Abstract
The mammalian bombesin (Bn) peptides, neuromedin B (NMB) and gastrin-releasing peptide (GRP), have widespread actions in many tissues, and their effects are mediated by two closely related G-protein-coupled receptors, the NMBR and GRPR. Little is known about the structural determinants of NMBR selectivity for NMB, in contrast to GRP selectivity for the GRPR, which has been extensively studied. To provide insight, chimeric NMBR-GRPR loss-of-affinity and gain-of-affinity mutants were made, as well as NH2-terminally truncated NMBR and point mutants using site-directed mutagenesis. Receptors were expressed in Balb-3T3-cells or CHOP cells, and affinities were determined. NMB had 115-fold greater affinity for NMBR than GRPR. Receptor-chimeric studies showed that NMBR selectivity for NMB was primarily determined by differences in the third extracellular (EC3) regions of GRPR-NMBR and adjacent upper-transmembrane-5 (TM5) region. In this region, 24 NMB gain-of-affinity GRPR mutants or NMBR loss-of-affinity point/combination mutants were made. Three gain-of-affinity mutant GRPRs [[A198I] (EC3), [H202Q] (EC3), [S215I] (upper TM5)] had increased NMB affinity (2.4–21-fold), and these results were confirmed with NMBR loss-of-affinity mutants [I199A,Q203H,I215S-NMBR]. The combination mutant [A198I,S215]GRPR had the greatest effect causing a complete NMB gain-of-affinity. The importance of differences at position 199NMBR or 203NMBR was studied by substituting amino acids with various properties. Our results show that NMBR selectivity for NMB is due to differences in the EC3 of NMBR-GRPR and the adjacent upper-TM5 region. Within these regions, isoleucines in NMBR [position 199 (EC3)] (instead of A198GRPR) and in 215NMBR (TM5) (instead of S214GRPR), as well as Q203NMBR (instead of H202GRPR) are responsible for high NMB-affinity/selectivity of NMBR. The effect at position 199 is primarily due to differences in hydrophobicity of the substitution, whereas steric factors and charge of the substitution at position 203 were important determinants of NMB selectivity.
The mammalian bombesin (Bn)-related peptides [neuromedin B (NMB) and gastrin-releasing peptide (GRP)] have high structural homology with 7 of 10 amino acid identities in their biologically active COOH terminus (Jensen et al., 2008). However, their different biological activities are mediated by two closely related receptors, the NMBR and GRPR, which share 55% amino acid identities (Jensen et al., 2008). GRP and NMB, as well as their receptor, GRPR/NMBR, are widely distributed in mammals. They occur in both the central nervous system (CNS) and peripheral tissues, including the gastrointestinal tract (Ladenheim et al., 1992; Moody and Merali, 2004; Jensen et al., 2008). Animal studies suggest that GRPR/NMBR may be involved in a broad spectrum of biological responses. These include actions in the CNS (circadian rhythm, thyrotropin secretion, behavior control, thermoregulation, and satiety) and in the immune system (effects on macrophages, lymphocytes, leukocytes, and dendritic cells) and endocrine effects (release of numerous hormones/neurotransmitters), effects in the gastrointestinal tract (motility, secretion, and growth), and effects in the urogenital tract and respiratory system (Moody and Merali, 2004; McCoy and Avery, 1990; Jensen et al., 2008). GRP and, to a lesser extent, NMB have important pathophysiological effects. These include having a prominent effect on the growth and/or differentiation of a number of important human tumors (colon, prostrate, lung, head/neck squamous cell, CNS, pancreatic, and some gynecologic cancers) and, in some cases, function as autocrine growth factors (Cuttitta et al., 1985; Jensen and Moody, 2006; Jensen et al., 2008). In addition, GRPR/NMBR is one of the G-protein-coupled receptor family most frequently expressed ectopically or overexpressed by a different tumors, including prostate cancer, small cell lung cancer, breast cancer, CNS tumors (glioblastomas), and carcinoids (intestinal, thymic, and bronchial) (Jensen et al., 2001, 2008; Reubi et al., 2002; Jensen and Moody, 2006).
The structural basis for the selectivity of GRPR agonists/antagonists and high affinity for the GRPR has been extensively studied (Akeson et al., 1997; Tokita et al., 2002; Nakagawa et al., 2005; Gonzalez et al., 2008). However, little is known about the molecular basis of selectivity and high affinity of NMB for the NMBR. One previous study (Fathi et al., 1993) provides evidence that differences between GRPR and NMBR in the upper fifth transmembrane region might be particularly important. However, in this study (Fathi et al., 1993), other NMBR/GRPR regions were not systematically examined. In this present study, we attempted to identify the amino acids responsible for the high affinity/selectivity of NMBR for NMB. We made an NH2-terminal truncated NMBR mutant and used a chimeric NMBR-GRPR receptor approach. The latter approach was used because it has been proven to be useful in elucidating the structural basis of other G-protein-coupled receptor interaction with their ligands (Fathi et al., 1993; Tokita et al., 2002; Gonzalez et al., 2008). The receptor chimeric approach was accomplished by exchanging extracellular NMBR and GRPR domains (NMB loss- and gain-of-affinity chimeras). This was combined with a site-mutagenesis approach to identify critical amino acid(s) in the important regions localized by the receptor-chimera studies. Our results show that the selectivity of NMBR for the NMB over the GRPR depends primarily on differences in the amino acids in the third extracellular (EC) domains of the receptors and in the adjacent upper fifth transmembrane (TM) region of these two receptors. Site-directed mutagenesis demonstrated that the presence of isoleucines 199 and 215 in NMBR and that of glutamine located in position 203 in NMBR, instead of an alanine 198, serine 216, and histidine 202, in the comparable positions of GRPR, respectively, are the crucial differences responsible for high affinity/ selectivity for NMB. Additional site-directed mutagenesis at these sites suggested an important role of hydrophobicity as well as steric factors and charge differences between the different substitutions in these two receptors at these positions in determining high affinity for NMB.
Materials and Methods
Materials
The mammalian expression vector, pcDNA3, custom primers, restriction endonucleases (BamHI, HindIII, XbaI, and EcoRI), penicillin-streptomycin, Lipofectamine Plus reagent, Geneticin-selective antibiotic (G418 sulfate), and essential amino acid solution were from Invitrogen (Carlsbad, CA). QuikChange site-directed mutagenesis kit, Seamless cloning kit, and ExSite polymerase chain reaction-based site-directed mutagenesis kit were from Stratagene (La Jolla, CA). Dulbecco's minimum essential medium (DMEM), phosphate-buffered saline, fetal bovine serum (FBS), and trypsin/EDTA (Versene) solution were from BioSource International (Camarillo, CA). Balb-3T3 cells (mouse embryo) were from American Type Culture Collection (Manassas, VA), and CHOP cells (Polyoma large T antigen-expressing Chinese hamster ovary cells) were a gift from James W. Dennis (Samuel Lunenfeld Research Institute, Toronto, ON, Canada). Bn and NMB were from Bachem California (Torrance, CA). PD168368 was from Tocris Bioscience (Ellisville, MO). [d-Phe6, β-Ala11,Phe13,Nle14]Bn-(6–14) and [d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) were a gift from David H. Coy (Peptide Research Laboratories, Tulane University Health Sciences Center, New Orleans, LA). Na125I (2200 Ci/mmol) was from GE Healthcare (Little Chalfont, Buckinghamshire, UK). 1,3,4,6-Tetrachloro-3α,6α-diphenylglycoluril(IODO-GEN) and dithiothreitol were from Pierce Biotechnology Inc. (Rockford, IL). Bovine serum albumin fraction V (BSA) and HEPES were from MP Biomedicals (Solon, OH). Soybean trypsin inhibitor type I-S and bacitracin were from Sigma-Aldrich (St. Louis, MO). All other chemicals were of the highest purity commercially available.
Methods
Construction of NH2-Terminal Truncated NMBR and Isolation of NMBR-Truncated Stably Transfected Cell Lines.
The cDNA of the rat NMBR was identical to that described previously (Wada et al., 1991; Benya et al., 1992). The cDNA of the wild-type NMBR was cloned into the EcoRI restriction enzyme site of pcDNA3. An NMB receptor, in which the NH2 terminus was completely truncated, was constructed by using the ExSite polymerase chain reaction-based site-directed mutagenesis kit following the manufacturer's instruction with minor modification. Thirty-eight amino acids in the NH2 terminus of NMBR (from proline at the position 2 of the amino acid sequence in NMBR through glutamic acid at the position 39 of amino acid sequence) were deleted. This truncated DNA plasmid was transfected into Balb-3T3 cells with Lipofectamine (“Cell Transfection”), and the transfected cells were selected with the selecting medium, which contains the Balb-3T3 growth medium with 800 mg/l G418. Selected colonies were maintained with medium containing 300 mg/l of G418. Nucleotide sequence analysis of the entire coding region was performed using an automated DNA sequencer (ABI Prism 377 DNA sequencer; Applied Biosystems Inc., Foster City, CA).
Construction of the GRPR/NMBR Chimeras.
The cDNA of the mouse GRPR was identical to that described previously (Battey et al., 1991; Benya et al., 1993, 1994). The cDNA of the wild-type GRPR was cloned between the HindIII and XbaI restriction enzyme sites of pcDNA3. Chimeric GRP and NMB receptors were made by exchanging the extracellular portions of each receptor as described previously (Tokita et al., 2001a, 2002). In brief, each chimera was constructed using the Seamless cloning kit as described previously (Tokita et al., 2002). Nucleotide sequence analysis of the entire coding region was performed using an automated DNA sequencer (ABI Prism 377 DNA sequencer; Applied Biosystems Inc.). The [e1NMBR]GRPR chimera was constructed by replacing residues Met1-His37 of GRPR with Met1-Ala38 of NMBR and the [e1GRPR]NMBR chimera by performing the opposite replacement. The [e2NMBR]GRPR chimera was constructed by replacing residues Asp98-Lys115 of GRPR with Asp100-Lys117 of NMBR and the [e2GRPR]NMBR chimera by performing the opposite replacement. The [e3NMBR]GRPR chimera was constructed by replacing residues Phe179-Ala214 of GRPR with Phe181-Leu215 of NMBR and the [e3GRPR]NMBR chimera by performing the opposite replacement. The [e4NMBR]GRPR chimera was constructed by replacing residues Arg288-Thr305 of GRPR with Arg289-Val306 of NMBR and the [e4GRPR]NMBR chimera by performing the opposite replacement.
