Abstract
Deubiquitinating enzymes (DUBs) are a family of proteases responsible for the specific removal of ubiquitin attached to target proteins and thus control the free cellular pools of this molecule. DUB activity is usually assayed using full-length ubiquitin, and these enzymes generally show low activity on small substrates that constitute the P4-P1 LRGG C-terminal motif of ubiquitin. To gain insight into the C-terminal recognition region of ubiquitin by DUBs we synthesized positional scanning libraries of fluorogenic tetrapeptides and tested them on three examples of human DUBs (OTU-1, Iso-T and UCH-L3) and one viral ubiquitin specific protease – Plpro from West Nile virus. In most cases the results show flexibility in the P4 position, very high specificity for Arg in P3 position and Gly at P2 – in accord with the sequence of the natural substrate ubiquitin. Surprisingly, screening of the P2 position revealed that UCH-L3 in contrast to all the other tested DUBs, demonstrates substantial tolerance of Ala and Val at P2, and a parallel analysis using the appropriate mutation of the full-length ubiquitin confirms this. We have also used an optimal tetrapeptide substrate, Ac-LRGG-AFC to investigate the activation mechanism of DUBs by ubiquitin and elevated salts concentration. Together, our results reveal the importance of the dual features of 1) substrate specificity and 2) the mechanism of ubiquitin binding in determining deubiquitination by this group of proteases.
Keywords: combinatorial chemistry, fluorogenic substrate, protease, DUB, ubiquitin
Introduction
The post-translational modification of proteins by ubiquitin (Ub) plays an important role in the regulation of a variety of biological processes[l, 2]. Besides the highly recognized pathway in which poly-ubiquitination targets proteins for removal by the proteasome, ubiquitin plays important roles in the maintenance of cellular events such as control of protein expression, gene silencing, cellular trafficking, and receptor internalization or down regulation[3]. Ubiquitin is selectively attached to target proteins by a cascade of enzymes involving ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), the latter of which primarily defines specificity for the target protein[4].
The ubiquitination process is reversible because ubiquitin can be selectively removed from target proteins by a family of hydrolases known collactively as DUBs (deubiquitinating enzymes)[5]. Deconjugation of ubiquitin by DUBs in the proteasome pathway is responsible for controlling the pool of free ubiquitin by recycling the protein for further rounds of conjugation[6]. Moreover, deconjugation of ubiquitin from cellular proteins is an important regulatory mechanism, which antagonizes ubiquitin conjugation and is involved in many cellular processes including cell cycle progression, tissue development and differentiation, memory and learning, oncogenesis, viral infection, and neurodegenerative disorders[7–12].
There are around one hundred DUBs in the human genome, and they are generally divided into five distinct subfamilies based on their sequence similarities and mechanisms of action. Four of the subfamilies are cysteine proteases and one family is represented by zinc-dependent metalloproteases. The most diverse structural subfamilies of DUBs are UBPs (Ubiquitin-specific processing proteases) - which are able to both deconjugate ubiquitin from target proteins and disassemble polyubiquitin chains, and UCHs (Ubiquitin carboxy-terminal hydrolases) - which are responsible for processing ubiquitin precursors that have C-terminal extensions[5]. These families are structurally distinct from the Ubiquitin-like proteases, which possess a mechanism related to DUBs, but which process Ub-like modifiers such as SUMOs[13].
A general feature of DUBs is the presence of two recognition regions, which are both thought to be required for interaction with conjugated proteins to gain the desired specificity. The first region binds the ubiquitin surface. This region interacts with the surface of the protease often leading to large conformational changes in the DUB required for optimal positioning of the catalytic machinery[14, 15]. The second region, the protease active site cleft, binds a linear epitope consisting of the sequence RLRGG (P5-P1), where the terminal Gly residue at the C-terminus of ubiquitin is conjugated to a Lys residue of a target protein. This linear sequence is highly conserved throughout evolution, implying a role in defining specificity for ubiquitin ligation, or ubiquitin deconjugation, or both. Peptide-based reporter substrates encompassing this region are cleaved by DUBs, but with poor catalytic rates[16, 17]. However, the sequence specificity requirements for recognition of this motif by DUBs have never been systematically analyzed[18]. The objective of this study was to define the sequence preference of representative unrelated DUBs for this C-terminal motif, by using positional scanning peptide libraries, to explore the relative importance of this region in DUB catalysis, and to examine the catalytic enhancement at the active site cleft produced by ubiquitin binding. Understanding these fundamental properties of DUBs is vital to delineating their function in the critical roles that this group of enzymes plays in cell fates and functions.
Experimental Section
Materials and methods
All the chemicals and solvents were obtained from commercial suppliers and used without further purification, unless otherwise stated. Anhydrous N,N-dimethyl formamide (DMF) was from Sigma-Aldrich. Rink amide and Safety Catch resins were purchased from Novabiochem. Column chromatography was performed using grade 60 silica gel (Fisher Scientific, 70– 230 mesh). 1H-MR spectra were obtained with the aid of the Burnham Structural Biology facility using a Varian 300 spectrometer in chloroform-d3 or dimethyl sulfoxide-d6 (Aldrich). 1H-MR (300 MHz) spectra are reported as follows: chemical shifts in ppm downfield from TMS, the internal standard; resonance signal description (s, singlet; d, doublet; t, triplet; m, multiplet), integration, and coupling constant (Hz). Analytical high performance liquid chromatography (HPLC) analysis were conducted on a Beckman-Coulter System Gold 125 solvent delivery module equipped with a Beckman-Coulter System Gold 166 Detector system by using a Varian Microsorb-MV C18 (250 × 4.8 mm) column. Preparative HPLC analysis were conducted on a Beckman-Coulter System Gold 126P solvent delivery module equipped with a Beckman-Coulter System Gold 168 Detector system by using a Kromasil 100-10 C18 (20 mm ID) column (Richard Scientific). Solvent composition system A (0.1%TFA in water) and system B (acetonitrile/water 80%/20% with 0.1% of TFA). Mass spectra were recorded in ESI mode with the aid of the Burnham Proteomics facility. The solid phase positional substrate library (SP-PSL) was synthesized using semiautomatic FlexChem Peptide Synthesis System (Model 202). Enzymatic kinetic studies were performed using a fMax fluorimeter (Molecular Devices) operating in the kinetic mode in 96-well plates. Human ubiquitin was purchased from Boston Biochem.
