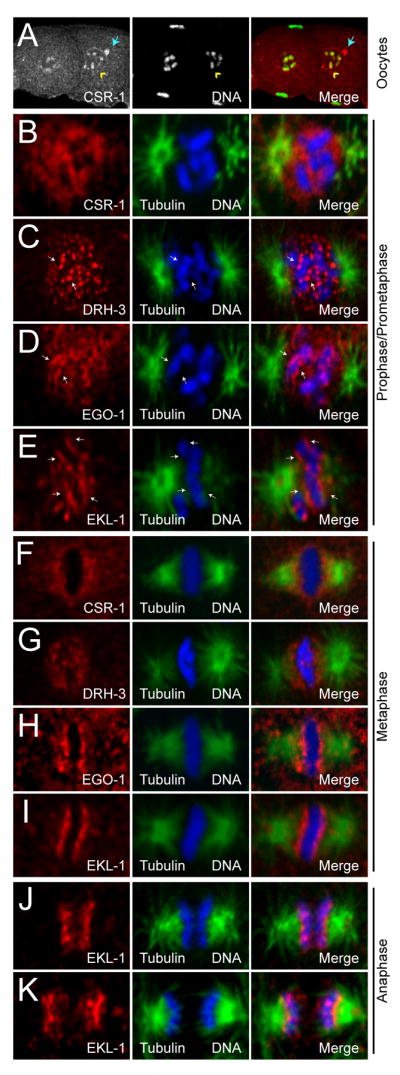
(A) Single confocal sections of CSR-1 (left, red) in wild type oocytes. CSR-1 is enriched on diakinetic chromosomes as oocytes mature (yellow arrowhead), and remains in some P Granules (blue arrow) (DNA, center, green; distal is to the left).
(B) to (E) Single confocal sections of CSR-1 (B), DRH-3 (C), EGO-1 (D), EKL-1 (E) (red) in wild type embryo prophase/prometaphase (tubulin, green; DNA, blue).
(F) to (I) Single confocal sections of CSR-1 (F), DRH-3 (G), EGO-1 (H), EKL-1 (I) (red) in wild type embryo metaphase (tubulin, green; DNA, blue).
(J) to (K) Single confocal sections of EKL-1 in wild type embryo early (J) and late (K) anaphase (tubulin, green; DNA, blue).
In the mitotic cells of embryos, each factor became enriched in prophase nuclei. As chromosomes condensed, DRH-3, EGO-1, and EKL-1 became enriched along the length of each chromosome, while CSR-1 remained nuclear (Figure 4B–E). All four proteins exhibited robust localization around the metaphase plate (Figure 4F–I). CSR-1 and DRH-3 displayed a pattern similar to cohesins (Mito et al., 2003), while EKL-1 (and to a lesser degree, EGO-1) appeared to be more closely associated with chromosomes in a pattern similar to kinetochore proteins. In fact, EKL-1 retained a robust association with chromosomes during anaphase, while the other RNAi factors became more difficult to detect (Figure 4J–K). Cytoplasmic localization was also detected for each protein (data not shown). Finally, all aspects of the localization patterns were absent in each respective mutant background (Figure S5).
We then asked whether DRH-3, EGO-1, CSR-1 and EKL-1 depend on each other’s wild type activities for their expression and localization. Consistent with the idea that these factors function together, we found a co-dependence for proper localization to both metaphase chromosomes and to the P granules. While Western blotting demonstrated that the expression of EGO-1, CSR-1 and EKL-1 was undiminished in drh-3 mutants (Figure S5) (Gu et al., cosubmitted), the localization of each protein to chromosomes at metaphase was nearly abolished (Table S1 and data not shown), and EGO-1 and CSR-1 lost their association with germline P granules. In ekl-1 and ego-1 RNAi-depleted embryos only CSR-1 exhibited greatly reduced association with the metaphase plate and with P granules. Finally, DRH-3, EGO-1, and EKL-1 localized to the disrupted metaphase plates in csr-1 depleted embryos, and DRH-3 and EGO-1 associated with mis-localized P granules in csr-1 RNAi-depleted germlines. Taken together, these data indicate a hierarchy in the RNAi/chromosome segregation pathway, in which the wild type activity of DRH-3 was necessary for the proper targeting of EKL-1, EGO-1 and CSR-1 to chromosomes.