Construction of Mutant GRP and NMB Receptors.
The GRPR and NMBR mutant receptors were constructed using the QuikChange site-directed mutagenesis kit following the manufacturer's instructions, with the exception that the DpnI treatment was 2 h instead of the 1-h time period recommended by the manufacturer. Nucleotide sequence analysis of the entire coding region of each mutant receptor was performed using an automated DNA sequencer (ABI Prism 377 DNA sequencer; Applied Biosystems Inc.).
Growth and Maintenance of Cells.
Balb-3T3 cells were grown in DMEM containing 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. CHOP cells were grown in DMEM containing 10% (v/v) FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 200 μg/ml G418. All cells were maintained at 37°C in a 5% CO2 atmosphere. Cells were split every 3 to 4 days at confluence after detaching the cells with trypsin/EDTA solution.
Cell Transfection.
Balb-3T3 or CHOP cells were seeded in a 10-cm tissue culture dish at a density of 106 cells/dish and grown overnight at 37°C in growth medium. The next morning, 5 μg of plasmid DNA was transfected by cationic lipid-mediated method by using 30 μl of Lipofectamine reagent and 20 μl of Plus reagent in Opti-MEM I reduced-serum medium for 3 h at 37°C. The transfection medium was then replaced with fresh growth medium as outlined above. Cells were maintained at 37°C in a 5% CO2 atmosphere and were used 48 h later for binding assays. Balb-3T3 cells were used in all transient transfection studies, with the exception of studies examining the effect of different substitutions for Ile199 in NMBR, because of low expression and binding when these mutant receptors were expressed in Balb-3T3 cells.
Preparation of 125I-[Tyr4]Bn and 125I-[d-Tyr6, β-Ala11,Phe13,Nle14]Bn-(6–14).
125I-[Tyr4]Bn and 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) at a specific activity of 2200 Ci/mmol were prepared by a modification of methods described previously (Mantey et al., 1997). In brief, 0.8 μg of IODO-GEN (in 0.01 μg/ml chloroform) was added to a vial, dried under a stream of nitrogen, and washed with 100 μl of KH2PO4 (pH 7.4). To the reaction vial, 20 μl of 0.5 M KH2PO4 (pH 7.4), 8 μg of peptide in 4 μl of water, and 2 mCi (20 μl) of Na125I were added, mixed gently, and incubated at room temperature for 6 min. The incubation was stopped by the addition of 100 μl of distilled water. In the case of 125I-[Tyr4]Bn, 300 μl of 1.5 M dithiothreitol was also added, and the iodination mixture was reincubated at 80°C for 60 min to reduce the oxidized methionines. Radiolabeled peptides were separated using a Sep-Pak (Waters, Milford, MA) and high-pressure liquid chromatography as described previously (Mantey et al., 1997; Tokita et al., 2001a). Radioligands were stored with 0.5% BSA at −20°C.
Whole-Cell Radioligand Binding Assays.
Binding studies were performed as described previously (Benya et al., 1994; Tokita et al., 2001a,b). In brief, disaggregated Balb-3T3 cells, which had been stably transfected with the NH2-terminal truncated NMBR, or Balb-3T3/CHOP cells were used in the binding assay 48 h after transient transfection with GRPR/NMBR mutants with Lipofectamine reagent. These cells were incubated for 1 h at room temperature in 250 μl of binding buffer [24.5 mM HEPES, pH 7.4, 98 mM NaCl, 6 mM KCl, 2.5 mM KH2PO4, 5 mM sodium pyruvate, 5 mM sodium fumarate, 5 mM sodium glutamate, 2 mM glutamine, 11.5 mM glucose, 0.5 mM CaCl2, 1.0 mM MgCl2, 0.01% (w/v) soybean trypsin inhibitor, 0.2% (v/v) essential amino acid mixture (Invitrogen), 0.2% (w/v) BSA, and 0.05% (w/v) bacitracin] with 50 pM 125I-[Tyr4]Bn (2200 Ci/mmol) or 50 pM 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) (2200 Ci/mmol) in the presence of the indicated concentration of unlabeled peptides. 125I-[Tyr4]Bn was used for all binding studies, with the exception of a few mutants with low binding in Balb-3T3 cells as indicated in the figure and table legends. 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) was used for binding studies with mutants with low 125I-[ Tyr4]Bn binding because this peptide is a universal Bn receptor ligand with high affinities for all NMBRs and GRPRs (Mantey et al., 1997; Pradhan et al., 1998; Reubi et al., 2002). Even though a previous study has shown that GRPR receptor expression levels over a >350-fold range do not alter agonist affinity (Tsuda et al., 1997), to correct for any possible degradation, the cell concentration was adjusted to be between 0.01 and 5 × 106 cells/ml so that <20% of the total added radioactive ligand was bound during incubation. Furthermore, the difference of the total added radioactive ligand percentage between the wild-type and mutant receptor was adjusted to be within 5%. After the incubation, 100-μl aliquots were added to 400-μl microcentrifuge tubes (PGC Scientific, Frederick, MD), which contained 100 μl of binding buffer to determine the total radioactivity. The bound tracer was separated from unbound tracer by pelleting the cells through the binding buffer by centrifugation at 10,000g in a microcentrifuge system (Microfuge E; Beckman Instruments, Palo Alto, CA) for 3 min. The supernatant was aspirated, and the pelleted cells were rinsed twice with a washing buffer that contained 1% (w/v) BSA in phosphate-buffered saline. The amount of radioactivity bound to the cells was measured in a Cobra II Gamma counter (PerkinElmer Life and Analytical Sciences, Boston, MA). Binding was expressed as the percentage of total radioactivity that was associated with the cell pellet. All binding values represented saturable binding (i.e., total binding minus nonsaturable binding). Nonsaturable binding was defined as the amount of binding that occurred with 1 μM NMB in the incubation solution. Nonsaturable binding was <15% of the total binding in all experiments. Each point was measured in duplicate, and each experiment was replicated at least three times. Calculation of affinity was performed by determining the IC50 [NMB concentration causing half-maximal inhibition of binding] using the curve-fitting program KaleidaGraph (Synergy Software, Reading, PA). Statistical analysis was performed with Statview version 4.02 (BrainPower, Inc., Calabasas, CA). The paired t test was used to determine the statistical significance.
Results
Comparison of the Affinity NMB and Bn for the NH2-Terminal Truncated NMBR [NMBR(1,40)-390] and the Wild-Type NMB Receptor.
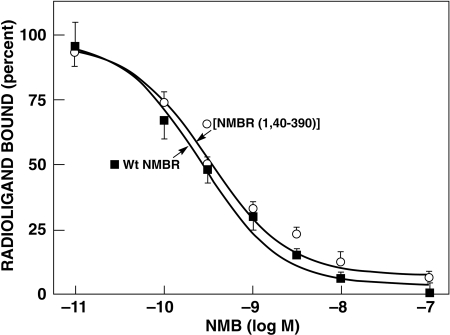
Previous studies with the GRPR show that its NH2 terminus is not essential for high-affinity agonist binding and that the selectivity of Bn peptide receptor agonists for GRPR was primarily determined by differences in extracellular receptor domains and, to a lesser extent, by differences in the adjacent upper TM regions (Akeson et al., 1997; Tokita et al., 2002; Nakagawa et al., 2005; Jensen et al., 2008). To investigate the importance of the NH2 terminus of NMBR for determining the affinity of NMB, we made an NMBR mutant in which the NH2 terminus was completely truncated. The N-terminal truncated NMBR showed the same affinity (IC50 = 0.3 nM) for NMB as the wild-type NMBR (IC50 = 0.3 nM) (Fig. 1; Table 1). Furthermore, no significant differences were found between the affinity of Bn for [NMBR(1-40)-390] and the wild-type NMBR (IC50 = 2.8 and 1.5 nM, respectively). NMB demonstrated a 115-fold higher affinity for NMBR (IC50 = 1 nM) compared with GRPR (115 nM) (Table 1; Figs. 2 and 3), whereas Bn demonstrated a 6-fold higher affinity for GRPR than NMBR (Table 1).
Fig. 1.
Affinities of NMB for wild-type and an N-terminal truncated NMBR mutant [NMBR(1-40)-390]. The dose-inhibition curves for NMB of the wild-type NMBR and the N-terminal truncated NMBR, [NMBR(1,40–390)], were determined on stably transfected Balb cells using 125I-[Tyr4]Bn as the ligand. Each point on the dose-inhibition curves is the mean ± S.E.M. from at least three separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present.
TABLE 1.