DUBs expression in E. coli
The plasmid encoding wild-type human UCH-L3 was subcloned into a pET28 expression vector bearing an N-terminal His-tag. Full-length UCH-L3 was expressed in BL21 E.coli cells and obtained by Ni-NTA column purification. Expression and purification of IsoT, Plpro and OTU-1 were done as described previously[19, 20].
Ubiquitin expression in E. coli
The cDNA for ubiquitin was cloned into pET21b vector using NdeI and HindIII restriction enzymes engineered to contain a C-terminal His-tag. The Gly96Val mutation of ubiquitin was produced by standard PCR using reverse primer carrying specific mutation. The cloning strategy used resulted in generation of C-terminal tail of ubiquitin composed of KLAAALEHHHHHH amino acids. Wild type and mutant ubiquitin were expressed in BL21 (DE3) E. coli strain (Novagen), purified using Ni-NTA agarose and eluted with a 20–200 mM gradient of imidazole in 50 mM Tris, pH 8.0, 100 mM NaCl.
Cleavage of full-length ubiquitin and Ub-P2-Val by DUBs
The enzymatic reaction was performed in a total of 30 µL. The indicated quantities of enzymes were incubated with 10µM substrate for 30 min. at 37°C in 20 mM Tris buffer, pH 7.6, 50 mM NaCl and 5 mM DTT, which is conventionally used for analyzing DUBs in vitro. The reaction was stopped by adding 10 µL of 4x loading buffer and cleavage products were analyzed by Laemmli SDS-PAGE (15% gel) followed by GelCode staining.
Synthesis of the diverse tetrapeptide-ACC PS-SCL
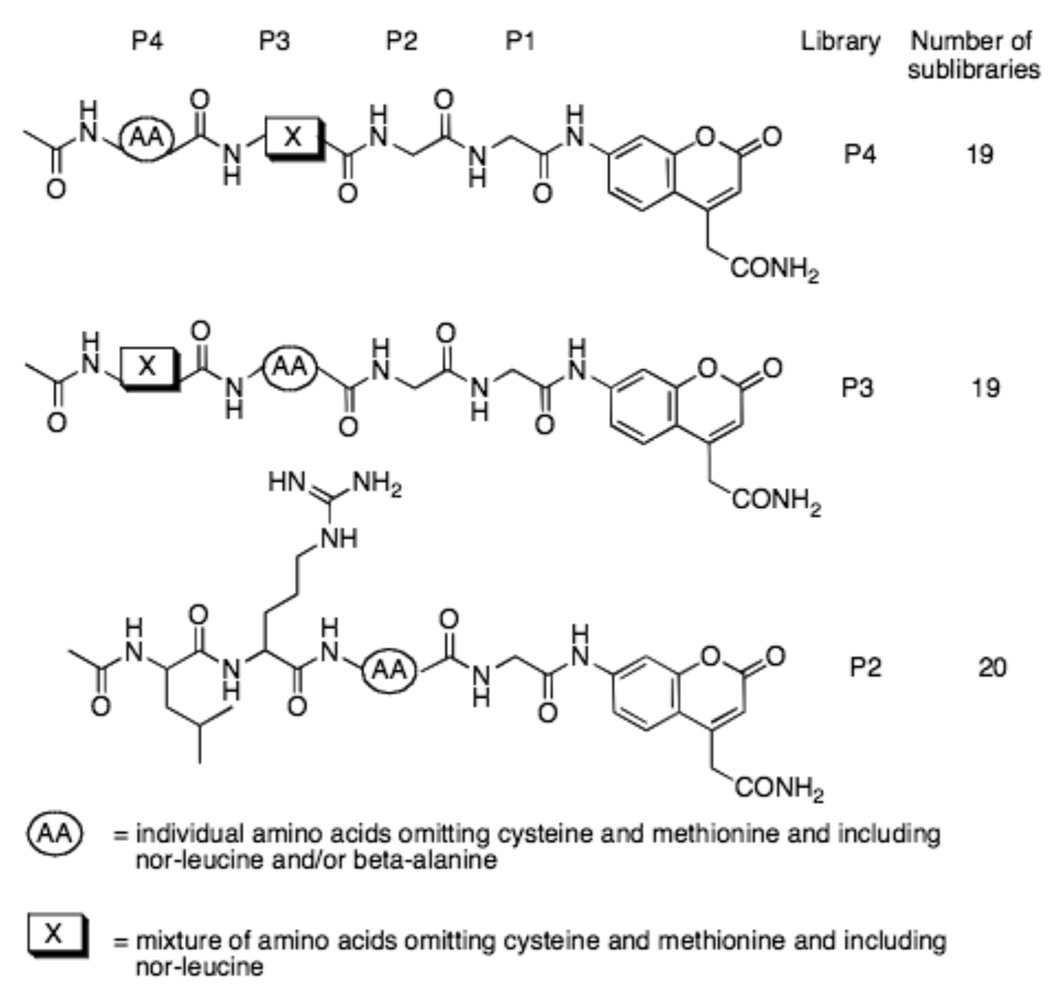
Synthesis of the positional scanning substrate combinatorial library was carried out as previously described (see Fig.l)[21]. After completing synthesis and analysis steps, each sub-library was dissolved in biochemical grade dried DMSO at a concentration of 10 mM and stored at −20°C until use.