CSR-1 associates with small RNAs that are antisense to germline-expressed genes
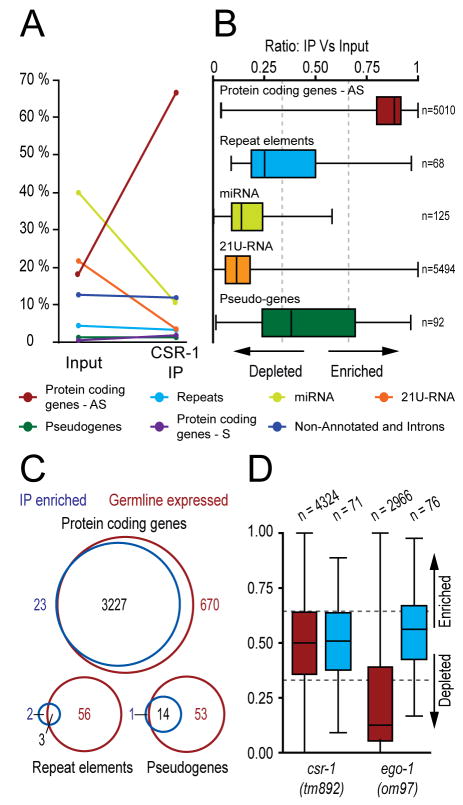
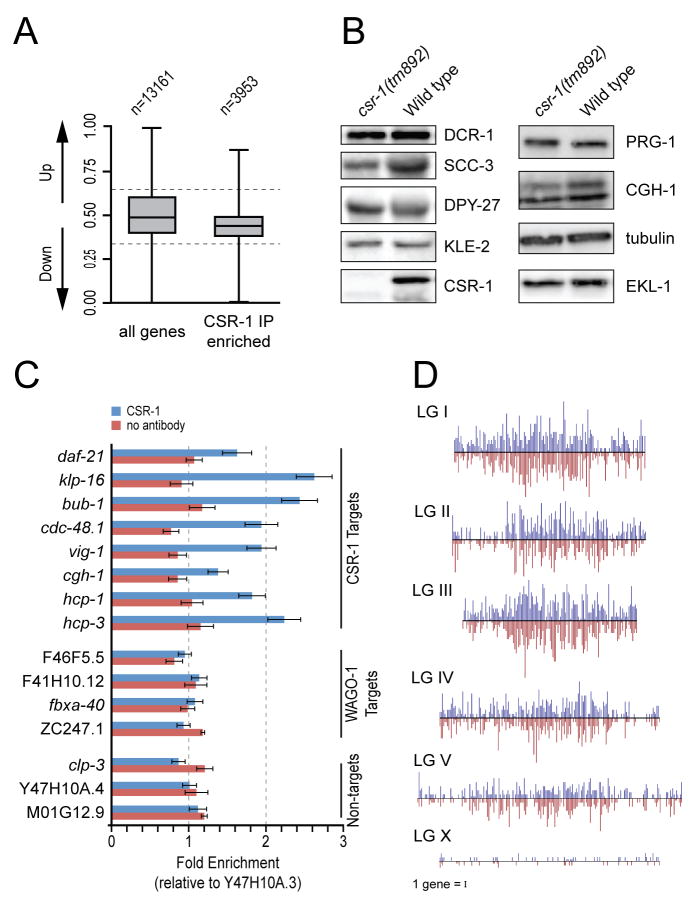
The targets of AGO proteins can be deduced by analyzing the sequences of the AGO-associated small RNAs. Therefore, we recovered CSR-1 complexes and analyzed the associated small RNAs using a deep-sequencing approach. CSR-1 complexes were enriched two-fold or greater for a class of drh-3, ego-1 and ekl-1-dependent 22G-RNAs that are antisense to at least 4191 protein-coding genes. These gene-targeted 22G-RNAs collectively represented greater than 99% of all 22G-RNA reads matching loci with a two-fold or greater increase in read count in the CSR-1 IP complex (Figure 5A–B, Figure S6 and Table S2). microRNAs, 21U-RNAs and nearly all other 22G-RNA species, including those targeting transposons and other repetitive sequences, pseudogenes, and intergenic or non-annotated regions were depleted in CSR-1 complexes (Figure 5A–B). The exceptions were 22G-RNAs targeting seven families of repetitive elements and 23 loci annotated as pseudogenes. Altogether, repeat-targeted 22G-RNAs accounted for only 0.25% of the total reads enriched in the CSR-1 IP complex, while pseudogene-targeted reads represented less than 0.5% (Table S3). 22G-RNAs corresponding to at least 80% of the CSR-1-targeted mRNAs were strongly depleted in the glp-4(bn2) mutant (Figure 5C) (Gu et al., cosubmitted) which lacks a germline, indicating that the CSR-1 22G-RNAs are expressed in the germline. Finally, consistent with the involvement of the β-nucleotidyl transferase CDE-1 in the uridylation of CSR-1-associated 22G-RNAs, approximately 40% of the 22G-RNA reads enriched in the CSR-1 IP were extended at the 3′ end with at least one uridine (Figure S6) (van Wolfswinkel et al., cosubmitted).
Figure 5. Analysis of small RNAs enriched in CSR-1 IP complexes.