Affinities of NMB and Bn for wild-type GRP and NMB receptors on N-terminal truncated NMBR [NMBR(1,40–390)], chimeric GRP, and NMB receptors
The affinities (IC50) were measured by competitive displacement of 50 pM 125I-[Tyr4]Bn by NMB or Bn in whole-cell radioligand binding assays as described under Materials and Methods. The top part of this table shows the comparison between WT NMBR, N-terminal truncated NMBR, [NMBR(1,40–390)], and stably transfected Balb-3T3 cells and are determined from the dose-inhibition curves shown in Fig. 1. The bottom part of this table shows the IC50 of NMB and Bn for wild-type GRPR, NMBR, four chimeric GRP receptors (gain-of-affinity for NMB), and four chimeric NMB receptors (loss-of-affinity for NMB) transiently expressed in Balb-3T3 cells. The chimeric NMBR and GRPR receptor were made by exchanging the extracellular domain of each receptor as shown in Figs. 2 and 3 and described in their legends. The significant decrease or increase in affinity from the wild type was calculated using the paired t test. Values are mean ± S.E.M. from at least four experiments, and in each experiment, each point was measured in duplicate.
| IC50 |
||
|---|---|---|
| NMB | Bn | |
| nM | ||
| Stably transfected | ||
| WT NMBR | 0.3 ± 0.1 | 1.5 ± 0.2 |
| [NMBR(1,40–390)] | 0.3 ± 0.1 | 2.8 ± 0.1 |
| Transiently transfected | ||
| WT GRPR | 115.1 ± 7.3 | 0.8 ± 0.1 |
| WT NMBR | 1.0 ± 0.2 | 4.5 ± 0.5 |
| Chimeric GRPRs (gain-of-affinity for NMB) | ||
| [e1NMBR]GRPR | 61.1 ± 16.3a | 0.9 ± 0.3 |
| [e2NMBR]GRPR | 128.2 ± 39.5 | 0.7 ± 0.1 |
| [e3NMBR]GRPR | 24.9 ± 4.9b | 0.6 ± 0.1 |
| [e4NMBR]GRPR | 62.8 ± 15.8a | 1.4 ± 0.2 |
| Chimeric NMBRs (loss-of-affinity for NMB | ||
| [e1GRPR]NMBR | 1.5 ± 0.2 | 4.0 ± 0.8 |
| [e2GRPR]NMBR | 4.0 ± 1.1 | 4.0 ± 0.7 |
| [e3GRPR]NMBR | 80.4 ± 14.5c | 5.2 ± 0.7 |
| [e4GRPR]NMBR | 2.8 ± 0.5 | 1.2 ± 0.2 |
a P < 0.03 vs WT GRPR for NMB.
b P < 0.004 vs WT GRPR for NMB.
c P < 0.0001 vs WT NMBR for NMB.
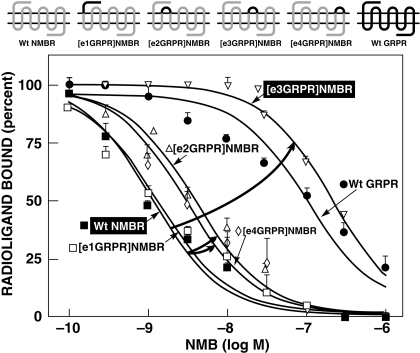
Fig. 2.
Affinities of NMB for wild-type NMBR, chimeric extracellular GRPRs (loss-of-affinity), and wild-type GRPR. Top, the diagrams of the chimeric receptors formed by replacing each of the extracellular domains of NMBR by the comparable domain of GRPR one at a time. Bottom, the dose-inhibition curves for NMB of each receptor. The curved arrows indicate large changes in affinity from the wild type. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand. Each point on the dose-inhibition curves is the mean ± S.E.M. from at least three separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present. [e1NMBR]GRPR refers to the GRPR chimera in which the first extracellular loop (NH2 terminus) of GRPR is replaced by the comparable domain from NMBR.
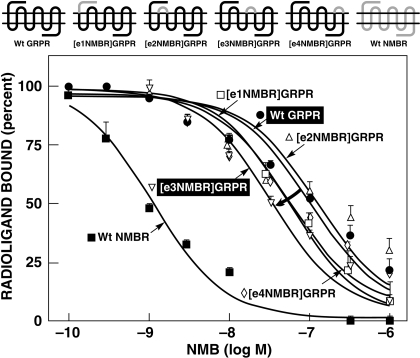
Fig. 3.
Affinities of NMB for wild-type NMBR, chimeric extracellular NMBRs (gain-of-affinity), and wild-type GRPR. Top, the diagrams of the NMB gain-of-affinity chimeric receptors formed by replacing each of the extracellular domains of GRPR by the comparable domain of NMBR one at a time. Bottom, the dose-inhibition curves for NMB for each receptor. The curved arrows indicate large changes in affinity from the wild type. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand. Each point on the dose-inhibition curves is the mean ± S.E.M. from at least three separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present. [e1GRPR]NMBR refers to the NMBR chimera in which the first extracellular loop (NH2 terminus) of NMBR is replaced by the comparable domain from GRPR.
Extracellular Chimeric Receptors.
To investigate the importance of differences in the NMBR and GRPR extracellular domains for determining NMB selectivity (Figs. 2 and 3), chimeric NMBR and GRPR were made by exchanging their extracellular domains (Figs. 2 and 3). Four potential NMB gain-of-affinity GRPR chimeras were made by substituting the extracellular domains of GRPR with the comparable domain of NMBR (Fig. 3). Furthermore, the reverse study was performed by making four NMB loss-of-affinity NMB chimeric receptors by substituting the extracellular domains of NMBR with the comparable domains of GRPR (Fig. 2).
With the NMB gain-of-affinity GRPR chimeras, the substitution of EC3 in GRPR by the comparable domain of NMBR showed the greatest effect by increasing NMB affinity 4.6-fold (from 115.1 ± 7.3 to 24.9 ± 4.9 nM) (Fig. 3; Table 1). In contrast, the substitution of their EC1 or EC4 extracellular domain of GRPR, with those from NMBR, had only a minimal effect on NMB affinity, increasing it by 1.8-fold (from 115.1 ± 7.3 to 61.1 ± 16.3 and 62.8 ± 15.8 nM, respectively) (Fig. 3; Table 1). When the EC2 domain from GRPR was substituted by the equivalent extracellular domains of NMBR, no difference in the affinity of that chimera for NMB was observed (128.2 ± 39.5 nM) (Fig. 3; Table 1).
The reverse study was performed by making NMB loss-of-affinity chimeras by substituting extracellular domains of GRPR into NMBR (Fig. 2). Substitution of the EC3 domain in NMBR by the comparable domain of GRPR decreased affinity for NMB by 80-fold (from 1.0 ± 0.2 to 80.4 ± 14.5 nM) (Fig. 2; Table 1). In contrast, the substitution of EC1, EC2, or EC4 in NMBR by the compare domains of GRPR only minimally altered the NMB affinity of these chimeras by causing a decrease of 0.5- to 4-fold (Fig. 2; Table 1). The differences seen with the loss-of-affinity and gain-of-affinity chimeric receptors were not due to a global alteration in receptor conformation, because each chimeric receptor retained high affinity for Bn (Table 1). These results show that differences in the third extracellular domain of the NMB and GRP receptors play an important role in determining the selectivity of the NMBR for NMB.
EC3 of GRPR and Adjacent TM Region Mutants (Gain-of-Affinity Point and Combination Mutants).
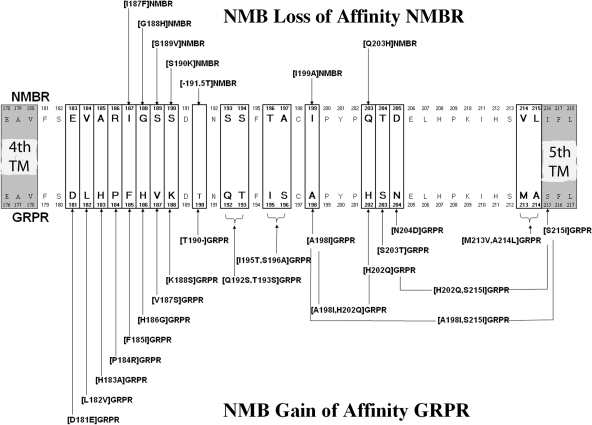
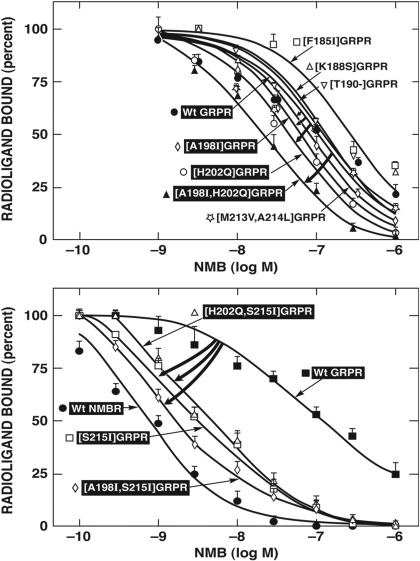
To investigate further the molecular basis for the selectivity of NMB, we attempted to determine which specific amino acid(s) differences in the EC3 region of GRPR/NMBR are responsible for the high affinity of NMB for NMBR. We did this by systematically studying the importance of individual amino acid(s) differences and similarities in the EC3 domain of these two receptors (Fig. 4). We also included in this analysis differences in the upper adjacent fifth TM region (Fig. 4), because a previous study (Fathi et al., 1993) demonstrated that differences between GRPR and NMBR in this region were important for NMB affinity. The NMBR and GRPR in this region differed in 20 amino acids (Fig. 4). In NMBR, of the 20 differences, 19 occurred in the EC3 region and one occurred in the upper adjacent fifth TM region. Specifically, the differences occurred at NMBR positions 183 to 190, 193 to 194, 196 to 197, 199, 203 to 205, and 214 to 216 (Fig. 4, top), which are comparable to GRPR positions 181 to 188, 192 to 193, 195 to 196, 198, 202 to 204, and 213 to 215 (Fig. 4, bottom). To study the 20 amino acid(s) differences, we first made 14 GRPR gain-of-affinity point mutants and three double GRPR mutants by substituting in GRPR the comparable different amino acids from NMBR (Fig. 4).
Fig. 4.
Alignment of amino acids sequences in the EC3 or adjacent TM regions of the NMB and GRP receptors. The boxes show divergent amino acids between these two receptors in the EC3 region. Shown above the diagrams are the seven-point and group NMBR mutants made by substituting, in the NMBR, corresponding different amino acid(s) in a similar position of the GRPR (loss-of-affinity mutants). Shown below the diagrams are the 20-point and six double GRPR mutants (gain-of-affinity point mutants) made by substituting, in the GRPR, the corresponding different amino acid(s) in a similar position in NMBR. The EC3 of GRPR was one amino acid longer (position 190.T of GRPR) than NMBR, and therefore, a GRPR mutant with 190T deleted was made ([T190-]GRPR) (bottom) and a NMBR with this amino acid (threonine) added in the comparable position to GRPR was made ([-191.5T]NMBR) (top). All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand.