Figure 1. Schematic representation of the fluorogenic Positional Scanning Library (PSL).
Each substrate library contains a mixture of 19 amino acids omitting cysteine and methionine and including nor-leucine in all sublibraries, and beta-alanine in the P2 sublibrary. The substrates were dissolved at 10 mM in DMSO and used at a final concentration of 250 or 500 µM (representing 13 or 26 µM per individual substrate sequence).
Synthesis of the fluorogenic substrates
Both solution and solid phase synthesis were carried out according to established methods. Boc-Gly-AFC was synthesized using the method of Alves, and subsequently the Boc group was deprotected using 4M HCl in dioxane[22]. Solid phase synthesis of the final substrate was performed using a Safety Catch resin exactly as described by Backes and Ellman[23]. Ac-Leu-Arg-Gly-Gly-AFCxTFA: 1H NMR (DMSO-d6): 0.83 (m, 6H), 1.40–1.73 (m, 7H), 1.85 (s, 3H), 3.11 (m, 2H), 3.81 (m, 2H), 3.98 (d, 2H, J=6.0 Hz), 4.25–4.28 (m, 2H), 6.94 (s, 1H), 6.82–7.43 (bs, 3H), 7.49 (m, 1H), 7.58 (d, 1H, J=9.5 Hz), 7.72 (d, 1H, J=7.5 Hz), 7.95 (d, 1H, J=1.8Hz), 8.04 (d, 1H, J=7.8Hz), 8.04 (d, 1H, J=7.8Hz), 8.18 (d, 1H, J=6.9Hz), 8.33-8–36 (m, 2H), 10.44 (s, 1H); Ac-Arg-Leu-Arg-Gly-Gly-AFCx2TFA: 1H NMR (DMSO-d6): 0.82 (d, 6H, J=6.6 Hz), 1.40–1.80 (m, 11H), 1.85 (s, 3H), 3.11 (m, 2H), 3.81 (d, 2H, J=5.4 Hz), 3.98 (d, 2H, J=6.0 Hz), 4.25–4.28 (m, 3H), 6.95 (s, 1H), 6.64–7.49 (bs, 6H), 7.52 (m, 1H), 7.58 (d, 1H, J=9.5 Hz), 7.70 (d, 1H, J=7.5 Hz), 7.95 (d, 1H, J=1.6Hz), 8.09 (d, 1H, J=7.8Hz), 8.11 (d, 1H, J=7.8Hz), 8.19 (d, 1H, J=6.3Hz), 8.33-8–36 (m, 2H), 10.47 (s, 1H)
Assay on the PS-SCL
Each DUB was assayed in a 50 mM HEPES, 0.5 mM EDTA, pH 7.5 containing 5mM DTT, a buffer used in previous biochemical characterization of DUBs [16, 17]. The buffer was made at 23°C and assays were performed at 37°C. All the enzymes were preincubated for 10 min at 37°C before adding to the wells containing substrate. Standard enzyme assay conditions for the P3 and P4 libraries were as follows (100 µL reaction), 250 or 500 µM total final substrate mixture concentration (giving around 13 or 26 µM per single substrate), and enzymes were between 2–6 µM. In the case of the P2 library each individual substrate was assayed at the concentration 50 or 100 µM final concentration. Release of fluorophore was monitored continuously with excitation at 355 nm and emission at 460 nm. Total assay time was 60 minutes and the linear portion of the progress curve (generally 15–30 min) was used to calculate velocity. All the experiments were repeated at least twice and the data presented here are the average. The difference between individual values was in every case less than 10%. Analysis of the results was based on total RFU (Relative Fluorescence Unit) for each sub-library setting the highest value to 100% and adjusting the other results accordingly.
Assay of DUBs using Ac-LRGG-AFC
Ac-LRGG-AFC was screened against DUBs at 37°C in the above assay buffer in the presence or absence of full-length ubiquitin at varied ratios, or in the presence of 0 – 1.2 M concentrations of the Hofmeister salt sodium citrate in 25 mM Tris, pH 8.0 containing 5mM DTT. Buffers were prepared at 22°C, and pH was adjusted after the addition of the Hoffmeister salt. Assays were performed at 37°C, at which temperature the pH of the buffer declines to about 7.6. Enzymes were preincubated for 10 min at 37°C before adding substrate to the wells of a 96-well plate reader operating in the kinetic mode. Enzyme assay conditions were as follows: (100 µL reaction), 100 µM final substrate concentration and enzymes at 1 – 4 µM. Release of AFC fluorophore was monitored continuously with excitation at 405 nm and emission at 510 nm. Each experiment was repeated at least twice and the results presented as an average. Final substrate concentrations for of kcat/Km determination ranged from 1 to 100 µM. Concentration of DMSO in the assay was less than 1% (v/v). To determine the catalytic efficiency of enzymes the initial velocities (vi) were measured as a function of [S0], When [S0]<<Km the plot of vi versus [S0] yields a straight line with slope representing Vmax/Km. The kcat/Km ratios were calculated using the expression kcat/Km=slope/E, where E is the total enzyme concentration.