(A) Line plot comparing the relative proportions of small RNA classes between wild type (N2) Input (left) and CSR-1 IP (right) samples. (AS=antisense, S=sense)
(B) Box and whisker plot of the relative proportion of small RNA reads for each locus targeted within each small RNA class, in the CSR-1 IP relative to Input. Loci with values closer to 1 indicate enrichment of small RNA reads in the IP, a value of 0.5 indicates equal proportions of reads in the IP and input, and values closer to 0 indicate loci depleted of small RNA reads in the IP. Boxes contain 50% of siRNA loci (between the 25th and 75th percentile), with the line inside each box representing the median value. Lines extending to the right of the box represent the most enriched value, and lines extending to the left of the box represent the most depleted value in the IP. X axis is relative proportion of reads (measured as IP value divided by Input plus IP values for any given locus). Dotted lines indicate the values corresponding to two-fold enrichment (a value of 0.66) or depletion (a value of 0.33). Calculations were made with small RNA cutoffs as described in Supplemental Experimental Procedures.
(C) Venn diagram depicting the proportion of loci that possess a two-fold or greater depletion of 22G-RNAs in the glp-4(bn2) mutant that are also enriched two-fold or more in the CSR-1 IP. Only loci present in both datasets with 25 reads per million or more are represented.
(D) Box and whisker plot of the relative proportion of small RNA reads for each locus in the csr-1(tm892) and ego-1(om97) relative to a congenic wild type strain (DA1316). Protein coding genes (red) and repeat elements (blue) are represented. drh-3 and ekl-1 small RNA analyses are described in (Gu et al., cosubmitted).
When factors involved in Argonaute-mediated small RNA biogenesis are absent or nonfunctional, the corresponding small RNAs are also depleted (Grishok et al., 2001; Yigit et al., 2006; Batista et al., 2008). Thus, we prepared small RNA libraries from csr-1(tm892) and ego-1(om97) mutants and compared them to libraries from drh-3(ne4253) and ekl-1(tm1599) mutant populations (Gu et al., cosubmitted). Consistent with the IP analysis described above, csr-1 and ego-1 mutants were depleted for a set of 22G-RNAs that are antisense to protein coding genes (Figures 5D, S7, and Tables S4–S5). To be scored as depleted in the mutants, an arbitrary cut off of 25 reads per million in the wild type data set was used. As a consequence, many loci for which read counts were significantly increased in the IP studies above were excluded from this analysis. Nevertheless, approximately 900 loci exhibited 22G-RNAs that were dependent on csr-1, as well as on ego-1, drh-3, and ekl-1 (Gu et al., cosubmitted). Consistent with a germline origin for these 22G-RNAs, the majority were depleted in glp-4(bn2) animals, which lack a germline (data not shown). While the proportion of 21U-RNAs was unaltered in the four mutants, microRNA populations, overall, appeared slightly decreased in csr-1 and ego-1, relative to the total read count, possibly due to a dearth of embryos in these mutant populations (Figure S7) (Gu et al., cosubmitted).
As expected, based on their broad role in 22G-RNA biogenesis, all 22G-RNAs, including those targeting repetitive elements, were depleted in drh-3 and ekl-1 samples (Gu et al., cosubmitted). 22G-RNAs targeting repeats (including those targeting the seven repeat families that were enriched in CSR-1 complexes) were unaltered in small RNA populations from the csr-1 and ego-1 mutants (Figure 5D). Furthermore, those 22G-RNAs which were not associated with, or dependent on CSR-1, were instead dependent on the activity of the ego-1 paralog, rrf-1, or on a combination of ego-1 and rrf-1 activities, but exhibited no other distinguishing biochemical properties (Gu et al., cosubmitted). These remaining CSR-1-independent 22G-RNAs, including those produced by RRF-1, engage a distinct family of Argonautes that mediate transposon silencing and other silencing activities unrelated to chromosome segregation (Gu et al., cosubmitted). These data are consistent with CSR-1 IP data, and suggest that csr-1 and ego-1 are specifically involved in the expression of a particular subset of gene-targeted 22G-RNAs.
CSR-1 targets are not mis-regulated in csr-1 mutants
The genes targeted by CSR-1 22G-RNAs include numerous genes whose mRNAs are expressed in the germline, oocytes and embryos. To determine if CSR-1 regulates its targets at the mRNA level, we performed transcriptional profiling on csr-1(tm892) mutant versus wild type N2 adult worms. Previous work demonstrated that CSR-1 is capable of degrading target mRNAs in vitro (Aoki et al., 2007). However, strikingly, the global profile of gene expression for csr-1, including the profile of CSR-1 22G-RNA targets, was very similar to that for N2 (Figure 6A and Table S6). Thus, CSR-1 does not down-regulate its target mRNAs. Similar results were reported for the expression of CSR-1 targets in transcriptional profiling studies performed on drh-3 (Figure S8) (Gu et al., cosubmitted) and cde-1 mutants (van Wolfswinkel et al., cosubmitted).