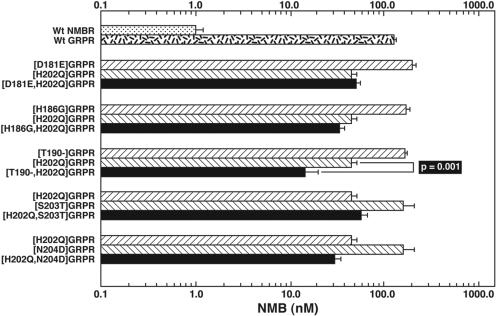
Three gain-of-affinity GRPR mutations had a prominent effect on NMB affinity (Fig. 5; Table 2). Specifically, the substitution of alanine in position 198 ([A198I]GRPR) in the EC3 of GRPR by isoleucine from the comparable position in NMBR, the substitution of histidine by glutamine in position 202 ([H202Q]GRPR), and the substitution of serine in position 215 in GRPR by isoleucine in a comparable position of NMBR ([S215I]GRPR) caused an increase in the affinity of 2.4-, 2.4-, and 21-fold for NMB (Fig. 5; Table 2). In contrast, in the other positions where there were amino acid differences in the EC3 region, the substitutions in the GRPR (i.e., [D181E], [L182V], [H183A], [P184R], [F185I], [H186G], [V187S], [K188S], [S203T], and [N204D]) by the comparable amino acid(s) of NMBR had no effect on NMB affinity (Fig. 5; Table 2). Because neither the substitution of [A198I] in GRPR nor [H202Q] in GRPR caused a gain-of-affinity equal to that seen with the substitution of the complete EC3 of NMBR in GRPR (Tables 1 and 2, 47.5 and 46.7 versus 24.9 nM), we explored the possible effect of multiple simultaneous amino acid substitutions in the EC3. We first made gain-of-affinity mutant GRPR with combinations of the point mutations that alone had an effect. The double mutant [A198I,H202Q] in the GRPR resulted in a 5-fold increase in the NMB affinity (23.4 ± 1.9 nM) compared with native GRPR (115.1 ± 7.3 nM). This result represented an additive effect because the increase in the NMB affinity in the double mutant is greater than the increase in NMB affinity seen with either [A198I] or [H202Q] alone. Furthermore, the increase in NMB affinity in the double mutant [A198I,H202Q] GRPR is equal to the increase in affinity caused by the substitution of the whole EC3 domain in GRPR by the comparable domain of NMBR (Fig. 5, top; Tables 1 and 2). A previous study (Fathi et al., 1993) reported that the presence at the EC3-fifth TM border of NMBR (position 216) of an isoleucine, instead of a serine at this position in GRPR (position 215), had an important effect on NMBR affinity. Therefore, we examined its effect alone or in combination with the important EC3 regions that we identified. The [S215I]GRPR mutant demonstrated a marked gain in affinity for NMB (from 115 to 5.4 nM) (Table 2). However, it did not completely restore NMB affinity to that seen with the wild-type NMBR (i.e., 1 nM) (Table 2). When this mutation was combined with the EC3 mutation, H202Q, in GRPR [H202Q,S215I]GRPR, no further increase in affinity was seen (Fig. 5, bottom; Table 2). However, when the EC3 mutation [A198I] was combined with the [S215I] mutation in GRPR, double mutant [A198I,S215I]GRPR had a marked effect on enhancing NMB affinity (increase 68-fold) (Fig. 5, bottom; Table 2). Furthermore, the double mutant [A198I,S215I]GRPR almost demonstrated a complete gain-of-affinity for NMB with an affinity (IC50 = 1.7 ± 0.2 nM) similar to NMB affinity for the native NMBR (IC50 = 1.0 ± 0.2 nM). None of the other double mutants in GRPR ([D181E,H202Q], [H186G,H202Q], [H202Q,S203T], or [H202Q,N204D]) had a higher affinity for NMB than the point mutant [H202Q] alone (Fig. 6; Table 2). Because the EC3 of NMBR is one amino acid shorter than the EC3 of GRPR [i.e., lacks an amino acid equivalent to Thr190 in GRPR (Fig. 4)], the effect of deletion of Thr190 from GRPR (Thr190-GRPR) on NMB affinity was determined. This deletion did not increase the affinity (150 ± 10 nM) for NMB (Fig. 5; Table 2). However, when the EC3 mutation [H202Q] was combined with the Thr190 deletion in GRPR, [T190-,H202Q], a greater gain in NMB affinity (12 ± 1 nM) was seen than the gain of NMB affinity with H202Q alone (Fig. 6). The differences seen with the gain-of-affinity chimeric receptors were not due to a global alteration of receptor conformation, because each mutated receptor retained high affinity for Bn and/or [d-Tyr6,βAla11,Phe13,Nle14]Bn-(6–14) (Table 2).
Fig. 5.
Affinities of NMB for wild-type GRPR and selected single and double amino acid(s) mutant GRP receptors (gain-of-affinity), with changes in the third EC region. These mutations were made by substituting, in GRPR, the different amino acids in the comparable positions of NMBR. The curved arrows indicate large gains in affinity of the mutant GRPR for NMB caused by the indicated mutation. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand. Each point on the dose-inhibition curves is the mean ± S.E.M. from at least four separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present
Table 2.
Affinities of NMB, Bn, and [d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) for wild-type GRPR, NMBR, and GRPR EC3 region of single or double gain-of-affinity mutants for NMB
Affinities of NMB, Bn, and [d-Phe6,ß-Ala11,Phe13,Nle14]Bn-(6–14) for wild-type GRP or NMB peptide and for selected single and double EC3 amino acid(s) mutant GRP receptors (gain-of-affinity). All mutants were from the EC3 regions (Fig. 4), with the exception of the [S215I]GRPR mutants, which contains a substitution of a serine by isoleucine in the upper TM5 (Fig. 4). The terminology [D181E]GRPR, for example, indicates that Asp, which occurs in position 181 of GRPR (Fig. 4) was substituted by Glu, which occurs in the comparable position in NMBR (Fig. 4). The affinities (IC50) were measured as measured by competitive displacement of 50 pM 125I-[Tyr4]Bn by NMB or Bn in transiently transfected Balb-3T3 cells. Cells were transfected as described under Materials and Methods. These data were calculated from the NMB dose-inhibitation curve for selected mutants shown in Fig. 5. Values are means ± S.E.M. from at least four experiments, and in each experiment, each point was determined in duplicate.
| IC50 |
|||
|---|---|---|---|
| NMB | Bn | [d-Tyr6,β-Ala11,Phe13,Nle14]Bn-(6–14) | |
| nM | |||
| WT NMBR | 1.0 ± 0.2 | 4.5 ± 0.5 | 0.7 ± 0.1 |
| WT GRPR | 115.1 ± 7.3 | 0.8 ± 0.1 | 0.6 ± 0.1 |
| [D181E]GRPR | 185.6 ± 18.7 | 4.5 ± 0.1 | |
| [L182V]GRPR | 259.3 ± 16.3 | 3.0 ± 0.1 | |
| [H183A]GRPR | 135.5 ± 29.9 | 2.7 ± 0.1 | |
| [P184R]GRPR | 188.0 ± 25.9 | 3.2 ± 0.1 | |
| [F185I]GRPR | 205.2 ± 65.9 | 120.7 ± 8.3d | 0.5 ± 0.1 |
| [H186G]GRPR | 159.1 ± 16.5 | 0.8 ± 0.1 | |
| [V187S]GRPR | 171.8 ± 52.1 | 0.8 ± 0.0 | |
| [K188S]GRPR | 153.7 ± 33.1 | 0.7 ± 0.1 | |
| [T190-]GRPR | 150.2 ± 10.4 | 1.0 ± 0.1 | |
| [Q192S,T193S]GRPR | 329.8 ± 47.4 | 3.2 ± 0.1 | |
| [I195T,S196A]GRPR | 442.1 ± 30.6 | 7.9 ± 0.3d | 0.4 ± 0.1 |
| [A198I]GRPR | 47.5 ± 9.9a, b | 7.9 ± 0.4d | 0.7 ± 0.2 |
| [H202Q]GRPR | 46.7 ± 4.8a, b | 0.9 ± 0.2 | |
| [A198I,H202Q]GRPR | 23.4 ± 1.9a | 0.8 ± 0.2 | |
| [S203T]GRPR | 140.5 ± 43.6 | 0.8 ± 0.2 | |
| [N204D]GRPR | 143.4 ± 30.7 | 1.2 ± 0.4 | |
| [M213V,A214L]GRPR | 79.5 ± 7.5 | 10.6 ± 0.2d | 0.5 ± 0.1 |
| [S215I]GRPR | 5.4 ± 0.4a | 1.7 ± 0.2 | |
| [A198I,S215I]GRPR | 1.7 ± 0.2a, c | 1.3 ± 0.2 | |
| [H202Q,S215I]GRPR | 4.8 ± 0.6a | 1.5 ± 0.1 | |
a P < 0.0001 vs WT GRPR for NMB.
b P < 0.02 vs [A198I,H202Q]GRPR for NMB.
c P < 0.0001 vs [S215I]GRPR for NMB.
d P < 0.03 vs WT NMBR for Bn.
Fig. 6.
Comparison of affinities of NMB for single amino acid GRPR gain-of-affinity mutants and GRPR mutants, with both amino acid mutations made in combination. The mutations were made by substituting, in the GRPR, the different amino acids in the comparable positions of NMBR. Each point on the dose-inhibition curves is the means ± S.E.M. from at least four separate experiments, and each point was determined in duplicate in each experiment. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present. The NMB affinity of the combination mutant was compared to the single amino acid mutant with the closest NMB affinity.
EC3 NMBR Mutants (Loss-of-Affinity Point and Combination Mutants).