Specificity Cluster Analysis
The results from the PS-SCL library assays for DUBs and SENPs were clustered using CLUSTER software. Each position (P4-P2) was analyzed separately. Maximum activity rates were set at 100%, and amino acids that showed no activity were assigned as 0%. The results were analyzed with the program CLUSTER and were visualized using TreeView software as heat map diagrams showing 100% activity as red and 0% activity as black[24].
Results
Design of the libraries
To determine the substrate sequence requirements in the enzymesubstrate complex around the active center in DUBs, we utilized a combinatorial approach by synthesizing three positional scanning tetrapeptide fluorogenic substrate libraries. As targets for our investigation of specificity we used examples of DUBs from three main groups of cysteine proteases, namely Iso-T (UBPs), UCH-L3 (UCH) and OTU-1 (OTU family). Additionally we extended the screening to SARS viral processing enzyme (SARS-CoV PLpro), which has recently been recognized as a deubiquitinylating enzyme, and which is considered a target in the discovery of antiviral drugs[20, 25]. Previous reports demonstrated that fluorogenic tetra- and pentapeptides based on the C-terminal sequence of ubiquitin are cleaved much less efficiently by the DUBs - IsoT and UCH-L3 than full-length ubiquitin-based substrates[16]. Because catalytic rates were expected to be low, we designed the positional scanning tetrapeptide fluorogenic substrate library so that a small number of individual fluorogenic sequences were in each sub-library, so as to reach the highest possible concentration during the assay. This was achieved by fixing three positions varying an equimolar mixture of 19 amino acids in a fourth position (Fig.1).
To avoid oxidation artifacts we omitted cysteine and replaced methionine by nor-leucine. In the case of the P3 and P4 libraries, positions P1 and P2 of the library were fixed as Gly because this represents the equivalent residues in the natural substrate ubiquitin. The P2 library was designed by fixing P1, P3 and P4 residues as the natural amino acids present in Ub. The amino acids in the P2 library included two unnatural amino acids: nor-leucine and beta-alanine (Fig.1). This approach gave two 19-possibility sub-libraries (each exploring P3 and P4), and a 20-possibility sub-library exploring the P2 position. As the fluorophore leaving group we employed 7-amino-4-carbamoylmethylcoumarin (ACC) because of its convenience in solid phase synthesis[26–28].
Results of library screening
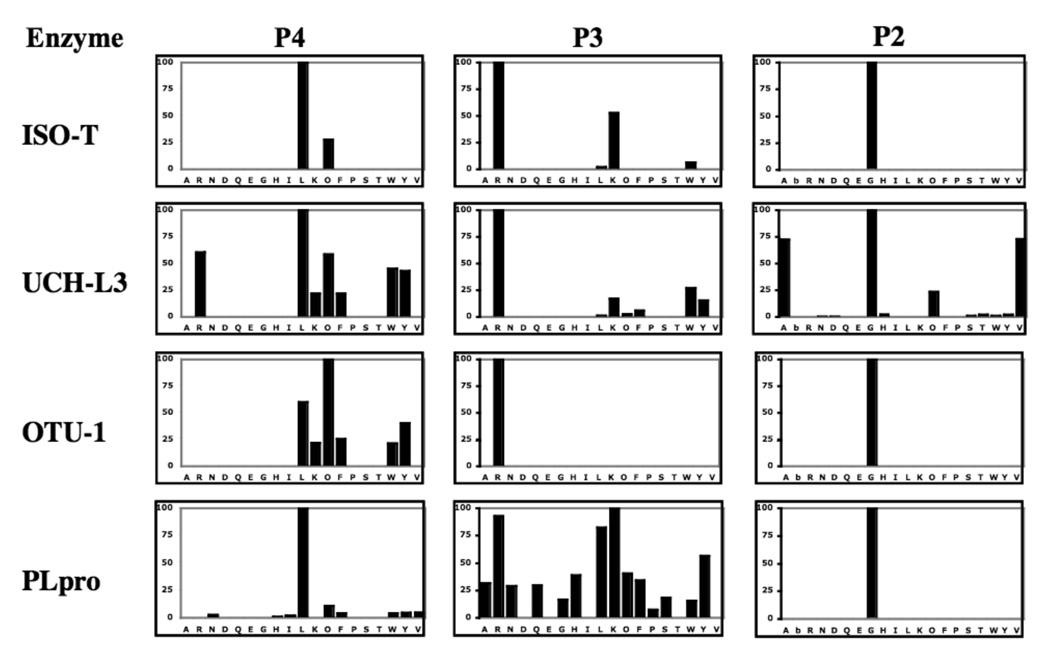
The signatures in the P2, P3 and P4 positions of the DUBs are summarized in Fig. 2, and we present an overview of the key findings for each enzyme.
Figure 2. Subsite preferences of DUBs – IsoT, UCH-L3, OTU-1 and PLpro.
The enzyme concentration was in the range 1–5 µM. ACC production was monitored using an fmax multi-well fluorescence plate reader (Molecular Devices) at excitation wavelength of 355 nm and an emission wavelength of 460 nm. Assay time 15–30 min. The x-axis represents the one letter code of natural amino acids (O - nor-leucine, bA-beta-alanine). The y-axis represents the average relative activity expressed as a percent of the best amino acid.
IsoT
This enzyme is the most “canonical” of the DUBS, showing high preference for the natural Leu-Arg-Gly motif in the P4-P2 positions. Only reactivity with Nle at P4 and Lys at P3 mar the exquisite specificity.
