Abstract
Background and Aims
The oriental forest ecosystem in Madagascar has been seriously impacted by fragmentation. The pattern of genetic diversity was analysed on a tree species, Dalbergia monticola, which plays an important economic role in Madagascar and is one of the many endangered tree species in the eastern forest.
Methods
Leaves from 546 individuals belonging to 18 small populations affected by different levels of fragmentation were genotyped using eight nuclear (nuc) and three chloroplast (cp) microsatellite markers.
Key Results
For nuclear microsatellites, allelic richness (R) and heterozygosity (He,nuc) differed between types of forest: R = 7·36 and R = 9·55, He,nuc = 0·64 and He,nuc = 0·80 in fragmented and non-fragmented forest, respectively, but the differences were not significant. Only the mean number of alleles (Na,nuc) and the fixation index FIS differed significantly: Na,nuc = 9·41 and Na,nuc = 13·18, FIS = 0·06 and FIS = 0·15 in fragmented and non-fragmented forests, respectively. For chloroplast microsatellites, estimated genetic diversity was higher in non-fragmented forest, but the difference was not significant. No recent bottleneck effect was detected for either population. Overall differentiation was low for nuclear microsatellites (FST,nuc = 0·08) and moderate for chloroplast microsatellites (FST,cp = 0·49). A clear relationship was observed between genetic and geographic distance (r = 0·42 P < 0·01 and r = 0·42 P = 0·03 for nuclear and chloroplast microsatellites, respectively), suggesting a pattern of isolation by distance. Analysis of population structure using the neighbor-joining method or Bayesian models separated southern populations from central and northern populations with nuclear microsatellites, and grouped the population according to regions with chloroplast microsatellites, but did not separate the fragmented populations.
Conclusions
Residual diversity and genetic structure of populations of D. monticola in Madagascar suggest a limited impact of fragmentation on molecular genetic parameters.
Keywords: Dalbergia monticola, genetic structure, fixation index, bottleneck, molecular markers, Bayesian methods, Madagascar
INTRODUCTION
The impact of human activities on tropical ecosystems has increased dramatically in recent decades leading to a global reduction of primary forests. For tree species, fragmentation of forests into patches has led to degradation of both their habitat and ecological processes. Reduction of patch size, increased patch isolation, habitat loss and reduction of tree density generally lead to effects such as bottlenecks, increased genetic drift, increased inbreeding, reduced gene flow and founder effects (Lowe et al., 2005). In some cases, it has also been demonstrated that fragmentation can reduce quantitative genetic variation and enhance changes due to selection (Willi et al., 2007). However, other authors stressed that the combination of individual longevity, high intra-population genetic diversity and the potential for high pollen flow rates should make tree species especially resistant to negative fragmentation effects (Hamrick, 2004).
The forest ecosystems of Madagascar are recognized as being among the most species-rich (Myers et al., 2000), but they have been seriously impacted by a combination of recent human activities and ancient climate changes. The plant species of Madagascar are about 80 % endemic and the island contains a wealth of fauna and flora. Present patterns of the Malagasy ecosystem were determined by many factors such as the last maximum glaciation, which is believed to have had a major impact on species distribution (Gasse and Van Campo, 2001) and within-species genetic structure (Andrianoelina et al., 2006). Although human presence in the island does not date back very long (2000 years bp), human activities have had a marked influence on the distribution of species, especially since the 15th century (Straka, 1996). The highlands are thought to have been burned and are today covered with grass. More recently, during the last two centuries, the extent of the eastern forests has decreased dramatically mainly due to agricultural practices such as ‘slash and burn’, livestock production and logging. Today, primary vegetation probably covers only about 10 % of the original area (Myers et al., 2000), and dense forest has been reduced to a fragmented landscape. In addition, forest exploitation has greatly increased over the last 50 years due to rising demand for wood, fuel and saw timber. Although numerous studies have focused on conservation in Madagascar, especially for the fauna, no empirical results are available to measure accurately the impact of fragmentation on tree species genetic diversity and structure.
The study was focused on Dalbergia monticola which belongs to a genus that plays a significant economic role in Madagascar. The wood of this tree is of high quality and is used for furniture and construction (it is one of the various species sold under the common name of rosewood or palissandre). It is representative of many tree species of the eastern forest that have experienced a similar impact and present the same biological features (Du Puy and Moat, 2002; Bosser and Rabevohitra, 2005). The results presented here can thus be extrapolated to other trees.
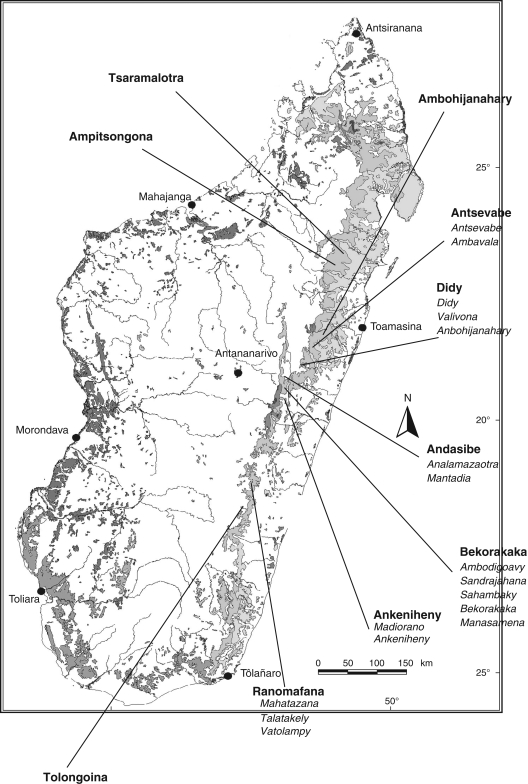
Dalbergia monticola is considered ‘vulnerable’ and is on the red list of the International Union for Conservation of Nature and Natural Resources (Du Puy, 1998). Its natural range extends from 15° to 22° latitude south including the entire primary forest on the eastern edge of the island forming a 1000 km long and 100 km wide range (Fig. 1). It is a long-lived tree species (over 200 years) and adults can reach 20 m in height and 1 m in diameter at breast height. The species is found in two main ecological zones: sub-montane evergreen seasonal forest and dense rainforest. The populations are located at altitudes ranging from 350 m to 1600 m, with mean temperatures ranging from 18 °C to 23 °C, and mean annual rainfall ranging from 750 mm to 2500 mm. Dalbergia monticola reproduces mainly sexually and is mainly insect pollinated. Flowering and fruiting extend from August to November, with some geographical variations. The species is mainly barochorous, but the seeds can also be dispersed by birds, monkeys and rodents, although no research has been conducted on this topic. As far as is known no experiment was conducted to study the ecological conditions of regeneration. However, field observations showed that seedling and juvenile are mainly present in open areas.
Fig. 1.
Map of Madagascar showing remaining primary vegetation (grey), location of the provenances (in bold) and populations (in italics) sampled throughout the eastern forest.
Chloroplast and nuclear microsatellite markers were used to analyse genetic diversity of Dalbergia monticola. This combination facilitates the understanding of both historical and recent events. It provides a complementary view of gene flow pattern because chloroplasts are transmitted by seeds in angiosperms (Petit et al., 2005). Chloroplast markers are also more sensitive to drift, one of the main effects of fragmentation. Although criticised for a homoplasy effect, i.e the same number of repeats may evolve in two different microsatellite lineages through independent mutational events (Navascués and Emerson, 2005), recent simulations have demonstrated that chloroplast microsatellites are efficient in studying genetic structure and gene flow (Hansen et al., 2005). Both marker types are expected to provide useful information for the implementation of a conservation strategy and restoration of populations in Madagascar as suggested by different studies (Crandall et al., 2000; Lhuillier et al., 2006; Muller et al., 2009).