Figure 6. CSR-1 22G-RNA complexes bind to target genomic loci.
(A) Box and whisker plot of mRNA expression from microarray experiments in wild type vs. csr-1(tm892) mutants. The analysis was done for all genes measured by the array (left), and the subset of only CSR-1 22G-RNA target genes (right).
(B) Western blot analysis of wild type and csr-1(tm892) protein lysates, probed for CSR-1 22G-RNA target proteins. EKL-1 is a loading control.
(C) ChIP/Quantitative real-time PCR analysis of CSR-1 enrichment at CSR-1 22G-RNA or WAGO-1 22G-RNA target loci. Fold enrichment is calculated relative to the Y47H10A.3 locus, which, like clp-3, Y47H10A.4, and M01G12.9, is not targeted by small RNAs. Data from a single, representative set of experiments is presented, error bars are standard deviation. (IP with CSR-1, blue; IP with beads only/no antibody, red)
(D) Density of CSR-1 22G-RNA target genes on each chromosome. Each bar represents the numbers of genes in a 100 kb bin. (Watson strand, blue; Crick strand, red). Chromosome number is as indicated. Scale bar represents one gene.
Using available antibodies for the protein products of several CSR-1 22G-RNA targets, we next examined protein expression levels in csr-1, drh-3, and cde-1 mutants by immunofluorescence and Western blotting. There were no significant changes in the protein levels of the CSR-1 22G-RNA targets we examined, including those of the small RNA pathway components PRG-1 and DCR-1; the P Granule factors PGL-1, CAR-1, and CGH-1; the cohesin SCC-3; and the dosage-compensation factors DPY-27 and CAPG-1 (Figure 6B, S8 and data not shown). Together, these data suggest that CSR-1 22G-RNA complexes do not act globally to significantly alter target gene expression.
CSR-1 is bound to chromatin at 22G-RNA target loci
In S. pombe, the Argonaute Ago1 associates directly with chromatin as a part of the RITS complex (Motamedi et al., 2004; Buhler et al., 2006). A large-scale proteomics study identified CSR-1 associated with fractions of sperm and oocyte chromatin (Chu et al., 2006). Using a similar method (Chu et al., 2006), we have determined that CSR-1 associates with chromatin in embryos (Figure S9). These observations led us to examine whether CSR-1 complexes directly bind to the genomic loci of the CSR-1 22G-RNA targets.
Using chromatin immunoprecipitation (ChIP), we found an enrichment of CSR-1 at target loci when compared to several other genomic loci that are not targeted by small RNAs. RNA Polymerase II was used as a positive control and consistently showed enrichment at many CSR-1 target loci (Figure S9). In contrast, negative control experiments using agarose beads alone (without CSR-1 antibody) never displayed enrichment (Figure 6C and S9). Of the twelve CSR-1 22G-RNA target loci examined, ten showed 1.5 fold or greater enrichment of CSR-1-binding in five or more independent experiments (Figure 6C and data not shown). Conversely, CSR-1 was never enriched at the targets of another germline-expressed Argonaute, WAGO-1 (Figure 6C). CSR-1 was not detected in chromatin fractions treated with RNase A (data not shown), nor did we detect CSR-1 enrichment by ChIP at target loci in the drh-3(ne4253) mutant, in which 22G-RNAs are depleted (Figure S9). These findings indicate that CSR-1 interacts with its target genomic loci in a 22G-RNA-dependent manner. Furthermore, CSR-1 22G-RNA target loci are distributed relatively uniformly along the chromosomes (Figure 6D), suggesting that the CSR-1 22G-RNA pathway could act in a genome-wide manner to influence chromosome segregation.
DISCUSSION
Here we have investigated the role of the C. elegans Argonaute CSR-1 in promoting proper chromosome segregation. We have shown that CSR-1 interacts with a class of 22-nucleotide RNAs, called 22G-RNAs, which are antisense to at least 4,191 protein-coding genes, seven repeat element families, and 23 pseudogenes distributed throughout the genome. A parallel study by Gu et al. (cosubmitted) has shown that a distinct Argonaute, WAGO-1, interacts with a non-overlapping set of 22G-RNAs that primarily target transposons, cryptic elements and pseudogenes (see below and Gu et al., cosubmitted). The biogenesis of both CSR-1- and WAGO-1-bound 22G-RNAs is dependent on a core set of factors, including DRH-3, EKL-1, an RdRP and a β-nucleotidyl transferase (Gu et al., cosubmitted). However, WAGO-1-associated 22G-RNAs appear to down-regulate their mRNA targets (Gu et al., cosubmitted), whereas CSR-1 22G-RNAs do not. Whole genome microarray studies showed that the mRNA targets of CSR-1 22G-RNAs are not mis-regulated in the csr-1, drh-3, and cde-1 mutant backgrounds (Gu et al. and van Wolfswinkel et al., cosubmitted) (Figure 6 and S8). In addition, immunoflourescence and Western blot analysis on the protein products of several CSR-1 targets revealed no change in expression (Figure 6 and S8). Based on these findings, it seems unlikely that perturbed expression of CSR-1 22G-RNA targets results in the observed chromosome segregation defects. Instead, our findings support a model in which the CSR-1 pathway may directly contribute to holocentric chromosome organization by ensuring that the expressed, euchromatic domains within the genome support the proper juxtaposition and alignment of the kinetochores, which must span these domains (Figure 7).