UCH-L3
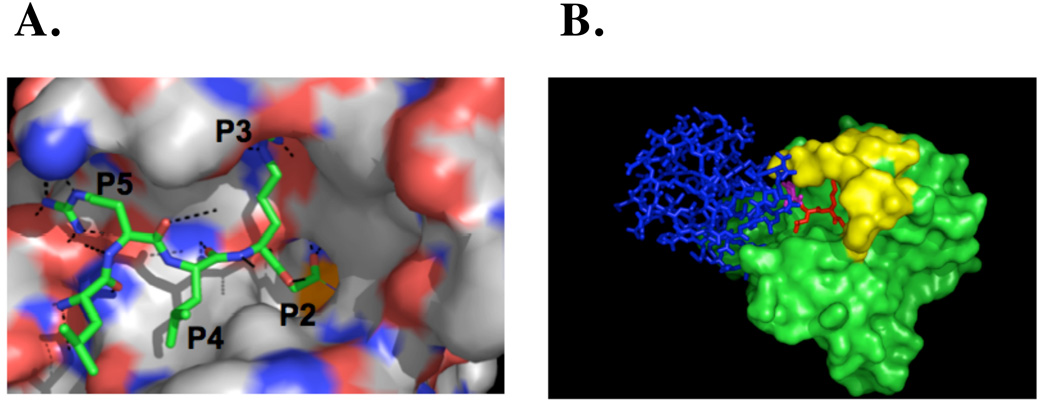
This enzyme shows a preference for Leu at P4, but also a broad tolerance of several other hydrophobic residues at this position. It also shows a surprising selectivity for Ala and Val at P2, in additional to the canonical Gly. Of the enzymes tested here it possesses the least selective, being quite catholic in its tastes at the linear epitope specificity site. The high preference at the P3 position and relatively low specificity in the P4 position can be explained by comparison with the published crystal structure of UCH-L3 bound with ubiquitin inhibitor (Fig.3). The side chain of Arg in the P3 position of ubiquitin is oriented toward the surface of UCH-L3 and is located in a deep pocket, constructed of the acidic residues Asp and Glu. These interact with the amine groups of Arg and explain the high specificity in this position. In contrast to the P3 Leu side chain, the P4 side-chain is oriented away from the surface of UCH-L3 and there is no clearly defined pocket that could be responsible for the tight binding of any amino acids. The restricted tolerance at P4 for hydrophobic residues, revealed in our library screen, is not explained by available crystal structures[29, 30]
Figure 3. (A) UCH-L3 ubiquitin aldehyde inhibitor complex.
Comparison of the binding mode of the ubiquitin inhibitor in the active site of UCH-L3 made on the basis of the available complex structure of these proteins from PDB accession 1XD3. For clarity, only the C-terminal ubiquitin residues are shown. (B) Two separate binding motifs of ubiquitin to DUB. Model showing binding of ubiquitin aldehyde to UCH-L3 using PDB accession 1XD3: blue - the size dominant exosite that binds epitopes on the ubiquitin surface, red - the smaller linear epitope corresponding to the C-terminal tetrapeptide (P2–P4) that binds the cleavage site cleft of the enzymes, purple - Arg at P5. The surface of UCH-L3 is shown in green, with the yellow highlight representing the loop that is disordered in the free enzyme structure (1UCH.PDB). Both views are in the standard orientation for purposes of comparison.
OTU-1
Similarly to UCH-L3, we observe high specificity in P2 and P3 and less specificity in the P4 position. The P4 position prefers Nle over Leu, but other bulky amino acids are also accepted, such as Tyr, Trp, Phe or Lys.
PLpro
This enzyme reveals very high specificity in the P4 position, where practically only Leu can be tolerated, and even higher specificity in the P2 position with only Gly being accepted. However, the P3 position can accommodate a number of different amino acids, with some preference for the hydrophobes Leu and Tyr, and also for the basic side-chains of Lys and Arg – which possess substantial hydrophobic character.
Activation by salts
In all cases, the optimal tetrapeptide sequence LRGG (P4-P1) matches the natural substrate ubiquitin. Kinetic rates on the consensus substrate Ac-LRGG-AFC were low, with kcat/Km values varying from about 2 – 400 M−1s−1 in the HEPES assay buffer (Table 1). These slow catalytic rates imply that additional interactions must take place upon ubiquitin binding to enhance catalysis. According to available crystal structures, most DUBs undergo substantial conformational changes upon binding ubiquitin. Depending on the particular DUB, this effect can result from distinct mechanisms. In one mechanism a ubiquitin molecule binds a distant pocket on the enzyme, resulting in a change that facilitates binding of a second ubiquitin (the one that will be cleaved) into the enzyme catalytic site (trans activation). In a second mechanism the ubiquitin molecule that will be cleaved docks directly with the catalytic site and causes a reorientation of catalytic residues (cis activation)[31]. Fluorogenic substrates are a convenient tool in the investigation of such changes, as demonstrated for the cleavage of full-length Ub-AMC or Z-LRGG-AMC by IsoT[17]. Importantly, by using small peptidyl substrates that occupy only the active site cleft of the enzyme, we can address the role of allosteric changes that affect only the active site of a DUB. This allows us to observe the role of ubiquitin binding, and thence derive the mode of ubiquitin-directed conformational changes that enhance activity in a systematic manner (see below).
Table 1. Comparison of catalytic rates on an optimal tetrapeptide substrate.