The aim of the present study was thus to evaluate, in the context of fragmented eastern Malagasy forest, the relationship between degree of fragmentation and genetic diversity level and structure of one representative forest tree species. To test for fragmentation effects, the genetic diversity and structure parameters between sets of patches located in fragmented zones were compared with patches that were present in untouched forests.
MATERIALS AND METHODS
Plant material
Due to the effect of logging and fragmentation and the consequent rarity of populations and individuals, sampling was laborious. In addition, the poor road network and the low quality of existing roads prevented exhaustive sampling (Table 1).
Table 1.
Main characteristics and coordinates of the populations sampled within the natural range of D. monticola
| Region | Nreg | Provenance | NProv | Population within provenance | NPop | Latitude (S) | Longiude (E) | Altitude | Min_max circ (cm)* | Area† | Landscape code‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| North | 19 | Tsaramolotra§ | 10 | Tsaramolotra | 10 | 16°57′ | 48°44′ | 900–1200 | – | ≈6 ha | 3 |
| Ampitsongona | 9 | Ampitsongona | 9 | 17°05′ | 48°42′ | 900–1200 | – | ≈4 ha | 3 | ||
| Centre-North | 59 | Didy | 33 | – | 18°10′ | 48°35′ | 800–900 | ||||
| Didy | 13 | – | 4·50 ha | 3 | |||||||
| Valivona | 20 | – | 5·10 ha | 2 | |||||||
| Anbohijanahary | 9 | Anbohijanahary | 9 | 17°45′ | 48°35′ | 800–900 | – | ≈3 ha | 3 | ||
| Antsevabe | 17 | – | 17°55′ | 48°32′ | 800–900 | ||||||
| Antsevabe | 11 | – | – | – | – | ≈3 ha | 3 | ||||
| Ambavala | 6 | – | – | – | – | ≈3 ha | 2 | ||||
| Centre | 243 | Ankeniheny | 8 | Madiorano/Ankeniheny | 8 | 19°10′ | 48°35′ | 800–1100 | 56–75 | ≈3 ha | 3 |
| Bekorakaka | 205 | – | 19°06′ | 48°21′ | 850–900 | ||||||
| Ambodigoavy | 12 | – | – | – | 24 ha | 3 | |||||
| Sandrajahana | 61 | – | – | – | 10–67 | 9·68 ha | 3 | ||||
| Sahambaky | 58 | – | – | – | 11–70 | 4·95 ha | 3 | ||||
| Bekorakaka | 56 | – | – | – | 10–71 | 16·84 ha | 3 | ||||
| Manasamena | 18 | ≈16 ha | 3 | ||||||||
| Andasibe | 30 | – | 18°56′ | 48°25′ | 800–1000 | ||||||
| Analamazaotra | 21 | – | – | – | – | 4·50 ha | 1 | ||||
| Mantadia | 9 | – | – | – | – | 2·20 ha | 1 | ||||
| South | 225 | Tolongoina | 18 | Tolongoina | 18 | 21°34′ | 47°32′ | 800–1200 | – | 1·60 ha | 2 |
| Ranomafana | 207 | – | 21°16′ | 47°26′ | 900–1000 | ||||||
| Mahatazana | 26 | – | – | – | – | 4 ha | 2 | ||||
| Talatakely | 121 | – | – | – | 12–213 | 10·92 ha | 1 | ||||
| Vatolampy | 60 | – | – | – | 16–109 | 3·56 ha | 1 | ||||
| Total | 546 |
Nreg is the number of individuals in each region, NProv is the number of individuals in each provenance, NPop is the number of individuals in each population.
* Minimum and maximum circumference of individuals sample in the population.
† Approximate area of total forest habitat occupied by population estimated by the product of the transect length and the width of the band prospected (20 m each side).
‡ Landscape code: 1, forest landscape presenting no fragmentation but some small zones (<0·5 ha) are clear cut within the forest; 2, recently fragmented forest, characterized by large fragments (several hundred hectares) separated by 500–1500 m; 3, forest strongly impacted by slash and burn leading to fragmentation, in which the landscape is characterized by small forest fragments (10–20 ha) separated by >2000 m (fragmentation occurred >20 years ago).
§ Provenance represented by a single population.
Leaves of Dalbergia monticola Bosser & R.Rabev. from 567 individuals of ten provenances (geographic zones; Fig. 1) scattered over four main regions of the eastern forest, north, centre-north, centre and south, were collected in March 2004 and March 2006. The regions were chosen to separate sampling areas but were not of any particular interest from an ecological point of view. Sampling covered most of the natural range. Within each provenance, a different number of populations was sampled depending on the abundance of the species. Each population was defined as a set of individuals located in the same zone which show no significant geographical disruption (Table 1). The number of populations varied from one provenance to another and some were separated by several kilometres. The number of trees varied between populations and when the number was smaller than eight, analyses were only conducted at the level of the provenance.
Five leaves were collected from each tree and immediately dried using silica gel. When the population size was large enough, trees separated by >10 m were selected to avoid selecting the same clone (assuming that resprouting by root suckers can create patches of the same clone). The choice of this distance to avoid clones was confirmed by analysis. The hypothetical presence of clones was tested with the procedure used in Lhuiller et al. (2006) (results not shown).
Populations were sampled from three landscapes, coded 1, 2 and 3 (Table 1), and differentiated by their disturbance level. Landscape 1 corresponded to forest landscapes presenting no fragmentation, but some small zones (<0·5 ha) were clear cut within the forest. Code 2 corresponded to recently created large forest fragments (several hundred hectares) separated by 500–1500 m of secondary forest, fallows and agriculture fields. The population coded 3 corresponded to forest strongly impacted by slash and burn agriculture and was characterized by small fragments (between 10 ha and 20 ha) separated by >2000 m of secondary forest, agriculture fields and fallows. According to the forest service, fragmentation of this type occurred >20 years ago.
The tree circumference was measured in some populations (Table 1). Within populations, the distribution of diameter classes showed that the sample was composed by a mixture of cohorts. Diversity parameter estimates were calculated without distinguishing cohorts, although cohort is considered in the Discussion.
DNA extraction
DNA was extracted from dried leaves according to a modified protocol derived from Bousquet et al (1990). The protocol is described in Andrianoelina et al (2006).
Molecular markers
Chloroplast microsatellite method
Seven universal microsatellite primers (Ccmp) described by Weising and Gardner (1999), and a set of 33 microsatellite primers (Ntcp) conserved within the Solanaceae family described by Bryan et al. (1999), were tested over a subset of the populations. Among these 40 primer pairs tested on a sample of eight individuals, three were polymorphic (ccmp4, ccmp6 and ccmp7), showing differences in mononucleotide repeats. The amplification protocol is described in Andrianoelina et al. (2006). The genotyping was performed on acrylamide gel and visualized by silver staining. The allele size has been determined by comparison to a DNA ladder.