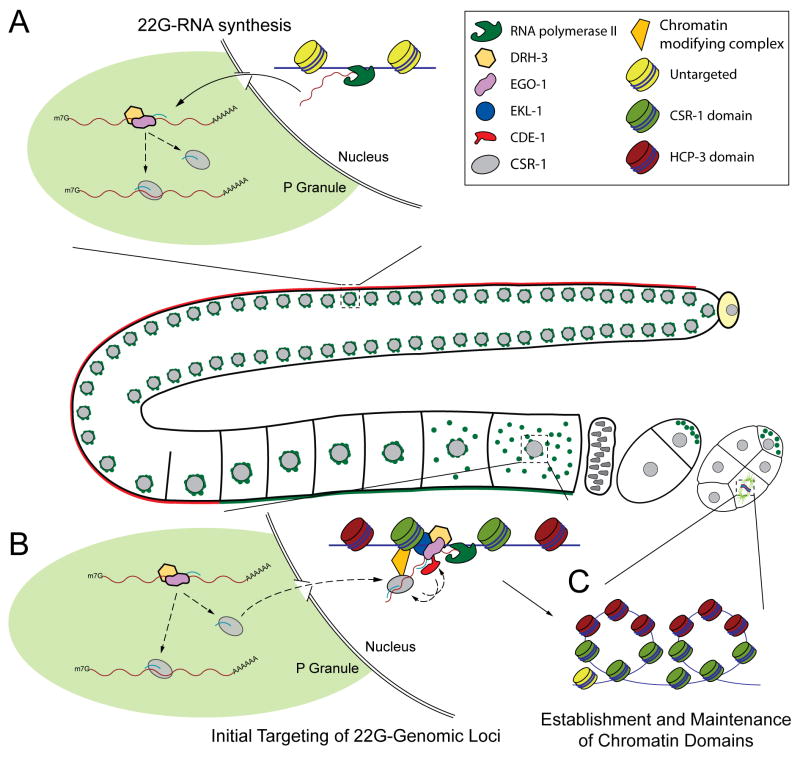
Figure 7. Model for the activity of the CSR-1 22G-RNA pathway in chromosome segregation.
(A) 22G-RNA synthesis: In the germline, DRH-3, EGO-1 and CSR-1 localize to perinuclear P Granules, where DRH-3 and EGO-1 initiate the synthesis of 22G-RNAs from transcripts that are important for germline development and early embryogenesis. These 22G-RNAs are loaded onto CSR-1 and can guide the complex to its targets.
(B) Initial targeting of genomic loci: In oocytes, CSR-1 22G-RNA complexes move into the nucleus where they target nascent transcripts, possibly by cleaving them. Chromatin modifying factors may associate with CSR-1 complexes to promote local modification of histones at and near CSR-1 target loci, establishing pericentromeric chromatin domains (green nucleosomes). A complex containing EGO-1, DRH-3, and possibly EKL-1 is proposed to amplify the signal in a positive feedback loop, by generating more 22G-RNAs in the nucleus with the CSR-1 22G-RNA-targeted nascent transcripts as the template.
(C) Establishment and maintenance of chromatin domains: The CSR-1 22G-RNA dependent chromatin domains containing modified histones (green nucleosomes) may promote the proper binding and organization of other components such as condensins and cohesins in embryo mitotic divisions. Furthermore, these chromatin domains could both help to recruit and restrict the incorporation of the centromeric Histone H3 variant, HPC-3/CENP-A (red nucleosomes) in chromatin domains adjacent to those targeted by CSR-1 22G-RNA complexes. Regions of the chromatin loop out and self-associate, permitting the assembly of a proper planar, rigid kinetochore on the poleward faces of condensed chromosomes. As cell divisions continue, chromatin domains could be maintained epigenetically, possibly even by EKL-1, thus becoming less reliant on CSR-1 22G-RNA activity throughout development.
How does CSR-1 influence chromosome segregation?