Data represent the mean and standard deviation of at least triplicate experiments. Substrate concentration 1–100 µM, final enzyme concentration 0.5–4 µM based on total protein concentration. Note that PLpro is substantially more active than the other DUBs on the peptide substrate.
| kcat/Km (M−1s1) Ac-LRGG-AFC | |||
|---|---|---|---|
| OTU-1 | PLpro | UCH-L3 | IsoT |
| 12.3 ± 0.5 | 408.2 ± 20 | 2.4 ±0.1 | 14.3 ± 3.2 |
In all DUBs tested here we observed an activation of the enzymes with increasing salt concentration, reaching a maximum above 0.8 M. However, even though activation takes place in three out of four cases, the magnitude of this effect is different for each DUB. The most effective activation is observed in the case of UCH-L3, where 1.1 M sodium citrate enhances catalysis on the synthetic substrate Ac-LRGG-AFC by about 24-fold, compared to low salt conditions (Fig 4 – left panels).
Figure 4. Activation of DUBs.
Alterations in activity in the presence of sodium citrate or ubiquitin was assayed using the Ac-LRGG-AFC substrate. Data are expressed as fold increase above the activity in 0.1M sodium citrate (left panels), or fold change in activity in the presence of ubiquitin (right panels). The y axis represents the activity as an average from individual assays. The standard deviation was +/−10%. The enzyme concentration was 0.5–4 µM (based on the total enzyme concentration), the substrate concentration was 50–100µM, and the assay time was 20 min. The sharp decrease in activity of OTU-1 above 0.8 M sodium citrate is due to enzyme precipitation in the assay.
Much lower activation is observed for OTU-1 and PLpro, where maximum activity in sodium citrate gives about a 7-fold increase in catalysis compared to low salt conditions. At very high sodium citrate concentrations we observed a drop in activity, most dramatic for OTU-1, which is due to enzyme precipitation in the assay. The weakest activation was observed in the case of IsoT, where there is almost no increase in catalysis compared to low salt conditions. Interestingly, at low sodium citrate concentrations the potency of the enzyme is even weaker than in the absence of salt suggesting some disorganization of the structure. The salt effect could, in principle, either enhance catalysis by optimizing the conformation of the enzyme or the tetrapeptide substrate.
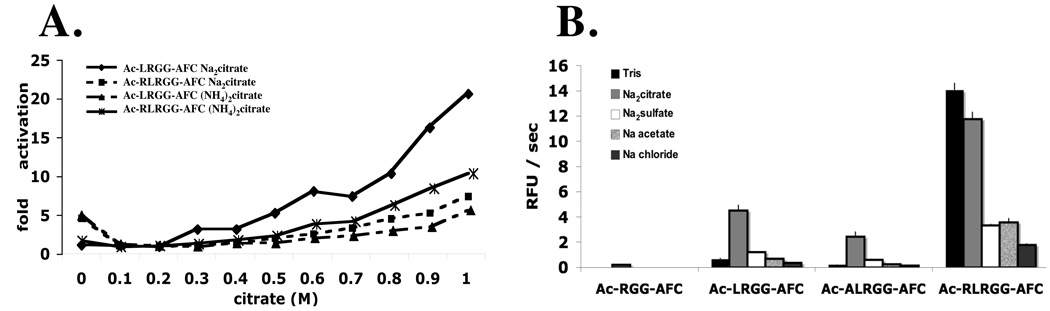
Because the magnitude of catalytic enhancement was different for each DUB tested, it is likely that the effect is enzyme specific, and to test this we analyzed the influence of salts and length of peptide substrate on catalysis by UCH-L3 (Fig 5). Enhancement followed the order citrate>sulfate>acetate>chloride (Fig 5). But the extent of activation was dependent on the length of substrate (Fig 5B). Interestingly, the tetrapeptide substrate Ac-LRGG-AFC and the pentapeptide substrate containing Ala at P5 – Ac-ALRGG-AFC – both demonstrated a salt effect, but the pentapeptide substrate containing Arg in P5 - Ac-RLRGG-AFC - did not demonstrate one (Fig 5B). Since Ac-ALRGG-AFC is more hydrophobic than Ac-RLRGG-AFC there is a possibility that the former substrate is activated for catalysis by a hydrophobic “salting in” onto the enzyme active site – a sort of concentration effect - whereas the latter is not, and that this accounts for the lack of enhanced cleavage of the latter substrate by UCH-L3. However, it is also likely that the Arg at P5 in Ac-RLRGG-AFC negates the salt-dependent activation of UCH-L3, possibly because it is in contact with the mobile loop of the enzyme (Fig 3B) and thus serves a similar function to the salt effect. This is supported by the higher basal activity on the Ac-RLRGG-AFC substrate, which in our hands is now the optimal short peptidyl substrate for DUBs.
Figure 5. Influence of salt concentration and substrate length on catalysis by UCH-L3.
A) Activity of UCH-L3 on Ac-LRGG-AFC and Ac-RLRGG-AFC in different Hofmeister buffers expressed as fold increase in the absence of salt. The data are an average of three experiments, and the range was less than 10%. The enzyme concentration was 4 µM (based on total protein), the substrate concentration was 100–200µM, the assay time was 15–20 min. B) Relationship of UCH-L3 activity to peptide length and sequence in different Hofmeister salts at pH 7.5 (final concentration of the given salt in the buffer was 0.8M). The non-Hofmeister salt NaCl was used as a control to rule out that the effect was due to alterations in ionic strength. Note that extending the substrate with an Arg in P5, but not Ala, negates the enhancement in activity afforded by the Hofmeister buffer. The enzyme concentration was 3 µM (based on the total enzyme concentration), the substrate concentration was 180 µM, and the assay time was 15 min. Error bars are the standard deviation from four independent experiments.