Nuclear microsatellite method
Genetic analysis was performed using eight nuclear microsatellites designed specifically for Dalbergia monticola Bosser & R. Rabev.: mDmCIRA04, mDmCIRA08, mDmCIRA12, mDmCIRB06, mDmCIRC02, mDmCIRC10, mDmCIRD01 and mDmCIRD02. The design of the primers, their characteristics, the PCR amplification and the genotyping are described in Favreau et al. (2007). Three loci (mDmCIRB06, mDmCIRA04 and mDmCIRC02) showed suspicious FIS values and a high percentage of missing data in a preliminary analysis. They gave systematically positive and high values of FIS which could be due to a null allele effect. Hence, it was decided to remove them from the analyses.
Statistical analyses
Diversity parameters
Allele frequencies, the number of alleles per locus (Na), observed heterozygosity (HO), expected heterozygosity (He; Nei, 1978), and the fixation index (FIS) per population were computed for each population, provenance and region. To check if diversity estimates were affected by the differences in the sample size, allelic richness per population was calculated. The dependence on sample size was taken into account by adapting the rarefaction index of Hurlbert (1971; El Mousadik and Petit, 1996), named ‘R’. The principle is to estimate the expected number of alleles in a sub-sample of 2n genes, given that 2N genes have been sampled (N > n). In FSTAT, n is fixed as the smallest number of individuals typed for a locus in a sample. The estimates of R were based on minimum sample size, which varied depending on the set of populations used for the estimation. All the estimates were obtained using FSTAT version 2.9.3.2 (Goudet, 2001).
A chlorotype was defined as a combination of the different alleles established at each locus. Because of the non-recombining nature of the chloroplast genome, chlorotypes were then treated as alleles at a single locus. Chlorotype diversity and genetic structure parameters were calculated using ARLEQUIN version 2.0 (Schneider et al., 2000). The gene diversity index Hcp was calculated using the Nei formula (Nei, 1987). The number of chlorotypes (Na,cp) and the effective chlorotype number (Ne,cp; Nei, 1987) were calculated for each population.
Mean comparisons
The differences among regions, provenances, populations and among level of fragmentation for genetic diversity parameters (heterozygosity, number of alleles, allelic richness) and fixation index were studied by one-way analysis of variance followed by a paired t-test for mean comparison (Nei, 1987; Tapio et al., 2003). Bonferroni corrections were applied during tests. These analyses were performed with the XLSTAT software version 2008.4.01 (Addinsoft, 2008).
Differentiation among populations
The global and pairwise FST among populations were estimated by the procedure of Weir and Cockerham (1984) and were calculated using FSTAT (Goudet, 2001). Population differentiation was tested by randomizing genotypes among samples. The test was based on 1000 randomizations, and probability values were determined according to the approach described by Excoffier et al. (1992), and compared with the P-value adjusted by the sequential Bonferroni procedure (Rice, 1989).
A complementary approach based on Bayesian theory was carried out with the STRUCTURE program (version 2β) (Pritchard et al., 2007) to estimate the number of clusters K and assign all individuals to the clusters (Pritchard et al., 2000, http://pritch.bsd.uchicago.edu). A non-admixture model with correlated allele frequencies was used and five independent runs of K were performed at 200 000 MCMC repetitions and a 100 000 burn-in period. The number of clusters was determined by the approach developed by Evanno et al. (2007).
The geographical distribution of cpDNA polymorphism was assessed with a Bayesian spatial analysis of population structure performed with the software BAPS 5.1 (Corander et al., 2004, 2008). The maximum number of clusters (K) was initially set to 20, and then the k-value exhibiting the minimum log-likelihood was selected in order to obtain the optimal partition of populations.
To illustrate the differentiation between populations, a distance tree was constructed using Darwin 5.0.84 software (Perrier et al., 2003). Pairwise genetic distances between pairs of populations were computed using Euclidian distance. The distance tree was constructed using the neighbor-joining method of Saitou and Nei (1987). The robustness of each node was assessed by bootstrapping data over loci and alleles with 1000 replications. A consensus tree using the method of strict consensus was elaborated with the two previous trees.
Spatial pattern among provenances
The spatial pattern of genetic diversity distribution was analysed using a Mantel test (Mantel, 1967). The test was used to check the correlation between the matrix of geographical distances, between provenances, and the genetic distances estimated by pairwise FST for nuclear and chloroplast microsatellites. This test was performed with the XLSTAT software version 2008.4.01 (Addinsoft, 2008).
Detection of recent bottlenecks
Test for the occurrence of a recent bottleneck was performed using the heterozygosity excess method described by Cornuet and Luikart (1996) and implemented in the BOTTLENECK software (Piry et al., 1999). The tests are based on the fact that populations that have experienced a recent reduction in effective size should exhibit a more rapid reduction in allelic diversity than observed heterozygosity. Hence, in a recently bottlenecked population, gene diversity is higher than the equilibrium heterozygosity estimated from the observed allele numbers, assuming mutation drift equilibrium (Luikart and Cornuet, 1998). Due to the small number of loci, Wilcoxon's signed-rank test was used to test the significance of the difference between observed and modelled heterozygosity. As recommended by Piry et al. (1999) in the case of microsatellite markers, the model used was a two-phase mutation model with 95 % stepwise mutations and 5 % multi-step mutations. However, the effect of 10 % multi-step mutations on the results was also tested.
RESULTS
After elimination of samples with >60 % of missing data, the total number of remaining individuals was 546. As a consequence, some populations had a very small number of individuals, and parameters were consequently calculated for populations containing more than eight individuals (Table 2).
Table 2.