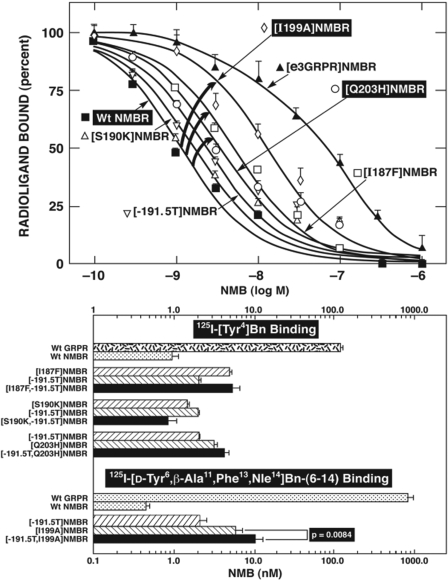
To attempt to confirm the findings from the GRPR gain-of-affinity point mutations and combination mutations and possibly provide additional insights, seven NMB loss-of-affinity NMBR point mutants were made in the EC3 of NMBR. These loss-of-affinity NMBR point mutants were made by substituting in NMBR the comparable different amino acids in the GRPR (Fig. 4). The substitution of isoleucine in position 199 in the EC3 of NMBR by an alanine ([I199A]NMBR), which is in the comparable position of GRPR, produced the most marked decrease (8.6-fold) in NMB affinity (from 1.0 ± 0.2 to 8.6 ± 1.5 nM) (Fig. 7). Point mutants [I187F] and [Q203H] in EC3 of the NMBR also showed a decrease in their affinity for NMB (3.2–4.9-fold), but to a less extent that than seen with [I199A] (Fig. 7, top; Table 3). However, the NMBR mutations [G188H], [S189V], or [S190K] did not produce changes in the affinity for NMB (Fig. 7, top; Table 3). Furthermore, the addition of a threonine after position 191 ([-191.5T]NMBR) in NMBR to make the EC3 of NMBR a size similar to that of GRPR (Fig. 4) did not produce changes in NMB for the mutant NMBR (Fig. 7, top; Table 3). The differences seen with these loss-of-affinity NMBR mutant receptors were not due to a global alteration of receptor configuration, because the [I187F] and [Q203H] mutant NMBR retained high affinity for Bn (Table 3). The point mutation [I199A]NMBR also showed a decrease in affinity for Bn (30.9 ± 7.2 nM) (Table 3) in addition to NMB; however, it demonstrated the same affinity for the potent and selective NMBR antagonist PD168368 (34.1 ± 5.7 nM) (Table 4) (Ryan et al., 1999) as observed in wild-type NMBR (31.9 ± 2.6 nM) (Table 4), demonstrating that these changes in affinity for NMB and Bn were not due to a global alteration in the receptor structure. None of the combinations of NMBR loss-of-affinity mutations [I187F,-191.5T], [S190K,-191.5T], or [-191.5T,Q203H] caused a greater loss-of-affinity for NMB than the point mutant [-191.5T] alone (Fig. 7, bottom). However, when the addition of threonine to the EC3 in NMBR to make a comparable length to the EC3 in GRPR ([-191.5T]NMBR) was combined with the EC3 mutation I199A ([-191.5T,I199A]NMBR) an additive decrease in affinity for NMB was observed (10 ± 1 nM) (Fig. 7, bottom). Because the combination gain-of-affinity GRPR mutation [A198I,H202Q]GRPR completely restored NMB affinity to GRPR, we made the equivalent combination of NMBR loss-of affinity mutant [I199A,I216S]NMBR. [I199A,I216S]NMBR demonstrated a complete loss of affinity for NMB, showing that this combination mutation had a greater effect than I199A alone (data not shown).
Fig. 7.
Affinities of NMB for various EC3 NMBR loss of affinity mutants alone (top) or in combination (bottom). Top, affinities of NMB for wild-type NMB, [e3GRPR]NMB, and selected single EC3 amino acid mutant NMB receptors (loss-of-affinity). The mutations were made by substituting, in NMBR, either the entire EC3 domain of GRPR or the different amino acids in comparable positions in the EC3 of GRPR. The curved arrows indicate large losses in affinity for NMB of the indicated mutant receptor. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor. Top, 50 pM 125I-[Tyr4]Bn was used as the ligand. Bottom, a comparison of affinities of NMB for single and double combination EC3 NMBR receptor mutants, in which the affinity of mutant NMB receptor for NMB was determined by performing binding studies using either 50 pM 125I-[Tyr4]Bn or 125I-[d-Tyr6,β-Ala11, Phe13,Nle14]Bn(6–14). 125I-[d-Tyr6,β-Ala11,Phe13,Nle14]Bn(6–14) was used for some of these mutants because of their very low level of binding of 125I-[Tyr4]Bn due to their loss-of-affinity for bombesin. Each point on the dose-inhibition curve is the means + S.E.M. from at least four separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present.
TABLE 3.
Affinities of NMB and Bn for wild-type GRPR, NMBR, and NMBR EC3 region of single loss-of-affinity mutants for NMB
Affinities of NMB and Bn for wild-type GRP or NMB receptors for selected single EC3 amino acid mutant NMB receptors (loss-of-affinity). The affinities (IC50) were determined as described in Table 1. Balb-3T3 cells were transfected as described under Materials and Methods, and binding was determined using 50 pM 125I-[Tyr4]Bn. Results for NMB for selected receptors were calculated from the NMB dose-inhibition curves shown in Fig. 7 (top). Significant decreases in affinity for NMB or Bn are shown. Values are mean ± S.E.M. from at least four experiments, and in each experiment, each point was measured in duplicate.
| IC50 |
||
|---|---|---|
| NMB | Bn | |
| nM | ||
| WT GRPR | 115.1 ± 7.3 | 0.8 ± 0.1 |
| WT NMBR | 1.0 ± 0.2 | 4.5 ± 0.5 |
| [E183D]NMBR | 1.0 ± 0.1 | 8.3 ± 1.6 |
| [I187F]NMBR | 4.9 ± 0.4b | 2.0 ± 0.2 |
| [G188H]NMBR | 1.1 ± 0.1 | 3.8 ± 0.3 |
| [S189V]NMBR | 0.8 ± 0.2 | 3.6 ± 0.7 |
| [S190K]NMBR | 1.5 ± 0.1a | 4.5 ± 1.1 |
| [-191.5T]NMBR | 2.1 ± 0.1a | 5.7 ± 1.6 |
| [I199A]NMBR | 8.6 ± 1.5b | 30.9 ± 7.2c |
| [Q203H]NMBR | 3.2 ± 0.3a | 6.4 ± 0.5 |
a P < 0.009 vs WT NMBR for NMB.
b P < 0.002 vs WT NMBR for NMB.
c P < 0.0001 vs WT NMBR for Bn.
TABLE 4 .
Importance of the position 199 amino acid substitution in NMBR for determining NMB affinity or affinity for selective NMBR antagonist, PD168368.
The affinities (IC50) were measured as described in Table 1. The wild-type and single amino position 199 of NMB mutant receptors was transiently expressed in CHOP cells for comparison because some demonstrated very low expression in Balb-3T3 cells. Values are mean ± S.E.M. from at least four experiments, and in each experiment, each point was measured in duplicate.
| IC50 |
||
|---|---|---|
| NMB | PD168368 | |
| nM | ||
| WT NMBR | 0.9 ± 0.3 | 31.9 ± 2.6 |
| WT GRPR | 98 ± 3 | 0.8 ± 0.1 |
| [I199A]NMBR | 10.2 ± 1.2a | 34.1 ± 5.7 |
| [I199V]NMBR | 1.2 ± 0.2 | 30.7 ± 0.8 |
| [I199L]NMBR | 2.3 ± 0.2 | 33.0 ± 1.1 |
| [I199M]NMBR | 1.5 ± 0.2 | 44.3 ± 1.6b |
| [I199E]NMBR | 48.9 ± 2.5a | 94.2 ± 18.0b |
| [I199K]NMBR | 32.4 ± 1.6a | 89.5 ± 16.0b |
a P < 0.0001 vs WT NMBR for NMB.
b P < 0.01 vs WT NMBR for PD168368.
NMBR Point Mutants for Isoleucine in Position 199.
The importance of isoleucine in position 199 in NMBR instead of an alanine in the comparable position in GRPR (position 198) (Fig. 4) for high-affinity NMB interaction suggests that hydrophobicity or steric effects of the substitution could be important. To explore these possibilities, additional NMBR point mutants were made by replacing isoleucine 199 of NMBR with different amino acids that show a wide range in hydrophobicity. Isoleucine 199 was replaced by the following amino acids: leucine or valine with high hydrophobicity; methionine, with a moderately high level; alanine, with intermediate hydrophobicity; and finally, glutamic acid and lysine, with a low hydrophobicity (Black and Mold, 1991). The substitution of isoleucine in position 199 of NMBR with amino acids with moderately high to high hydrophobicity [leucine, valine, and methionine] had either no effect or only a minimal effect on NMB affinity [i.e., IC50 =1.2–2.3 nM; Table 4]. The substitution with alanine with intermediate hydrophobicity caused a 10-fold decrease in NMB affinity (Table 4). Moreover, the substitution of isoleucine 199 in NMBR by amino acids with low hydrophobicity, such as glutamic acid or lysine, produced a marked loss-of-affinity (36–54-fold) for NMB, which was even greater than the decrease in NMB affinity, with the substitution of the comparable amino acid in GRPR [i.e., alanine (10-fold)] (Table 4). These differences seen with the loss-of-affinity position 199 NMBR mutant receptors were not due to a global alteration of receptor conformation, because each mutant receptor retained high affinity for [d-Tyr6,βAla11,Phe13,Nle14]Bn-(6–14) (data not shown) and/or PD168368 (Table 4).
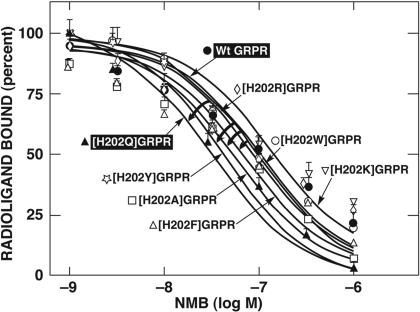
GRPR Point Mutants for Histidine in Position 202.