Activation by full-length ubiquitin
In the case of IsoT we observed a 20-fold enhancement of the cleavage of Ac-LRGG-AFC in the presence of equimolar ubiquitin (Fig 4), as shown earlier using full-length Ub-AMC as substrate[31]. This is interpreted as binding of a proximal ubiquitin molecule in the ZnF UBP domain of IsoT, which facilitates the processing of the second ubiquitin molecule in trans. Increasing the ubiquitin concentration resulted in a decrease in the cleavage efficiency, presumably as a result of competition between the second (substrate) ubiquitin molecule and the fluorogenic substrate binding to the enzyme active center (Fig 4). This would be simple substrate competition. In the case of the UCH-L3 we observed inhibition of the fluorogenic substrate by elevated full-length ubiquitin concentrations. This is likely due to the presence of only one ubiquitin binding domain in the UCH-L3, which results in the competition of ubiquitin for Ac-LRGG-AFC at the active center, similar to elevated concentrations of ubiquitin on IsoT. In the case of OTU-1 and PLpro, full-length ubiquitin did not result in any substantial decrease or increase of the processing of the fluorogenic substrate by the ubiquitin concentrations used in this study (Fig 4). Most likely, this is the result of relatively weak binding of ubiquitin. Thus it is possible to distinguish the effect of ubiquitin, which activates DUBs by exosite binding, from the Hofmeister salts, which we suggest activate DUBs by altering the mobility of loops in the active site cleft, as discussed below.
P2 specificity in the context of full-length ubiquitin
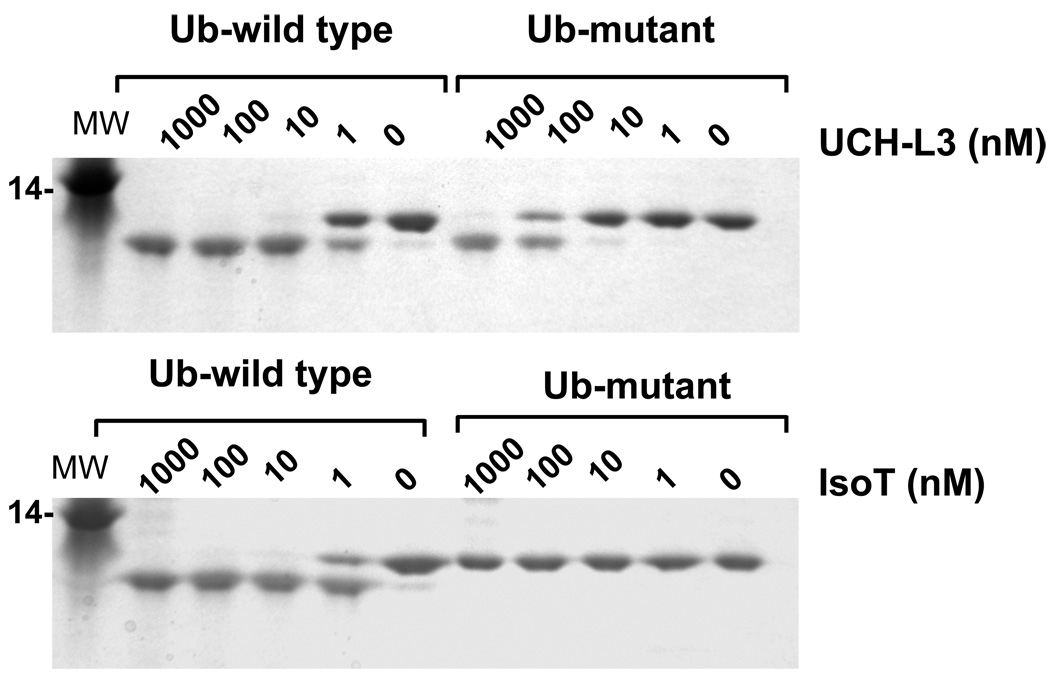
Our screening of P2 position for the UCH-L3 revealed that, besides the canonical P2 Gly, Ala and Val are also tolerated at this position. To validate this unexpected discovery in the context of a more natural substrate we generated a mutant of full-length ubiquitin where Gly in the P2 position was substituted by Val. Both natural and P2-Val ubiquitin were equipped with a His tag at the C-terminus to allow for visualization of cleavage efficiency by SDS/PAGE (Fig. 6). UCH-L3 was able to process both substrates, however the natural ubiquitin variant was cleaved about 20–50 times more efficiently than the P2-Val mutant, whereas in the context of a tetrapeptide there was much less discrimination between Gly and Val at P2 (Fig 2). One possibility for the enhanced discrimination of Gly over Val at P2 in the context of the natural substrate is that ubiquitin binding to UCH-L3 may help to position its P2 residue for optimal catalysis. Further experiments would be required to test this possibility. Control experiments using Iso-T show that this enzyme can tolerate only Gly at the P2 position in the context of the natural substrate (Fig. 6), confirming the PS-SCL library specificity. No processing of the P2-Val mutant of ubiquitin by Iso-T was observed even at very high concentrations of the enzyme. These results confirm that hits from the tetrapeptide library translate to the context of the natural protein substrate, and demonstrate that UCH-L3 possesses much broader tolerance in P2 than other tested DUBs.
Figure 6. Influence of P2 Val in the context of a full-length ubiquitin substrate.
The indicated concentrations of Iso T and UCH-L3 enzymes were incubated with wild type (LRGG) or mutant (LRVG) forms of ubiquitin containing a C-terminal extension. The processing products were analyzed by SDS-PAGE and visualized by staining with GelCode. Note that UCH-L3 is able to process the LRVG mutant, but IsoT is unable to, even at 1 µM enzyme concentration.