Diversity parameters assessed with nuclear microsatellite markers in the populations of D. monticola
| Nuclear microsatellites |
Chloroplast microsatellites |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Provenance | Population | Landscape† | Nnuc | Na,nuc | R‡ | He,nuc | Honuc | FIS | Ncp | Na,cp | Ne,cp | Hcp |
| North | 19 | 8·20 | 6·43 | 0·76 | 0·63 | 0·27** | 13 | 2 | 1·16 | 0·14 | |||
| Tsaramolotra | Tsaramolotra | 3 | 10 | 4·60 | – | 0·71 | 0·65 | 0·20 n.s. | 7 | 2 | 1·32 | 0·24 | |
| Ampitsongona | Ampitsongona | 3 | 9 | 5·20 | – | 0·64 | 0·58 | 0·25* | 6 | 1 | 1·00 | 0·00 | |
| Centre-north | 59 | 12·00 | 6·78 | 0·81 | 0·64 | 0·18** | 47 | 7 | 3·65 | 0·73 | |||
| Didy | 33 | 9·60 | 7·71 | 0·79 | 0·64 | 0·17* | 26 | 5 | 2·70 | 0·63 | |||
| Didy | 3 | 13 | 6·60 | – | 0·74 | 0·68 | 0·10 n.s. | 15 | 5 | 2·10 | 0·52 | ||
| Valivona | 2 | 20 | 8·00 | 7·14 | 0·79 | 0·62 | 0·19* | 11 | 1 | 1·00 | 0·00 | ||
| Ambohijanahary | Ambohijanahary | 3 | 9 | 5·00 | – | 0·67 | 0·50 | 0·34* | 10 | 6 | 5·00 | 0·80 | |
| Antsevabe | 17 | 8·60 | 7·53 | 0·75 | 0·68 | 0·05 n.s. | 11 | 3 | 2·44 | 0·59 | |||
| Antsevabe | 3 | 11 | 4·40 | – | 0·71 | 0·70 | –0·07 n.s. | 11 | 2 | 1·86 | 0·46 | ||
| Ambavala | 2 | 6 | – | – | – | – | – | – | – | – | |||
| Centre | 243 | 16·20 | 6·61 | 0·79 | 0·60 | 0·17** | 38 | 12 | 5·78 | 0·83 | |||
| Ankeniheny | Madiorano /Ankeniheny | 3 | 8 | 5·80 | – | 0·71 | 0·68 | 0·07 n.s. | 8 | 4 | 2·28 | 0·56 | |
| Bekorakaka | 205 | 15·20 | 8·62 | 0·79 | 0·68 | 0·18* | 9 | 7 | 4·54 | 0·78 | |||
| Ambodigoavy | 3 | 12 | 7·20 | – | 0·77 | 0·71 | 0·10 n.s. | 9 | 6 | 3·85 | 0·74 | ||
| Sandrajahana | 3 | 61 | 10·80 | 6·74 | 0·77 | 0·59 | 0·15* | – | – | – | – | ||
| Sahambaky | 3 | 58 | 11·60 | 7·06 | 0·76 | 0·58 | 0·18* | – | – | – | – | ||
| Bekorakaka | 3 | 56 | 11·40 | 6·53 | 0·76 | 0·56 | 0·19* | – | – | – | – | ||
| Manasamena | 3 | 18 | 6·60 | 6·43 | 0·70 | 0·66 | 0·06 n.s. | – | – | – | – | ||
| Andasibe | 30 | 8·60 | 7·36 | 0·73 | 0·66 | 0·05 n.s. | 21 | 9 | 6·01 | 0·83 | |||
| Analamazaotra | 1 | 21 | 8·00 | 6·64 | 0·73 | 0·68 | 0·03 n.s. | 14 | 6 | 3·76 | 0·73 | ||
| Mantadia | 1 | 9 | 6·20 | – | 0·66 | 0·62 | 0·07 n.s. | 7 | 5 | 4·45 | 0·78 | ||
| South | 225 | 19·20 | 7·28 | 0·79 | 0·70 | 0·08** | 58 | 5 | 2·64 | 0·62 | |||
| Tolongoina | Tolongoina | 2 | 18 | 9·00 | 7·27 | 0·77 | 0·74 | 0·04 n.s. | 15 | 3 | 2·27 | 0·56 | |
| Ranomafana | 207 | 18·40 | 9·55 | 0·79 | 0·70 | 0·08* | 43 | 4 | 2·46 | 0·59 | |||
| Mahatazana | 2 | 26 | 9·60 | 7·40 | 0·79 | 0·72 | 0·12 n.s. | 26 | 3 | 1·26 | 0·21 | ||
| Talatakely | 1 | 121 | 14·80 | 7·44 | 0·77 | 0·68 | 0·07 n.s. | 17 | 2 | 1·71 | 0·41 | ||
| Vatolampy | 1 | 60 | 12·80 | 7·08 | 0·74 | 0·68 | 0·03 n.s. | – | – | – | – | ||
| Total | 546 | 22·00 | 21·62 | 0·82 | 0·68 | 0·17 | 156 | 18 | 9·87 | 0·90 | |||
N, number of individuals genotyped; Na, number of alleles per locus; Ho, observed heterozygosity; He, expected heterozygosity; R, corrected allelic richness; FIS, fixation index.
P-values: n.s., P > 0·05;*, P < 0·05;**, P < 0·01;***; P < 0·001 (P-values were adjusted using the sequential Bonferroni procedure; Rice, 1989) [the indicative adjusted nominal level (5 %) is 0·0021, 0·00084, 0·00042 for region, provenance and population, respectively].
† Landscape code: 1, forest landscape presenting no fragmentation but some small zones (<0·5 ha) are clear cut within the forest; 2, recently fragmented forest, characterized by large fragments (several hundred hectares) separated by 500–1500 m; 3, forest strongly impacted by slash and burn leading to fragmentation, in which the landscape is characterized by small forest fragments (10–20 ha) separated by >2000 m (fragmentation occurred >20 years ago).
‡ Allelic richness per locus and population is based on minimum sample size of seven diploid individuals for regions, 14 diploid individuals for provenances and 15 diploid individuals for populations.
Genetic diversity across the natural range
Nuclear microsatellites
The diversity parameters varied across regions (Table 2). The estimates of diversity parameters of the north region were lower than those of other regions but the statistical test for mean comparison did not detect significant differences at the 5 % level (P = 0·017 after Bonferroni correction). An analogous range of genetic parameters variation was observed among the ten provenances (Table 2) Only Na,nuc presented significant differences at the 5 % level (P = 0·001 after Bonferroni correction), the Ranomafana provenance exhibiting a significantly higher estimate (Na,nuc = 18·40) than the nine others (ranging from 4·60 to 15·20). As for the populations, the range of variation was smaller for Na,nuc (4·40–14·80), which can be explained by the smaller sample size, but similar for He,nuc which varied from 0·64 to 0·79. Allelic richness was assessed with only 13 populations, as the estimation was not possible with populations below 13 individuals, and presented a small range of variation (6·43–7·44).
The fixation index FIS was significantly different from zero for all regions with relatively high positive values, but the difference among regions were not significant at the 5 % level (P = 0·017 after Bonferroni correction). Six out of ten provenances did not present a significant FIS and their differences were not significant. Similarly, 12 populations out of 18 did not exhibit a significant FIS and the differences among FIS were not significant at the 5 % level.
Chloroplast microsatellites
Based on the combination of the three universal chloroplast probes, 18 chlorotypes could be identified (results not shown). One chlorotype was particularly frequent (20·13 %), three others showed a frequency close to 10 %. The frequencies of the remaining chlorotypes varied between 0·63 % and 5·66 %. The diversity parameters varied markedly among the regions with highest estimates for centre region (Table 2), but no test can be used to statistically confirm the differences. The same pattern was observed for the ten provenances. The provenances originating from the centre region exhibited highest variability, e.g. Bekorakaka and Andasibe (Table 2). The diversity parameters varied greatly among the 13 populations, e.g. the number of chlorotypes (Na,cp = 1·00 in Ampitsongona to Na,cp = 6·00 in Ambodigoavy). The pattern of variation of these parameters was not linked to the sample size of the population, e.g. Ambohijanahary presented a high variability, but included only ten individuals.
Effect of fragmentation on diversity parameters
For the nuclear microsatellites, the results of the weighted ANOVA test for the fragmentation effect, based on diversity parameters, showed a significant difference among the three levels of disturbance for the mean number of alleles Na,nuc and the fixation index FIS (Table 3). Na,nuc was higher and FIS lower in non-fragmented populations at the 5 % level (1·17 % after Bonferroni correction). Although the non-fragmented forests presented higher estimates of R and He,nuc, the differences were not significant. For chloroplast microsatellites, a higher diversity was observed in non-fragmented forest for Na,cp, Ne,cp and Hcp, but there were no significant differences among the three levels of fragmentation (Table 3). In order to reduce the confounding effects of sample size, region and fragmentation level, the same analyses were conducted with samples restricted to the south and centre regions. Only FIS presented a significant difference and was lower in non-fragmented populations at the 5 % level (1·17 % after Bonferroni correction; Table 3).
Table 3.