Several lines of evidence, including ChIP, chromatin isolation, and immunolocalization studies, indicate that CSR-1 pathway components associate directly with chromatin in an RNA-dependent manner. These data support a direct role for CSR-1 22G-RNA complexes in promoting chromosome segregation, perhaps through a mechanism that is similar to the Ago1 pathway that regulates centromere formation in S. pombe. Indeed, both the CSR-1 and the Ago1 pathways utilize similar components for small RNA biogenesis. These include a helicase, an RdRP and a β-nucleotidyl transferase. However, these pathways target dramatically different loci: the Ago1 system targets repetitive, pericentromeric heterochromatin, whereas the CSR-1 pathway overwhelmingly targets protein-coding euchromatic domains distributed throughout the genome. Despite this difference, perhaps the small RNAs produced in both systems perform analogous functions. The targeting of CSR-1 22G-RNA complexes to chromosomal loci in the germline could recruit chromatin modifiers that mark CSR-1 22G-RNA targeted domains and provide boundaries that define the adjacent centromeric domains of HCP-3 incorporation. Consistent with this notion, a preliminary comparison indicates that the domains targeted by CSR-1 22G-RNAs are, in large part, mutually excluded from regions that are enriched for the conserved, centromeric histone variant HCP-3/CENP-A (R. Gassmann and A. Desai, personal communication). Thus, like the S. pombe Ago1 system, the CSR-1 pathway may help to define adjacent domains of HCP-3 incorporation, but does so by targeting protein-coding genes rather than repetitive heterochromatin.
CSR-1 22G-RNA targets are distributed relatively uniformly on each chromosome, as would be expected if these targets serve in the positioning or alignment of kinetochores along the length of each chromosome. The one notable exception is the X chromosome, which is depleted of genes expressed in the germline (Reinke et al., 2000), and which possesses fewer CSR-1 targets than the autosomes (~70 versus 500–900 per autosome). It is not clear how this lower number of CSR-1 target sites might impact X chromosome segregation. The X chromosome is the only chromosome whose loss is tolerated by the organism (resulting in spontaneous males within hermaphrodite populations). Indeed, the stability of X chromosome is more than an order of magnitude lower than that of the autosomes (the loss of which is generally not detected in wild type populations) (Meneely et al., 2002). Whatever the explanation for the reduced fidelity of X chromosome segregation, clearly the limited number of CSR-1 targets are sufficient, or there are other pathways governing segregation of the X chromosome.
P granules and 22G-RNA biogenesis
CSR-1 and the other protein components of the 22G-RNA pathway localize to P granules. P granules are found in close apposition on the cytoplasmic face of nuclear pores (Pitt et al., 2000), and are thought to be sites of accumulation for many mature poly-adenylated mRNAs (Schisa et al., 2001). The nuclear association of P granules is lost in csr-1, ego-1, ekl-1 and drh-3 mutant backgrounds, suggesting that the association of CSR-1 and its co-factors with mRNA targets emerging from the nuclear pore may help to drive the perinuclear association of P granules (Figure 7A). Perhaps consistent with this idea, P granules also lose their perinuclear association in transcriptionally quiescent or nearly quiescent germ cells, e.g. oocytes and early embryo germ cells.
If their initial biosynthesis occurs in P granules, 22G-RNAs may subsequently guide CSR-1 back to chromatin or to chromatin-associated nascent transcripts (Figure 7B). Because CSR-1 targets are robustly expressed in the maternal germline, it is possible that CSR-1 complexes initially engage nascent transcripts during gametogenesis. Once established, these hypothetical CSR-1-chromatin domains could be preserved throughout embryogenesis, perhaps even in the absence of additional transcription (Figure 7C). Consistent with this idea, we found that CSR-1-chromatin localization was most prominent in the two or three most mature oocytes in each gonad arm (Figure 4A and data not shown). The retention of CSR-1 complexes at target loci could occur through direct binding to other chromatin components, possibly even through EKL-1, as tudor domains have been shown to interact with the methyl-arginine and -lysine moieties of histone tails (reviewed in Taverna et al., 2007).
Distinct roles for Argonautes in RNAi and 22G-RNA pathways
Our studies indicate that at least two distinct germline 22G-RNA pathways with several overlapping core components exist in C. elegans: the CSR-1 and WAGO-1 pathways (Gu et al., cosubmitted). Like CSR-1, WAGO-1 prominently localizes to P granules. However, the perinuclear distribution of P granules and chromosome segregation are not altered by the loss of wago-1, even within the context of a 12-fold WAGO mutant (composed of null alleles of wago-1 and eleven related WAGO Argonautes) (Gu et al., cosubmitted). How are these Argonautes loaded with distinct 22G-RNA species, despite their shared localization and reliance on upstream factors? One attractive scenario is that mRNA targets are sorted into distinct P granule sub-compartments, wherein the amplification of 22G-RNAs takes place. Additional protein factors, such as CDE-1, and/or structural elements within target transcripts may be involved in the recognition and compartmentalization of target mRNAs (see the Discussion in Gu et al., cosubmitted).