The importance of glutamine in position 203 in NMBR for NMB affinity rather than a histidine in the similar position in GRPR (i.e., [H202Q]GRPR) (Figs. 4 and 5) was investigated in more detail. To explore the basis for the differences in more detail, additional possible GRPR gain-of-affinity for NMB point mutants were made by replacing histidine 202 of GRPR with related amino acids with different properties: three amino acids with different aromatic rings (phenylalanine, tryptophan, and tyrosine), with an uncharged amino acid with an aliphatic backbone substitution (i.e., alanine), or with two other basic amino acids (lysine and arginine) (Fig. 8). The substitution of histidine in position 202 in GRPR by tyrosine caused the most prominent effect with a 4-fold gain-of-affinity of NMB (Fig. 8; Table 5) and had a greater effect than the replacement of histidine by glutamine [H202Q] (2.5-fold) or by alanine (2.0-fold) (Fig. 8; Table 5). However, a phenylalanine substitution for histidine in position 202 caused only a small increase in the affinity of NMB (1.7-fold). Furthermore, no changes in the affinity for NMB were observed when lysine, arginine, or tryptophan was substituted for histidine in position 202 (Fig. 8; Table 5).
Fig. 8.
Affinities of NMB for wild-type, [H202Q]GRPR, and seven single mutant GRPRs (gain-of-affinity) where the His located in position 202 in GRPR was replaced with different amino acids, with different properties. The curved arrows shows the alteration of the affinity of NMB for single mutants [H202Y], [H202A], [H202F], and [H202Q]GRPR from the wild-type GRP receptor. All studies were performed in Balb-3T3 cells transiently transfected with the indicated wild-type or mutant receptor by using 50 pM 125I-[Tyr4]Bn as the ligand. Each point on the dose-inhibition curve is the mean ± S.E.M. from at least three separate experiments, and each point was determined in duplicate in each experiment. Data are expressed as the percentage of saturable binding when no unlabeled peptide was present. Curved arrows indicate gain-in-affinity for NMB compared with the wild-type GRPR.
TABLE 5.
Importance of the amino acid substitution in position 202 of GRPR for determining affinity for NMB
The affinities (IC50) were determined as described in Table 1 and, for NMB, were from the dose-inhibition curves shown in Fig. 8. Studies were performed in transiently transfected Balb-3T3 cells as described in Table 1 legend. Significant increases in affinity from the wild type are shown. Values are means ± S.E.M. from at least four experiments, and in each experiment, each point was measured in duplicate.
| IC50 |
||
|---|---|---|
| NMB | Bn | |
| nM | ||
| WT NMBR | 1.0 ± 0.2 | 4.5 ± 0.5 |
| WT GRPR | 115.1 ± 7.3 | 0.8 ± 0.1 |
| [H202Q]GRPR | 46.7 ± 4.8b | 0.86 ± 0.15 |
| [H202A]GRPR | 56.1 ± 12.1b | 0.80 ± 0.04 |
| [H202F]GRPR | 69.4 ± 7.9a | 0.89 ± 0.15 |
| [H202K]GRPR | 167.2 ± 46.2 | 1.17 ± 0.24 |
| [H202R]GRPR | 98.9 ± 28.3 | 1.04 ± 0.23 |
| [H202W]GRPR | 92.3 ± 8.6 | 1.28 ± 0.08 |
| [H202Y]GRPR | 31.2 ± 5.8b | 1.47 ± 0.34 |
a P < 0.02 vs WT GRPR for NMB.
b P < 0.005 vs WT GRPR for NMB.
Discussion
This study was undertaken to provide insights into the molecular basis for selectivity/high affinity of the NMBR for its natural agonist, NMB. The NMBR interacts with high affinity with one member (i.e., NMB) of a family of closely related, naturally occurring peptides (von Schrenck et al., 1989; Jensen et al., 2008). Despite the importance of NMBR in mediating many physiological/pathological processes (Sun et al., 2000; Jensen et al., 2008), there is little known about the molecular basis for its selectivity. This is in marked contrast to the interaction of GRP with GRPR, which has been extensively studied (Akeson et al., 1997; Tokita et al., 2001b; Nakagawa et al., 2005; Jensen et al., 2008). One study (Sainz et al., 1998) that used comparative amino acid analysis of the structures of various Bn receptors identified four amino acids (Arg127, Ser205, His294, and Ser315) in BRS-3, which determined its low NMB affinity. However, these four amino acids are conserved in the NMBRs and GRPRs from different species, and they do not provide any information about the differences in the affinity of NMB for NMBR over GRPR (von Schrenck et al., 1989; Sainz et al., 1998; Tokita et al., 2002; Jensen et al., 2008). Only one study (Fathi et al., 1993) examined the selectivity of NMBR for NMB. In that study, changing an isoleucine in position 216 in the upper TM5 domain of NMBR with the comparable amino acid in GRPR (serine 215) decreased affinity for NMB (Fathi et al., 1993). Other receptor regions were not systematically evaluated (Fathi et al., 1993).
In our study, we systematically examined the molecular basis of high affinity/selectivity of NMBR for NMB by using a combined chimeric/site-directed mutagenesis approach. A number of our results support the conclusion that differences in extracellular receptor domains and adjacent upper TM domains of the GRP/NMB receptors play an important role in NMB selectivity. First, with GRPR gain-of-affinity chimeras constructed by replacing GRPR EC domains by those from NMBR, differences in EC3 were the important determinants of NMB selectivity. Second, when the reverse study was performed with NMBR loss-of-affinity chimeras, the EC3 replacement had the most prominent effect. Third, the importance of an isoleucine in the upper TM5 domain of NMBR (Ile216), instead of serine in GRPR (Ser215), was confirmed to be important for NMB selectivity by making a GRPR gain-of-affinity mutant (i.e., [S215I]GRPR), as proposed in NMBR loss-of-affinity studies (Fathi et al., 1993). Compared with GRPR/BRS-3, our results are similar in that both extracellular and upper TM regions are important for selectivity of Bn, GRP, and some BRS-3-selective peptide agonists for GPRR or BRS-3 (Akeson et al., 1997; Tokita et al., 2002; Nakagawa et al., 2005; Gonzalez et al., 2008; Jensen et al., 2008). Compared with other G-protein-coupled receptors, our results are similar to the interaction of the peptide agonists, substance P with NK-1 receptors (Li et al., 1996) and CCK-8 with CCK-B receptors (Silvente-Poirot et al., 1998), where both the extracellular and TM domains play a marked role in selectivity. However, the affinity of CCK-8 for the CCK-A receptor (Silvente-Poirot et al., 1998), secretin for secretin receptors (Holtmann et al., 1995), or the GRPR peptide antagonist JMV641 (Tokita et al., 2001b) for GRPR is determined by extracellular domains, whereas differences in only TM regions are crucial for affinity of nociceptin for orphanin-FQ receptors (Meng et al., 1996) or the peptoid antagonist PD168368 for NMBR (Tokita et al., 2001a).
Our finding that important differences in determining NMB affinity/selectivity for NMBR/GRPR were in EC3 and that the upper TM5 region has both similarities and differences from studies with other Bn receptors or non–Bn G-protein-coupled receptors. In contrast, differences in EC2 are primarily important for the high affinity of some BRS-3-selective peptide agonists (Gonzalez et al., 2008), and differences in EC4 are important for GRPR selectivity of the highly selective GRPR peptide antagonists, JMV641 and JMV594 (Tokita et al., 2001b). Likewise, the high affinity of GRP or Bn for GRPR is primarily determined by differences from other Bn receptors in the EC3 region (Akeson et al., 1997; Tokita et al., 2002). In addition, with rat CCK-B receptor, EC3 plays a critical role for high-affinity interaction with gastrin (Silvente-Poirot and Wank, 1996) and CCK (Silvente-Poirot et al., 1999). Likewise, with the human corticotropin-releasing factor (CRF) receptor-1, both EC3 and TM5 were important for the peptide ligand rat/human CRF affinity (Liaw et al., 1997). Furthermore, with the vasoactive intestinal peptide receptor 1, high affinity for the peptide agonist peptide histidine isoleucinamide is determined by residues in EC2 and TM3 (Couvineau et al., 1996). In contrast, only the TM5 of NMBR and the neurokinin-1 receptor is critical for the high selectivity of the peptoid antagonists PD168368 (Tokita et al., 2001a) and CP96345 (Fong et al., 1992).
Within EC3 domain, 20 amino acids differ between NMBR and GRPR, with two differences [Ile199, Gln203 in NMBR instead of Ala198, His202 in GRPR] causing EC3 to be responsible for the high NMB selectivity of NMBR. These results demonstrated that NMBR and GRPR, which share 51% homology, have both similarities and differences in the location/nature of the key determinants of their selectivity for their respective naturally occurring agonist ligand, NMB or GRP. They are similar in that differences in EC3 play a key role in selectivity of NMBR for NMB and GRPR for GRPR (Tokita et al., 2002). In addition, with both receptors, the presence of the amino acid in the same position in the EC3 (Ile199 in NMBR or Ala198 in GRPR) plays a major role (Tokita et al., 2002). The basis of the agonist selectivity of these two receptors differs in that Gln203 in EC3 and Ile216 in TM5 of NMBR, instead of His202 and Ser215 in GRPR, respectively, are crucial for determining in NMBR affinity but not for GRPR affinity for GRP (Tokita et al., 2002). Not only was the presence of either an isoleucine in position 199 or 216 of NMBR important for NMB affinity, their presence in combination had a greater effect than either one alone. The fact that these two mutations together had a potentiating effect for NMB affinity could be the result of additive interaction with the ligand, a global change in the receptor conformation, or the fact that the two amino acids are necessary for a specific binding site conformation. Our results showing a synergetic effect on agonist affinity of multiple substitutions are similar to findings reported for the selectivity of peptide histidine isoleucinamide for the human vasoactive intestinal peptide receptor 1 (Couvineau et al., 1996), GRP for GRPR over human BRS-3 (Nakagawa et al., 2005), the BRS-3-selective agonist Ac-Phe-Trp-Ala-His(tBzl)-Nip-Gly-Arg-NH2 for BRS-3 over GRPR (Gonzalez et al., 2008), and the peptide antagonists JMV594/JMV641 for GRPR over NMBR (Tokita et al., 2001b) and the nonpeptide antagonist CP96345 for human over the rat neurokinin-1 receptor (Fong et al., 1992).