Results of one-way weighted analysis of variance testing the effect of fragmentation on the diversity parameters for nuclear and chloroplast microsatellites (P: probability values associated with the Fisher test) and paired t-test for mean comparisons (values followed by the same letter are not significantly different at the 5 % level after Bonferroni correction, P = 0·017)
| Nuclear microsatellites |
Chloroplast microsatellites |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Landscape | Npop (Nind) | Na,nuc | R* | He,nuc | FIS | Npop (Nind) | Na,cp | Ne,cp | Hcp |
| All populations | |||||||||
| P = 0·029 | P = 0·065 | P = 0·282 | P = 0·018 | P = 0·364 | P = 0·202 | P = 0·130 | |||
| Code 1: no fragmentation | 4 (211) | 13·18a | 7·25a | 0·75a | 0·05a | 3 (38) | 4·03a | 2·97a | 0·60a |
| Code 2: weak fragmentation | 3 (64) | 8·93ab | 7·28a | 0·78a | 0·12ab | 4 (54) | 2·52a | 1·48a | 0·26a |
| Code 3: strong fragmentation | 11 (265) | 9·41b | 6·75a | 0·75a | 0·15b | 7 (66) | 3·99a | 2·58a | 0·51a |
| Populations from South and Centre regions | |||||||||
| P = 0·179 | P = 0·091 | P = 0·564 | P = 0·006 | P = 0·531 | P = 0·374 | P = 0·319 | |||
| Code 1: no fragmentation | 4 (211) | 13·19a | 7·25a | 0·75a | 0·06a | 3 (38) | 4·03a | 2·97a | 0·60a |
| Code 2: weak fragmentation | 2 (44) | 9·36a | 7·35a | 0·78a | 0·09ab | 2 (41) | 3·00a | 1·63a | 0·34a |
| Code 3: strong fragmentation | 6 (203) | 10·43a | 6·75a | 0·76a | 0·16b | 2 (17) | 5·05a | 3·11a | 0·66a |
Npop, Number of populations in each level of fragmentation; Nind, number of individual trees corresponding to the populations.
* ANOVA based on the comparison of ten populations with sample sizes greater than 15 individuals (N = 3, N = 3 and N = 4 for no fragmentation, weak fragmentation and strong fragmentation, respectively).
Genetic differentiation among populations
Nuclear microsatellites
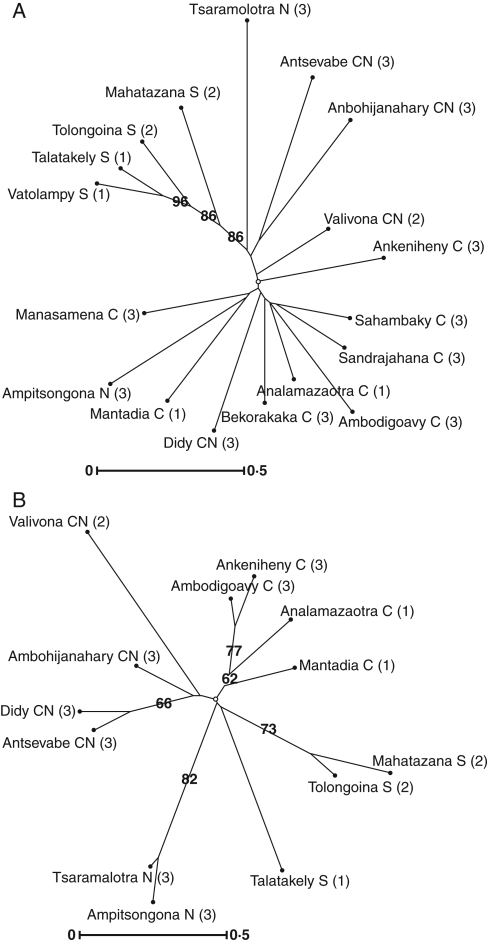
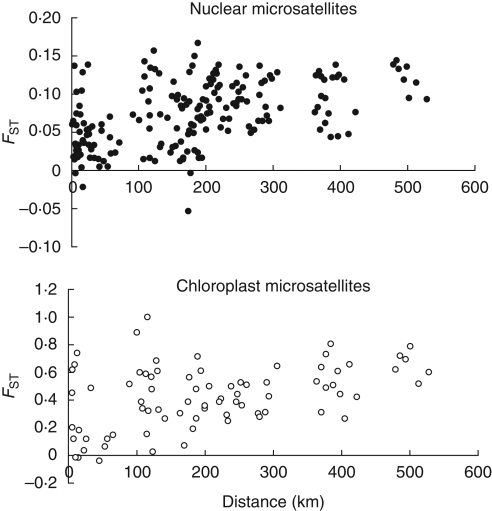
Overall estimation of population differentiation showed a moderate but significant FST,nuc (FST,nuc = 0·08, P < 0·0001). Pairwise FST,nuc among provenances presented values ranging from 0·02 to 0·15. Twenty-two pairwise FST,nuc out of 45 were significantly different from zero after Bonferroni correction [the indicative adjusted nominal level (5 %) for multiple comparisons was 0·0011]. With values ranging from −0·04 to 0·15, a lower proportion of significant pairwise FST,nuc was observed among populations: 57 out of 190 were significant [adjusted nominal level (5 %) for multiple comparisons equal to 0·000263]. The genetic relationship among populations is illustrated in the neighbor-joining tree (Fig. 2A). Many branches presented a bootstrap value lower than 50, showing weak differentiation among populations in the centre and centre-north regions. Only populations from the south formed a significant cluster (bootstrap value higher than 80). The Bayesian approach (STRUCTURE program) to detect the number of clusters led to a clear solution for K = 2 with a marked modal value of ΔK = 900 defined in Evanno et al. (2007). Two faint modes were observed for K = 3 and K = 9 corresponding to ΔK = 50 and ΔK = 20, respectively. The clearly visible mode at K = 2 corresponded to the separation of southern populations from the set of central and northern populations. The scattered points between the genetic pairwise FST,nuc and the geographic distances among populations presented a linear relationship (Fig. 3A). This observation was confirmed by the Mantel test, which revealed a significant coefficient of correlation between the genetic and geographic distances (r = 0·42, P < 0·01).
Fig. 2.
Unrooted neighbor-joining tree drawn with Darwin 5.0.84 software (Perrier et al., 2003), with the matrix of genetic distances calculated using the Euclidian distance. Numbers at the base of the branches are percentages corresponding to the bootstrap values after 1000 replications (only bootstraps higher than 50 % are shown). (A) Nuclear microsatellites; (B) chloroplast microsatellites. Region abbreviations: C, Centre; N, North; CN, Centre-North; S, South. For landscape codes (1, 2, 3) see Table 1.
Fig. 3.
Relationship between genetic and geographical distances among populations of Dalbergia monticola. Matrices of genetic distances were calculated using pairwise FST. (A) Nuclear microsatellites; (B) chloroplast microsatellites.
Chloroplast microsatellites
As expected, differentiation among provenances and populations was more pronounced when based on chloroplast microsatellites. The overall estimation of differentiation among the ten provenances was high and significant FST,cp (FST,cp = 0·48, P < 0·0001). Among the 45 pairwise comparisons for provenances, 39 FST,cp were significant. Regarding the 13 populations, the global FST,cp presented a slightly higher significant estimate (FST,cp = 0·49, P < 0·0001). Twenty-three per cent of the variability was distributed among regions and 26 % among populations within regions. The pairwise FST,cp among 13 populations presented values ranging from −0·04 to 1·00. Seventy pairwise FST,cp out of 78 were significantly different from zero after Bonferroni correction [the indicative adjusted nominal level (5 %) for multiple comparisons was 0·0011]. The genetic relationship among populations was illustrated by the neighbor-joining tree (Fig. 2B). Many branches presented a bootstrap value higher than 50, demonstrating the strong differentiation among populations within and among regions. The Bayesian approach implemented by the BAPS 5.1 software confirmed this clustering. The number of clusters in optimal partition was seven (probability of 0·87) and the best partition was: Cluster 1 {Tsaramolotra, Ampitsongoina}; Cluster 2 {Valivona}, Cluster 3 {Didy, Ambohijanahary, Antsevabe}, Cluster 4 {Ankeniheny, Ambodigoavy, Mantadia}, Cluster 5 {Analamazaotra}, Cluster 6 {Tolongoina, Mahatazana} and Cluster 7 {Talatakely}. Although the Mantel test revealed a moderately high and significant coefficient of correlation between the genetic and geographic distances (r = 0·42, P < 0·03) (Fig. 3B), the linear relationship between genetic and geographic distance was not marked among pairs of populations located between 0 and 100 km (Fig. 3B).