Recombinant CSR-1 protein has been shown to exhibit Slicer activity in vitro (Aoki et al., 2007), and CSR-1 has been implicated in down-regulating genes in response to foreign dsRNA (Yigit et al., 2006). However, endogenous CSR-1 22G-RNA targets do not appear to be down-regulated by CSR-1 (see above). CSR-1 22G-RNAs are expressed at low levels relative to WAGO-1 22G-RNAs (Figure S9) (Gu et al., cosubmitted), perhaps below a threshold to trigger mRNA turnover. Consistent with this idea, not all WAGO-1 22G-RNA targets exhibit mRNA silencing, but those that do typically exhibit the highest levels of corresponding 22G-RNA accumulation (Gu et al., cosubmitted).
It is tempting to speculate that the incompletely penetrant effects of csr-1 mutants on RNAi are indirect, perhaps arising as a consequence of the dramatic disruption of P granules in csr-1 mutants. There are already two distinct AGO systems implicated in the RNAi pathway, RDE-1 (Tabara et al., 1999) and the WAGO system (Yigit et al., 2006), and at least the WAGO-1 protein is localized to P granules (Gu et al., cosubmitted). In csr-1 mutants, perhaps the dissociation of P granules from germ nuclei disrupts access to target mRNAs or other activities required for the full activity of the germline RNAi response.
Our findings together with those of Gu et al. (cosubmitted) indicate that the majority of the genome is targeted by Argonaute systems that provide diverse surveillance functions. Expressed genes are targeted by CSR-1, while classically heterochromatic domains including transposons and pseudogenes are targeted by WAGO-1. Both of these systems contribute to the physical maintenance of the genome by promoting, respectively, (i) chromosome segregation and (ii) the suppression of mobile or otherwise potentially deleterious elements.
Correlates of these pathways are likely to function in other nematodes, and indeed, could help explain the classic observations in Parascaris made by Theodor and Marcella Boveri more than 100 years ago (reviewed in Pimpinelli and Goday, 1989; Satzinger, 2008). By targeting heterochromatic domains, a system analogous to the WAGO-1 pathway could promote chromosome fragmentation and the elimination of the heterochromatin in Parascaris. This could occur via an Argonaute pathway similar to that which mediates chromosome fragmentation during macronuclear formation in Tetrahymena (reviewed in Yao and Chao, 2005). By targeting genes, correlates of the CSR-1 22G-RNA system could ensure the proper higher-order assembly of the holocentric kinetochores found in diverse nematode species, and could provide this function even after fragmentation and the elimination of heterochromatin as in the tiny somatic chromosomes of Parascaris. Additional insights into the underlying molecular mechanisms through which Argonaute systems promote the higher-order structure of chromosomes will require further study. The observation that such pathways, however different, exist in nematodes and fungi suggests that similar activities are likely to be ubiquitous in eukaryotes.
EXPERIMENTAL PROCEDURES
Worm Strains
Bristol N2 was the wild type strain used in these studies. All other alleles used in this study, including csr-1(tm892) rescued strains can be found in Supplemental Experimental Procedures.
RNAi
1mg/ml dsRNA targeting drh-3, csr-1, or ekl-1 was injected into young adult Bristol N2 worms. After 36–48 hours at 20°C, worms containing embryos were dissected and fixed for immunostaining.
Antibody generation
A rabbit antibody, used in immunostaining, was generated against the CSR-1 polypeptide from amino acids E462 to E987 (containing the PAZ and most of the PIWI domain) (Capralogics, Inc.). Additional rabbit antibodies, used in IP experiments, were generated and purified by Anaspec using the peptides: VDYNAPKDPEFRQKYPNLKFP and QRCKDKGMHIGSYSMDQHNGERGSENFL. A GST-fusion protein containing an EKL-1 N-terminal fragment (L58 to S309) was used to generate rabbit antisera. DRH-3 and EGO-1 antibodies are described in (Gu et al., cosubmitted)
Immunostaining
Gonads and embryos were excised in 1x sperm salts, frozen and cracked on dry ice for 10 minutes, and fixed at −20 °C for 5 minutes each in methanol, 1:1 methanol:acetone, and acetone respectively. Blocking (one hour at 20 °C) and antibody incubations (primary, overnight at 4 °C; secondary one hour at 20 °C) were performed in 1xPBS/0.1% Tween-20/3%BSA. Washes were performed with 1xPBS/0.1% Tween-20 (PBST). DNA was stained with DAPI. For details, see Supplemental Experimental Procedures.