Our finding of the importance of an isoleucine in NMBR instead of serine in GRPR in TM5 (Ile216 NMBR) or Ala in EC3 (Ile199) has both similarities and differences from findings with a number of other G-protein-coupled receptors (Greenfeder et al., 1999). Isoleucine in TM5 in the neurokinin-2 receptor is necessary for high affinity for the agonist NKA as well as that in neurokinin-1 receptors for the antagonist MDL103,392 (Greenfeder et al., 1999). In contrast, substitution of methionine in TM5 of the CRF type 1 with isoleucine markedly (75-fold) decreased affinity for the nonpeptide antagonist NBI35965 (Hoare et al., 2006). Likewise, isoleucine in TM3 of the melanocortin-4 receptor was important for high affinity for melanocortin-4 receptor ligands (tetrahydroisoquinoline, M10, and adrenocorticotropin hormone) (Nickolls et al., 2003). Our results are similar to findings with both the CCK-1 and NK-2 receptors, where replacement of isoleucine in TM5 or TM7, respectively, with alanine markedly decreased agonist affinity (Greenfeder et al., 1999; Escrieut et al., 2002). There is almost no information on the molecular mechanisms that determine the importance of the presence of an isoleucine in the different G-protein-coupled receptors for high-affinity ligand interaction. Isoleucine is particularly important for hydrophobic interactions, which could contribute either to receptor protein folding and/or conformational receptor change or to receptor-ligand interaction (Mosebi et al., 2003). In NMBR, the isoleucine 199 is located adjacent to Cys198 in EC3, which is presumed to form a disulfide bridge with Cys116 in EC2, which is important in determining EC3 conformation. The possibility that the substitution of isoleucine 199 in NMBR, with the equivalent amino acid alanine in GRPR, could be effecting the global conformation of NMBR or the receptor structure is supported by studies that show that the presence of isoleucine promotes formation of Cys-Cys bridges (Kim, 2006). Our data suggest that it is unlikely that the presence of an alanine instead of isoleucine 199 is altering the global structure of the mutated receptor because the mutated receptor retained affinity for other peptide ligands. One of the principal differences in substituting alanine for isoleucine 199 is a marked change in hydrophobicity. We explored the importance of this change by making several NMB affinity point mutants with 199 replacements with variable hydrophobicity. Substitution of isoleucine 199 in NMBR, with either glutamic acid or lysine, both having much lower hydrophobicity, caused a marked decrease in affinity for NMB, as did the substitution of alanine, which also has reduced hydrophobicity compared with isoleucine. However, when isoleucine 199 was substituted by amino acids with high hydrophobicity (leucine and valine) or with a moderately high hydrophobicity (methionine), either no difference or only a slight decrease in the affinity of NMB for NMBR was observed. These results support the conclusion that the hydrophobicity of the substitution in position 199 of NMBR plays a crucial role for NMBR affinity for NMB and suggest that the substitution of isoleucine with alanine creates a difference in the transfer-free energies that could influence hydrophobicity and the thermodynamics of protein folding (Escrieut et al., 2002).
We also found that the presence of glutamine in NMBR (position 203) rather than histidine in GRPR (position 202) in the EC3 domain was important for NMB high affinity/selectivity. This result is similar to the peptide agonist PL017 for the μ-opioid receptor where a [H297Q] receptor replacement altered affinity 5-fold (Spivak et al., 1997). To provide possible insight into the molecular basis of this change, we made several GRPR (position 202) point mutants. Our results demonstrated that the substitution of histidine by either arginine or lysine, two other basic amino acid(s), did not improve the NMB affinity. In contrast, replacement of histidine by uncharged amino acids (Ala202,Tyr202) or glutamine all increased affinity, suggesting that the lack of basicity of the substitution in position 202 of GRPR was an important determinant of NMB affinity. Our results also supported the conclusion that neither differences in hydrophobicity of the amino acid backbone substitution nor the presence of an aromatic ring substitution per se were major determinants of this change of affinity; however, steric factors could play a minor role because insertion of a large aromatic group did not improve affinity.
In conclusion, our study demonstrates that three amino acid differences in the EC3 region of NBMBR and GRPR are critical for determining NMB affinity/selectivity. The change in NMB affinity/selectivity mediated by these differences is largely due to differences in hydrophobicity and to a lesser extent charge and steric factors with the different substitutions.
This research was supported by the Intramural Research Program of the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
- Bn
- bombesin
- BRS-3
- bombesin receptor subtype-3
- BSA
- bovine serum albumin fraction V
- DMEM
- Dulbecco's minimum essential medium
- EC
- extracellular
- TM
- transmembrane
- FBS
- fetal bovine serum
- GRP
- gastrin-releasing peptide
- GRPR
- gastrin-releasing peptide receptor
- NMB
- neuromedin B
- NMBR
- neuromedin B receptor
- CRF
- corticotropin-releasing factor
- CNS
- central nervous system
- WT
- wild type
- NKA
- neurokinin A
- CCK
- cholecystokinin
- CHOP
- polyoma large T-antigen Chinese hamster ovary fibroblasts
- M10
- cyclo(6β→10ϵ)(succinyl6)-d-(2′)Nal7-Arg8-Trp9-Lys10)-NH2
- PL017
- H-Tyr-Pro-(N-Me))Phe-d-Pro-NH2
- PD168368
- 3-(1H-indol-3-yl)-2-methyl-2-[3(4-nitrophenyl)-ureido]-N-(1-pyridin-2-yl-cyclohexylmethyl)-propionamide
- MDL103,392
- (R,S)-1-[3-[3,4-dichlorophenyl]-1-(3,4,5-trimethoxybenzoyl)-pyrrolidin-3-yl]ethyl-4-phenylpiperidine-4-carboxamide
- CP96345
- (2S,3S)-cis-2-(diphenylmethyl)-N-((2-methoxyphenyl)-methyl)-1-azabicyclo(2.2.2)-octan-3-amine
- JMV641
- (d-Phe-Gln-Trp-Ala-Val-Gly-His-Leu psi(CHOH-CH2))-(CH2)(2)-CH3
- JMV594
- [d-Phe6Stat13]Bn(6-14)
- NBI35965
- (S)-6-cyclopropylmethyl-2-(2,4-dichlorophenyl)-7-ethyl-4-methyl-7,8-dihydro-6H-1,3,6a-tetraazaacenaphthylene mesylate.
References
- Akeson M, Sainz E, Mantey SA, Jensen RT, Battey JF. ( 1997) Identification of four amino acids in the gastrin-releasing peptide receptor that are required for high affinity agonist binding. J Biol Chem 272: 17405–17409 [DOI] [PubMed] [Google Scholar]
- Battey JF, Way JM, Corjay MH, Shapira H, Kusano K, Harkins R, Wu JM, Slattery T, Mann E, Feldman RI. ( 1991) Molecular cloning of the bombesin/gastrinreleasing peptide receptor from Swiss 3T3 cells. Proc Natl Acad Sci U S A 88: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benya RV, Fathi Z, Battey JF, Jensen RT. ( 1993) Serines and threonines in the gastrin-releasing peptide receptor carboxyl terminus mediate internalization. J Biol Chem 268: 20285–20290 [PubMed] [Google Scholar]
- Benya RV, Fathi Z, Kusui T, Pradhan T, Battey JF, Jensen RT. ( 1994) Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization and growth: possible role for cyclic AMP. Mol Pharmacol 46: 235–245 [PubMed] [Google Scholar]
- Benya RV, Wada E, Battey JF, Fathi Z, Wang LH, Mantey SA, Coy DH, Jensen RT. ( 1992) Neuromedin B receptors retain functional expression when transfected into BALB 3T3 fibroblasts: analysis of binding, kinetics, stoichiometry, modulation by guanine nucleotide-binding proteins, and signal transduction and comparison with natively expressed receptors. Mol Pharmacol 42: 1058–1068 [PubMed] [Google Scholar]
- Black SD, Mould DR. ( 1991) Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem 193: 72–82 [DOI] [PubMed] [Google Scholar]
- Couvineau A, Rouyer-Fessard C, Maoret JJ, Gaudin P, Nicole P, Laburthe M. ( 1996) Vasoactive intestinal peptide (VIP)1 receptor. Three nonadjacent amino acids are responsible for species selectivity with respect to recognition of peptide histidine isoleucineamide. J Biol Chem 271: 12795–12800 [DOI] [PubMed] [Google Scholar]
- Cuttitta F, Carney DN, Mulshine J, Moody TW, Fedorko J, Fischler A, Minna JD. ( 1985) Bombesin-like peptides can function as autocrine growth factors in human small-cell lung cancer. Nature 316: 823–826 [DOI] [PubMed] [Google Scholar]
- Escrieut C, Gigoux V, Archer E, Verrier S, Maigret B, Behrendt R, Moroder L, Bignon E, Silvente-Poirot S, Pradayrol L, et al. ( 2002) The biologically crucial C terminus of cholecystokinin and the non-peptide agonist SR-146,131 share a common binding site in the human CCK1 receptor. Evidence for a crucial role of Met-121 in the activation process. J Biol Chem 277: 7546–7555 [DOI] [PubMed] [Google Scholar]