Bottleneck
The search for a recent bottleneck was conducted within populations containing more than ten individuals, which is the minimum required to run the test. For each population, the Wilcoxon sign-rank test did not veer from the mutation drift-equilibrium and recent bottlenecks were not detected (results not shown), whatever the percentage of multi-step mutations (5 or 10 %). The probability associated with the one tail for heterozygosity excess was considerably higher than 0·05. The juvenile and adult cohorts in the Ranomafana and Bekorakaka populations (trees smaller than 10 cm diameter in the juvenile cohort and higher in the adult cohort) were also separated to detect a possible effect of generation on a possible bottleneck. No sign of a recent bottleneck was observed in the youngest cohorts (results not shown).
DISCUSSION
Effect of fragmentation on genetic diversity
Although this type of comparison must be considered with caution due to the different number of markers and the sample size effect on estimations, the genetic diversity within D. monticola is one of the highest among tropical tree species assessed with nuclear microsatellites (see the review in Muller et al., 2009). The present study reveals that the highest He,nuc values (around 0·80) correspond to species distributed in a continental zone that has not yet been overexploited and are distributed in a non-fragmented forest, e.g. Swietenia macrophylla (Lemes et al., 2003; Novick et al., 2003) or Caryocar brasiliense (Collevatti et al., 2003).
Comparison of diversity parameters among fragmented (codes 2 and 3) and non-fragmented populations (code 1) reveals significant differences for Na,nuc. This parameter is strongly influenced by population size as shown by the same analyse performed only with populations from the south and centre regions. It was observed that the differences for Na,nuc were not significant (P = 0·179) (Table 3). For He,nuc the difference is not significant with both samples. In addition, in Bekorakaka and Ranomafana provenances that include juvenile and adult cohorts (defined arbitrarily with diameter smaller and greater than 10 cm, respectively) no significant difference was observed for He,nuc (results not shown). This result is in agreement with simulations conducted by Lowe et al. (2005). They have shown that a change in genetic diversity occurs after fragmentation when the real population size is markedly reduced, and after a considerable number of generations. For example, in their simulation, heterozygosity (H) decreased by 0·1 after ten generations in isolated populations of 50 individuals. The absence of differences among populations of D. monticola for He,nuc could mean that fragmentation is too recent to have a strong impact. FIS was significantly lower in non-fragmented forest with both samplings (all populations versus populations from the south and centre regions) (Table 3). The increase of the fixation index in fragmented populations can be explained by higher inbreeding due to the isolation of adult trees after logging, which favours self-pollination. In the fragmented zones, logging was very common and a lower tree density was noticed in theses stands. Similar results were observed in other studies but for young cohorts, e.g. in juveniles (Aldrich and Hamrick, 1998) and progenies of other species (Doligez and Joly, 1997; Dick et al., 2003; Fuchs et al., 2003; André et al., 2008).
For chloroplast microsatellites, the genetic diversity within D. monticola is one of the highest among tropical tree species assessed with chloroplast microsatellites (see the review in Muller et al., 2009). Although the higher estimates were observed for non-fragmented forest, Table 3 stresses the absence of significant difference among populations. There are various possible explanations for this. First, the small sample size from the non-fragmented forest prevents sampling of the total diversity. Secondly, some artefacts related to sampling have biased the comparison. For example, the present data stressed the high diversity in the fragmented population of Ambodigoavy (Na,cp = 7), although the number of individuals is small (ten trees). This could be explained by the wide zone used for sampling (24 ha) compared with other populations. Thirdly, there is still a high variability in fragmented populations due to the recent fragmentation and the weak effect of drift.
The heterozygosity excess method suggested that none of the populations had undergone a recent bottleneck. According to Cornuet and Luikart (1996), the Wilcoxon test can be used with as few as four polymorphic loci and any number of individuals. However, 15–40 individuals and 10–15 polymorphic loci are recommended to achieve high power. The effect of fragmentation was not observed with the present design, but the use of more loci may change these results.
Fixation index within populations
Positive and significant values of FIS were observed for regions and some provenances. This result was expected due to the aggregation of different populations exacerbating the Wahlund effect.
At the population level, it was observed that six populations among 18 presented positive and significant FIS values (Ampitsongona, Valivona, Ambohijanahary, Sandrajahana, Sahambaky and Bekorakaka) (Table 2). As a consequence, significantly higher estimates of FIS were noticed in fragmented forests than in untouched forest (Table 3). This result is expected in tree populations experiencing fragmentation due to the reduction in population size and the isolation of trees (Savolainen and Kuittinen, 2000; Henry and Gouyon, 2003), although it was not systematically observed in previous studies (Honnay and Jacquemyn, 2007; Aguilar et al., 2008; Kramer et al., 2008). Aguilar et al. (2008) noticed that significant increases in inbreeding coefficient of the fragmented habitats were only observed in studies analysing progenies. He explained the absence of overall significant effects on inbreeding coefficients because most sampled trees were adults (and were probably established before fragmentation took place). In the case of D. monticola, although the mode of insect pollination with this species is not known, selfing may contribute to higher FIS in disturbed forests. A more pronounced isolation of mature trees in fragmented zones was noticed during sampling activities as these zones have also undergone selective logging, reducing the density of adult trees. For example, in the provenance of Bekorakaka, selected trees are separated by 200–1000 m, whereas they were separated by 50–100 m in the untouched provenance of Ranomafana, thus facilitating cross-pollination. Other researchers have found an increase of FIS linked to a reduction in tree density, e.g. Carapa procera (Doligez and Joly, 1997), Symphonia globulifera (Aldrich and Hamrick, 1998), Dinizia excelsa (Dick et al., 2003), Pachira quinata (Fuchs et al., 2003), Swietenia macrophylla (André et al., 2008) and Prunus africana (Farwig et al., 2008). In addition, distribution of diameter class in fragmented populations (Table 1) showed that a mixture of generations was sampled which suggests that trees that appeared after fragmentation were selected and genotyped, thus contributing to higher estimates of FIS. However, calculation of FIS in juvenile and adult cohorts (defined arbitrarily with diameter smaller and greater than 10 cm, respectively) gave highly positive significant values in three populations of Bekorakaka (FIS = 0·17* and FIS = 0·15* in Sandrajahana, FIS = 0·21* and FIS = 0·18* in Sandrajahana and FIS = 0·23* and FIS = 0·16* in Bekorakaka for both adult and juvenile cohorts, respectively). It gave smaller values in two populations of Ranomafana (FIS = 0·10* and FIS = −0·03 in Talatakely and FIS = 0·04 and FIS = 0·02 in Vatolampy for both adult and juvenile cohorts, respectively). These results can be explained by the presence in adult cohorts of trees that appeared after forest disturbance. However, other explanations can be suggested. This result may reflect selection for local adaptation which has been disrupted in juvenile trees. It is also possible that juveniles were the product of more distant mate pairs because fragmentation opened the forest and pollinators could carry pollen from more distant trees.