FISH
Embryos were dissected in egg salts with 0.1% Tween-20, followed by brief 2% formaldehyde fixation, permeabilization by freeze crack and fixation for one minute in −20 °C methanol. Slides were washed in PBST and gradually transferred to 100% ethanol. Slides were dried and incubated in 2xSSC/50% formamide at 37°C for 1 hour. The probe was sealed on the slide, DNA was denatured at 95°C for 3 minutes, and hybridization was performed overnight at 37°C. Slides were washed in 2xSSC/50% formamide, 2xSSC, 1xSSC, and PBST respectively, then counterstained with DAPI.
Western blot analysis
Is as described in (Batista et al., 2008), with the exception that proteins were resolved by SDS-PAGE on Criterion Precast gradient gels (4–15%, Biorad).
Small RNA Cloning and Data Analysis
Is as described in (Batista et al., 2008) for Terminator exonuclease (Epicentre Technologies) treated samples. For small RNA cloning of CIP/PNK (New England Biolabs) and TAP (Tobacco Acid Pyrophosphatase, Epicentre Technologies) libraries, the procedure is described in Supplemental Experimental Procedures.
Tiling Microarray Procedures
Is as described in (Batista et al., 2008). For details, see Supplemental Experimental Procedures.
Chromatin Immunoprecipitation
ChIP procedures were based on the technique of (Whittle et al., 2008), except that live embryos were treated, when indicated, (Figure 6) with 10mM dimethyl 3, 3′-dithiobispropionimidate (DTBP, Thermo-Fisher Scientific) in M9 buffer (in 50ml total), for 30 minutes at room temperature. DTBP was quenched with 2.5ml of 2.5M glycine for 5 minutes, washed with M9, and then embryos were incubated with 2.6% formaldehyde. ChIP samples were analyzed by quantitative real-time PCR. Details are found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
JMC was an HHMI fellow of the LSRF. PJB and DAC were supported by predoctoral fellowships from Fundação para Ciência e a Tecnologia, Portugal (SFRH/BD/11803/2003 for PJB; and SFRH/BD/17629/2004/H6BM for DAC). CCM is an HHMI investigator. Thanks to K. Hagstrom, J. Carey and T. Tabuchi for discussion and reagents. Thanks to M. Hammell for bioinformatics assistance, and to M. Stoltz and P. Furcinitti for assistance with confocal microscopy. We thank D. Moazed, M. Papamichos-Chronakis, R. Gassman, and A. Desai for discussion. Thanks to A. Ensminger, H.Y. Tsai, and E. Youngman for critical reading of the manuscript. Thanks to the UMMS C. elegans community, the CGC, E. Kittler and the UMMS CFAR. This work was made possible by grant GMO58800 from the NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. Embo J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Straight AF. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, Yates JR, 3rd, Meyer BJ. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol. 2001;153:F33–38. doi: 10.1083/jcb.153.6.f33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Goday C, Gonzalez-Garcia JM, Esteban MR, Giovinazzo G, Pimpinelli S. Kinetochores and chromatin diminution in early embryos of Parascaris univalens. J Cell Biol. 1992;118:23–32. doi: 10.1083/jcb.118.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essntial for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- Maine EM, Hauth J, Ratliff T, Vought VE, She X, Kelly WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely PM, Farago AF, Kauffman TM. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics. 2002;162:1169–1177. doi: 10.1093/genetics/162.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Sugimoto A, Yamamoto M. Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol Biol Cell. 2003;14:2399–2409. doi: 10.1091/mbc.E02-09-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Kashihara K, Murata M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell. 2005;17:1886–1893. doi: 10.1105/tpc.105.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ando R, Nakazawa T, Yudazono T, Tsutsumi N, Hatanaka N, Ohgake T, Hanaoka F, Eki T. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells. 2007;12:997–1010. doi: 10.1111/j.1365-2443.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Goday C. Unusual kinetochores and chromatin diminution in Parascaris. Trends Genet. 1989;5:310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Cullison K, Huang K, Bernstein Y, Spilker AC, Sundaram MV. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics. 2008;178:1431–1443. doi: 10.1534/genetics.107.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Satzinger H. Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nat Rev Genet. 2008;9:231–238. doi: 10.1038/nrg2311. [DOI] [PubMed] [Google Scholar]
- Schisa JA, Pitt JN, Priess JR. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development. 2001;128:1287–1298. doi: 10.1242/dev.128.8.1287. [DOI] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos LJ, Famulski JK, Chan GK. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol. 2006;84:619–639. doi: 10.1139/o06-078. [DOI] [PubMed] [Google Scholar]
- Vought VE, Ohmachi M, Lee MH, Maine EM. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 2005;170:1121–1132. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.