- Fathi Z, Benya RV, Shapira H, Jensen RT, Battey JF. ( 1993) The fifth transmembrane segment of the neuromedin B receptor is critical for high affinity neuromedin B binding. J Biol Chem 268: 14622–14626 [PubMed] [Google Scholar]
- Fong TM, Yu H, Strader CD. ( 1992) Molecular basis for the species selectivity of the neurokinin-1 receptor antagonists CP-96,345 and RP67580. J Biol Chem 267: 25668–25671 [PubMed] [Google Scholar]
- Gonzalez N, Hocart SJ, Portal-Nuñez S, Mantey SA, Nakagawa T, Zudaire E, Coy DH, Jensen RT. ( 2008) Molecular basis for agonist selectivity and activation of the orphan bombesin receptor subtype 3 receptor. J Pharmacol Exp Ther 324: 463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S, Cheewatrakoolpong B, Billah M, Egan RW, Keene E, Murgolo NJ, Anthes JC. ( 1999) The neurokinin-1 and neurokinin-2 receptor binding sites of MDL103,392 differ. Bioorg Med Chem 7: 2867–2876 [DOI] [PubMed] [Google Scholar]
- Hoare SR, Brown BT, Santos MA, Malany S, Betz SF, Grigoriadis DE. ( 2006) Single amino acid residue determinants of non-peptide antagonist binding to the corticotropin-releasing factor 1 (CRF1) receptor. Biochem Pharmacol 72: 244–255 [DOI] [PubMed] [Google Scholar]
- Holtmann MH, Hadac EM, Miller LJ. ( 1995) Critical contributions of amino-terminal extracellular domains in agonist binding and activation of secretin and vasoactive intestinal polypeptide receptors. Studies of chimeric receptors. J Biol Chem 270: 14394–14398 [DOI] [PubMed] [Google Scholar]
- Jensen JA, Carroll RE, Benya RV. ( 2001) The case for gastrin-releasing peptide acting as a morphogen when it and its receptor are aberrantly expressed in cancer. Peptides 22: 689–699 [DOI] [PubMed] [Google Scholar]
- Jensen RT, Battey JF, Spindel ER, Benya RV. ( 2008) International Union of Pharmacology. LXVIII. Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling and functions in normal and disease states. Pharmacol Rev 60: 1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RT, Moody TW.( Kastin AJ. ed) ( 2006) Bombesin-related peptides and neurotensin: effects on cancer growth/proliferation and cellular signaling in cancer, in Handbook of Biologically active peptides, pp 429–434, Elsevier, Amsterdam [Google Scholar]
- Kim H. ( 2006) Activity of human dihydrolipoamide dehydrogenase is largely reduced by mutation at isoleucine-51 to alanine. J Biochem Mol Biol 39: 223–227 [DOI] [PubMed] [Google Scholar]
- Ladenheim EE, Jensen RT, Mantey SA, Moran TH. ( 1992) Distinct distributions of two bombesin receptor subtypes in the rat central nervous system. Brain Res 593: 168–178 [DOI] [PubMed] [Google Scholar]
- Li H, Hsu P, Sachais BS, Krause JE, Leeman SE, Boyd ND. ( 1996) Identification of the site in the substance P (NK-1) receptor for modulation of peptide binding by sulfhydryl reagents. J Biol Chem 271: 1950–1956 [DOI] [PubMed] [Google Scholar]
- Liaw CW, Grigoriadis DE, Lorang MT, De Souza EB, Maki RA. ( 1997) Localization of agonist- and antagonist-binding domains of human corticotropin-releasing factor receptors. Mol Endocrinol 11: 2048–2053 [DOI] [PubMed] [Google Scholar]
- Mantey SA, Weber HC, Sainz E, Akeson M, Ryan RR, Pradhan TK, Searles RP, Spindel ER, Battey JF, Coy DH, et al. ( 1997) Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates that it has a unique pharmacology compared with other mammalian bombesin receptors. J Biol Chem 272: 26062–26071 [DOI] [PubMed] [Google Scholar]
- McCoy JG, Avery DD. ( 1990) Bombesin: potential integrative peptide for feeding and satiety. Peptides 11: 595–607 [DOI] [PubMed] [Google Scholar]
- Meng F, Taylor LP, Hoversten MT, Ueda Y, Ardati A, Reinscheid RK, Monsma FJ, Watson SJ, Civelli O, Akil H. ( 1996) Moving from the orphanin FQ receptor to an opioid receptor using four point mutations. J Biol Chem 271: 32016–32020 [DOI] [PubMed] [Google Scholar]
- Moody TW, Merali Z. ( 2004) Bombesin-like peptides and associated receptors within the brain: distribution and behavioral implications. Peptides 25: 511–520 [DOI] [PubMed] [Google Scholar]
- Mosebi S, Sayed Y, Burke J, Dirr HW. ( 2003) Residue 219 impacts on the dynamics of the C-terminal region in glutathione transferase A1–1: implications for stability and catalytic and ligandin functions. Biochemistry (Mosc) 42: 15326–15332 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Hocart SJ, Schumann M, Tapia JA, Mantey SA, Coy DH, Tokita K, Katsuno T, Jensen RT. ( 2005) Identification of key amino acids in the gastrin-releasing peptide receptor (GRPR) responsible for high affinity binding of gastrin-releasing peptide (GRP). Biochem Pharmacol 69: 579–593 [DOI] [PubMed] [Google Scholar]
- Nickolls SA, Cismowski MI, Wang X, Wolff M, Conlon PJ, Maki RA. ( 2003) Molecular determinants of melanocortin 4 receptor ligand binding and MC4/MC3 receptor selectivity. J Pharmacol Exp Ther 304: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Pradhan TK, Katsuno T, Taylor JE, Kim SH, Ryan RR, Mantey SA, Donohue PJ, Weber HC, Sainz E, Battey JF, et al. ( 1998) Identification of a unique ligand which has high affinity for all four bombesin receptor subtypes. Eur J Pharmacol 343: 275–287 [DOI] [PubMed] [Google Scholar]
- Reubi JC, Wenger S, Schmuckli-Maurer J, Schaer JC, Gugger M. ( 2002) Bombesin receptor subtypes in human cancers: detection with the universal radioligand (125)I-[d-Tyr(6), beta-Ala(11), Phe(13), Nle(14)] bombesin(6–14). Clin Cancer Res 8: 1139–1146 [PubMed] [Google Scholar]
- Ryan RR, Katsuno T, Mantey SA, Pradhan TK, Weber HC, Coy DH, Battey JF, Jensen RT. ( 1999) Comparative pharmacology of the nonpeptide neuromedin B antagonist PD 168368. J Pharmacol Exp Ther 290: 1202–1211 [PubMed] [Google Scholar]
- Sainz E, Akeson M, Mantey SA, Jensen RT, Battey JF. ( 1998) Four amino acid residues are critical for high affinity binding of neuromedin B to the neuromedin B receptor. J Biol Chem 273: 15927–15932 [DOI] [PubMed] [Google Scholar]
- Silvente-Poirot S, Escrieut C, Galès C, Fehrentz JA, Escherich A, Wank SA, Martinez J, Moroder L, Maigret B, Bouisson M, et al. ( 1999) Evidence for a direct interaction between the penultimate aspartic acid of cholecystokinin and histidine 207, located in the second extracellular loop of the cholecystokinin B receptor. J Biol Chem 274: 23191–23197 [DOI] [PubMed] [Google Scholar]
- Silvente-Poirot S, Escrieut C, Wank SA. ( 1998) Role of the extracellular domains of the cholecystokinin receptor in agonist binding. Mol Pharmacol 54: 364–371 [DOI] [PubMed] [Google Scholar]
- Silvente-Poirot S, Wank SA. ( 1996) A segment of five amino acids in the second extracellular loop of the cholecystokinin-B receptor is essential for selectivity of the peptide agonist gastrin. J Biol Chem 271: 14698–14706 [DOI] [PubMed] [Google Scholar]
- Spivak CE, Beglan CL, Seidleck BK, Hirshbein LD, Blaschak CJ, Uhl GR, Surratt CK. ( 1997) Naloxone activation of mu-opioid receptors mutated at a histidine residue lining the opioid binding cavity. Mol Pharmacol 52: 983–992 [DOI] [PubMed] [Google Scholar]
- Sun B, Schally AV, Halmos G. ( 2000) The presence of receptors for bombesin/GRP and mRNA for three receptor subtypes in human ovarian epithelial cancers. Regul Pept 90: 77–84 [DOI] [PubMed] [Google Scholar]
- Tokita K, Hocart SJ, Coy DH, Jensen RT. ( 2002) Molecular basis of the selectivity of gastrin-releasing peptide receptor for gastrin-releasing peptide. Mol Pharmacol 61: 1435–1443 [DOI] [PubMed] [Google Scholar]
- Tokita K, Hocart SJ, Katsuno T, Mantey SA, Coy DH, Jensen RT. (2001a) Tyrosine 220 in the 5th transmembrane domain of the neuromedin B receptor is critical for the high selectivity of the peptoid antagonist PD168368. J Biol Chem 276: 495–504 [DOI] [PubMed] [Google Scholar]
- Tokita K, Katsuno T, Hocart SJ, Coy DH, Llinares M, Martinez J, Jensen RT. (2001b) Molecular basis for selectivity of high affinity peptide antagonists for the gastrin-releasing peptide receptor. J Biol Chem 276: 36652–36663 [DOI] [PubMed] [Google Scholar]
- Tsuda T, Kusui T, Hou W, Benya RV, Akeson MA, Kroog GS, Battey JF, Jensen RT. ( 1997) Effect of gastrin-releasing peptide receptor number on receptor affinity, coupling, degradation, and modulation. Mol Pharmacol 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Von Schrenck T, Heinz-Erian P, Moran T, Mantey SA, Gardner JD, Jensen RT. ( 1989) Neuromedin B receptor in esophagus: evidence for subtypes of bombesin receptors. Am J Physiol 256: G 747–758 [DOI] [PubMed] [Google Scholar]
- Wada E, Way J, Shapira H, Kusano K, Lebacq-Verheyden AM, Coy D, Jensen R, Battey J. ( 1991) cDNA cloning, characterization, and brain region-specific expression of a neuromedin B-preferring bombesin receptor. Neuron 6: 421–430 [DOI] [PubMed] [Google Scholar]