Although this change in mating system can explain the higher FIS in disturbed forests, other causes were examined. First, the effect of the sampling variance could be high because some samples were small. Secondly, although three suspected loci were removed from the analyses, the presence of null alleles at very low frequencies cannot be discounted and may have biased FIS upwards. The third explanation is the Wahlund effect, due to the presence of spatial or temporal structure in the sampled population (Henry and Gouyon, 2003). The Bayesian clustering method (Pritchard et al., 2000) was used to detect sub-structure in the six populations. The results (the details of cluster definitions, posterior probabilities and figures are not shown) showed that only Sandrajahana showed a substructure with two sub-populations presenting a lower fixation index (FIS = 0·156 *** FIS = 0·087 n.s.). These analyses suggest that a positive FIS in fragmented populations could be due to selfing resulting from reduction of adult tree density.
Genetic structure across the natural range
The FST,nuc among the populations representing the major part of the natural range exhibited a moderate value for nuclear microsatellites (FST,nuc = 0·08), showing that differentiation is weak within this species. The FST,nuc is close to that of species with a continental distribution such as Swietenia macrophylla (Lemes et al., 2003; Novick et al., 2003; FST,nuc = 0·07) or Caryocar brasiliense (Collevatti et al., 2003; FST,nuc = 0·10), and lower than for species distributed in a fragmented area such as Grevillea macleayana (England et al., 2002; FST,nuc = 0·22) or distributed in isolated islands such as Santalum austrocaledonicum (Bottin et al., 2005) or S. insulare (Lhuillier et al., 2006; FST,nuc = 0·23 and FST,nuc = 0·33, respectively). In addition, the Bayesian approach and the neighbor-joining tree (Fig. 2A) presented a separation between southern provenances and the rest of populations, and confirmed a weak differentiation among populations within these two groups. For nuclear microsatellites, the genetic structure analysis did not show marked isolation of fragmented populations.
As expected (Petit et al., 2005), chloroplast microsatellites showed a higher differentiation among populations (FST,cp = 0·49). This could be explained by the very limited gene flow by seeds. Field observation confirmed that barochory is the main mode of dispersion and that most of the seeds fall within a circle of 20–30 m radius around the fruiting tree. Although the overall FST,cp estimated for D. monticola is high, it remains smaller than FST,cp assessed in island species such as Santalum austrocaledonicum (Bottin et al., 2005), Santalum insulare (Butaud et al., 2005) and Pterocarpus officinalis (Müller et al., 2008; FST,cp = 0·60 FST,cp = 0·67 and FST,cp = 0·58, respectively) where important genetic drift occurs. The neighbor-joining and Bayesian analyses showed that populations were grouped according to provenances except for the southern ones, but did not display a specific isolation of fragmented populations.
Even if nuclear and chloroplast microsatellites displayed a different level of genetic structure, both exhibited a significant relationship between geographic and genetic distances (Fig. 3) demonstrating a pattern of isolation by distance. With marked and old fragmentation we would have expected a noticeable effect of drift (Willi et al., 2007) and a pattern different from the isolation by distance model, especially with chloroplast markers very sensitive to drift (Petit et al., 2005). The present results demonstrate that fragmentation has not yet intensified the genetic drift of populations. As for the diversity parameter, the simulations run by Lowe et al. (2005) have shown that FST increased by 0·09 with isolated (no gene flow) and small populations (50 individuals) and after ten generations. The present structure analyses confirmed that the effect of fragmentation is not yet perceptible for genetic structure parameters.
Conclusions
This study of D. monticola is the first analysis of the effect of fragmentation on genetic diversity and structure in a Madagascar forest tree species. Based on populations experiencing various degrees of fragmentation, the present results show that its effect is weak. It is not known precisely when fragmentation began in the eastern forest of Madagascar, but the major impact took place not much >100 years ago, i.e. corresponding to two generations for D. monticola. As stressed by Kramer et al. (2008) and Aguilar et al. (2008), with long-lived tree species, this weak genetic signal of fragmentation could be due to the small number of generations since its establishment (at most two). These first results should help in designing operational measures to manage the eastern forest ecosystem and to reduce the risk of disappearance of numerous threatened tree species having similar ecological attributes.
ACKNOWLEDGEMENTS
These results are part of O. Andrianoelina's PhD thesis on the impact of human activity on the genetic structure and adaptation of Dalbergia monticola in Madagascar. This study was one of the tasks of INCO's project FOREAIM funded by the European Union for ‘Bridging restoration and multi-functionality in degraded forest landscape of Eastern Africa and Indian Ocean Islands’. Lolona Ranaivoson is responsible for the National Forest Seed Centre in Madagascar where the study was undertaken. It was one of the sub-contractors of the INCO project along with CIRAD, through the research unit on forest tree genetics headed by J.-M. Bouvet. The laboratory work was partly done by B. Favreau. We thank Stephen Cavers for his valuable comments on the first version of this article and the suggestions of two anonymous reviewers.
LITERATURE CITED
- Addinsoft. XLSTAT software version 7.5.2. 2008. http://www.xlstat.com. (accessed 13 February 2009)
- Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J. Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Molecular Ecology. 2008;17:5177–5188. doi: 10.1111/j.1365-294X.2008.03971.x. [DOI] [PubMed] [Google Scholar]
- Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G. Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Molecular Ecology. 1998;7:933–944. doi: 10.1046/j.1365-294x.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- Andre T, Lemes MR, Grogan J, Gribel R. Post-logging loss of genetic diversity in a mahogany (Swietenia macrophylla King, Meliaceae) population in Brazilian Amazonia. Forest Ecology and Management. 2008;255:340–345. [Google Scholar]
- Andrianoelina O, Rakotondraoelina H, Ramamonjisoa L, Maley J, Danthu P, Bouvet J-M. Genetic diversity of Dalbergia monticola (Fabaceae) an endangered species in the fragmented oriental forest of Madagascar. Biodiversity Conservation. 2006;15:1109–1128. [Google Scholar]
- Bosser J, Rabevohitra R. Espèces nouvelles dans le genre Dalbergia (Fabaceae, Papilionoideae) à Madagascar. Adansonia. 2005;27:209–216. [Google Scholar]
- Bottin L, Verhaegen D, Tassin J, Vaillant A, Bouvet J-M. Genetic diversity and population structure of an insular tree, Santalum austrocaledonicum in New Caledonian archipelago. Molecular Ecology. 2005;14:1979–1989. doi: 10.1111/j.1365-294X.2005.02576.x. [DOI] [PubMed] [Google Scholar]
- Bryan GJ, McNicoll J, Ramsay G, Meyer RC, De Jong WS. Polymorphic simple sequence repeat markers in chloroplast genomes of solanaceous plants. Theoretical and Applied Genetics. 1999;99:859–867. [Google Scholar]
- Butaud JF, Rives F, Verhaegen D, Bouvet J-M. Phylogeography of eastern Polynesian sandalwood (Santalum insulare), an endangered tree species from the Pacific: a study based on chloroplast microsatellites. Journal of Biogeography. 2005;32:1763–1774. [Google Scholar]
- Bousquet J, Simon L, Lalonde M. DNA amplification from vegetative and sexual tissue of trees using polymerase chain reaction. Canadian Journal of Forestry Research. 1990;20:254–257. [Google Scholar]
- Collevatti RG, Grattapaglia D, Hay JD. Evidence for multiple lineages of Caryocar brasiliense populations in the Brazilian Cerrado based on the analysis of chloroplast DNA sequence and microsatellite haplotype variation. Molecular Ecology. 2003;12:105–115. doi: 10.1046/j.1365-294x.2003.01701.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Corander J, Sirén J, Arjas E. Bayesian spatial modelling of genetic population structure. Computational Statistics. 2008;23:111–129. [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Dick CW, Etchelecu G, Austerlitz F. Pollen dispersal of tropical trees (Dinizia excelsa, Fabaceae) by native insects and African honeybees in pristine and fragmented Amazonian rainforest. Molecular Ecology. 2003;12:753–764. doi: 10.1046/j.1365-294x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Doligez A, Joly HI. Mating system of Carapa procera (Meliaceae) in the French Guiana tropical forest. American Journal of Botany. 1997;84:461–470. [PubMed] [Google Scholar]
- Du Puy D. Dalbergia monticola. In: The 2006 IUCN Red List of Threatened Species. Geneva, Switzerland: The World Conservation Union; 1998. www.iucnredlist.org. (accessed 13 February 2009) [Google Scholar]
- Du Puy DJ, Moat J. Ecology of the Leguminosae in Madagascar. In: Du Puy DJ, Labat JN, Rabevohitra R, Villiers JF, Bosser J, Moat J, editors. The Leguminosae of Madagascar. Kew: Royal Botanic Gardens, Kew; 2002. [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theoretical Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- England PR, Usher AV, Whelan RJ, Ayre DJ. Microsatellite diversity and genetic structure of fragmented populations of the rare, fire-dependent shrub Grevillea macleayana. Molecular Ecology. 2002;11:967–977. doi: 10.1046/j.1365-294x.2002.01500.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2007;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwig N, Braun C, Böhning-Gaese K. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae) Conservation Genetics. 2008;9:317–326. [Google Scholar]
- Favreau B, Andrianoelina O, Nunez P, et al. Characterization of microsatellite markers in the rosewood (Dalbergia monticola Bosser and R Rabev) Molecular Ecology Notes. 2007;7:774–776. [Google Scholar]
- Fuchs EJ, Lobo JA, Quesada M. Effects of forest fragmentation and flowering phenology on the reproductive success and mating patterns of the tropical dry forest tree Pachira quinata. Conservation Biology. 2003;17:149–157. [Google Scholar]
- Gasse F, Van Campo E. Late quaternary environmental changes from a pollen and diatom record in the southern tropics (lake Tritivakely, Madagascar) Paleogeography, Paleoclimatology, Paleoecology. 2001;167:287–308. [Google Scholar]
- Goudet J. FSTAT: a program to estimate and test gene diversity and fixation indices (version 2.9.3.2) 2001 [updated from Goudet J. 1995. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity86: 485–486] [Google Scholar]
- Hamrick JL. Response of forest trees to global environmental changes. Forest Ecology and Management. 2004;197:323–335. [Google Scholar]
- Hansen OK, Kjær ED, Vendramin GG. Chloroplast microsatellite variation in Abies nordmanniana and simulation of causes for low differentiation among populations. Tree Genetics & Genomes. 2005;1:116–123. [Google Scholar]
- Henry JP, Gouyon . Paris: Dunod; 2003. Précis de génétique des populations: cours, exercices et problèmes résolus (Sciences Sup) [Google Scholar]
- Honnay O, Jacquemyn H. Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conservation Biology. 2007;21:823–831. doi: 10.1111/j.1523-1739.2006.00646.x. [DOI] [PubMed] [Google Scholar]
- Hurlbert SH. The non-concept of species diversity, a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Kramer AT, Ison JL, Ashley MV, Howe HF. The paradox of forest fragmentation genetics. Conservation Biology. 2008;22:878–885. doi: 10.1111/j.1523-1739.2008.00944.x. [DOI] [PubMed] [Google Scholar]
- Lemes MR, Gribel R, Proctor J, Grattapaglia D. Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon based on variation at microsatellite loci, implications for conservation. Molecular Ecology. 2003;12:2845–2883. doi: 10.1046/j.1365-294x.2003.01950.x. [DOI] [PubMed] [Google Scholar]
- Lhuiller E, Butaud JF, Bouvet J-M. Extensive clonality and strong differentiation in the insular Pacific tree Santalum insulare: implications for its conservation. Annals of Botany. 2006;98:1061–1072. doi: 10.1093/aob/mcl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe AJ, Bosier D, Ward M, Bacles CFE, Navarro C. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity. 2005;95:255–273. doi: 10.1038/sj.hdy.6800725. [DOI] [PubMed] [Google Scholar]
- Luikart G, Cornuet JM. Empirical evaluation of a test for identifying recent bottleneck populations from allele frequency data. Conservation Biology. 1998;12:228–237. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Muller F, Voccia M, Ba A, Bouvet J-M. Genetic diversity and gene flow in a Caribbean tree Pterocarpus officinalis Jacq, a study based on chloroplast and nuclear microsatellites. Genetica. 2009;135:185–198. doi: 10.1007/s10709-008-9268-4. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Navascués M, Emerson BC. Chloroplast microsatellites: measures of genetic diversity and the effect of homoplasy. Molecular Ecology. 2005;14:1333–1341. doi: 10.1111/j.1365-294X.2005.02504.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Novick RR, Dick CD, Lemes MR, Navarro C, Caccon A, Bermingham E. Genetic structure of Mesoamerican population of big-leaf mahogany (Swietenia macrophylla) inferred from microsatellite analysis. Molecular Ecology. 2003;12:2885–2893. doi: 10.1046/j.1365-294x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- Perrier X, Flori A, Bonnot F. Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC, editors. Genetic diversity of cultivated tropical plants. Montpellier: Enfield Science Publishers; 2003. pp. 43–76. [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet J-M. BOTTLENECK, a computer program for detecting recent reductions in effective population size from allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Falush D, Wen X. Documentation for structure software, Version 22. 2007 http://pritch.bsdu.chicago.edu. (accessed 13 February 2009) [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Kuittinen H. Small population processes. In: Young A, Boshier D, Boyle T, editors. Forest conservation genetics, principles and practice. Wallingford: CSIRO Publishing; 2000. pp. 91–100. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin version 2000: a software for population genetics data analysis. Geneva, Switzerland: Genetics and Biometrics Laboratory, Department of Anthropology, University of Geneva; 2000. [Google Scholar]
- Straka H. Histoire de la végétation de Madagascar oriental dans les derniers 100 millénaires In. In: Lourenço WR, editor. Biogéographie de Madagascar. Paris: Editions de l'ORSTOM; 1996. pp. 37–47. [Google Scholar]
- Tapio M, Miceikiene I, Vilkki J, Kantanen J. Comparison of microsatellite and blood protein diversity in sheep, inconsistencies in fragmented breeds. Molecular Ecology. 2003;12:2045–2056. doi: 10.1046/j.1365-294x.2003.01893.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Weising K, Gardner RC. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome. 1999;42:9–19. [PubMed] [Google Scholar]
- Willi Y, Van Buskirk J, Schmid B, Fischer M. Genetic isolation of fragmented populations is exacerbated by drift and selection. Journal of Evolutionary Biology. 2007;20:534–542. doi: 10.1111/j.1420-9101.2006.01263.x. [DOI] [PubMed] [Google Scholar]