Discussion
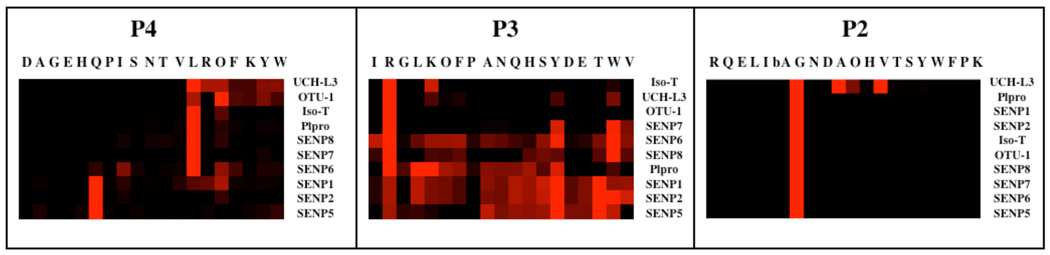
The primary function of DUBs is to remove ubiquitin from a target protein and/or to dismantle polyubiquitin chains. These enzymes reveal high specificity toward ubiquitin, and different enzymes are responsible for different cleavage events. Although some DUBs contain secondary ubiquitin binding sites that influence catalysis in trans, the general feature that all DUBs share is the binding and cleavage of the ubiquitin molecule by the catalytic unit of the enzyme. In this respect it is necessary to recognize two separate binding motifs: (i) the size dominant exosite that binds epitopes on the ubiquitin surface, and (ii) the smaller linear epitope corresponding to the C-terminal tetrapeptide that binds the active site cleft of the enzymes (Fig 3). Seeking to dissect the specificity requirements of the active site cleft we explored positions P2-P4. We did not explore P1 specificity because available crystal structures of DUBs reveal P1 occupies a restricted tunnel at the active center, and that only a Gly residue can be tolerated at this position. Fig.2 reveals that each enzyme has a different tolerance for individual amino acids within the active site cleft, with the viral protease PLpro demonstrating a broad specificity at P3. The high flexibility at P3 in part may explain why this enzyme can process protein substrates other than ubiquitin[33, 34]. UCH-L3 demonstrated unexpected tolerance for residues other than the canonical Gly at P2, compared to the extreme specificity of the other three tested DUBs. Despite these differences, the subsite pockets of all four enzymes are optimal for recognition of the LRGG sequence corresponding to the C-terminus of ubiquitin. Thus we show for the first time that DUBs from four distinct groups have evolved to optimally recognize the conserved linear motif. A comparison of the preferred residues in the P4-P2 subsites between the DUBs tested in this work, and the SENPs tested previously, is shown in Fig 7[21].
Figure 7. Comparison of DUB and SENP specificity.
Individual specificity profiles for P2, P3 and P4 were clustered separately. The results obtained by hierarchical clustering reveal the degree of similarity in each position for SENPs and DUBs. The most preferred positions are displayed in bright red, whereas a complete lack of activity is in black, with intermediate values represented by intermediate shades of red. Natural amino acids are shown in standard one letter mode with unnatural ones shown as O for nor-leucine and bA for beta-alanine.
SENPs are thought to be specific for ubiquitin-like modifiers such as SUMO and Nedd8, and are only very distantly related to DUBs. In terms of their P4 preferences, the DUBs cluster with SENPs 6, 7 and 8, whereas SENPs 1, 2 and 5 form a clearly defined group of their own. This group clustering is maintained at the P3 position, although the diversity of residues accommodated at this position by PLpro make it more like SENPs 6, 7 and 8 than the other DUBs. Most importantly, the cluster analysis demonstrates a preference of the ubiquitin sequence LRG (P4-P2) for all DUBs, and SENPs 6–8, and the SUMO sequence QTG (P4-P2) for SENPs 1, 2 and 5. This makes biological sense for the DUBs, SENPs 1, 2 and 5, as well as SENP8, whose natural target Nedd8 contains the same sequence as ubiquitin within the catalytic cleft. Only SENPs 6 and 7 seem to be outliers in this analysis, unless their primary substrates are not SUMOs, as had earlier been thought [35]. As previously demonstrated, the turnover values for short peptidyl substrates are several orders smaller than for substrates based on full-length ubiquitin[17, 29]. Clearly DUBs have evolved to recognize epitopes on the ubiquitin surface, and this is a major component that enhances catalysis. But it is also apparent that pockets in the active site cleft are important in recognition of ubiquitin, as revealed by the inability of IsoT to cleave a full-length ubiquitin substrate with P2 mutated from Gly to Val, a feat that UCH-L3 can accomplish (Fig.6). Consequently, it appears that a combination of interactions developed at both the ubiquitin binding exosite and the active site cleft are required for optimal activity and stringent recognition of substrates by DUBs.
By using the DUB consensus tetrapeptide substrate Ac-LRGG-AFC we were able to investigate the activity requirements of the four enzymes that represent the different DUB families. Sodium citrate, at high concentrations, activated OTU-1, UCH-L3 and PLpro, and this could be due to an ionic effect or a Hofmeister effect. Certain salts at high concentrations enhance the activity of a broad variety of enzymes, including deSUMOylating enzymes that are somewhat related to DUBs [13], through the Hofmeister effect. Sodium citrate at 0.8M enhanced activity of UCH-L3 by 12-fold versus 0.8M NaCl for Ac-LRGG-AFC and 20-fold for Ac-ALRGG-AFC (Fig. 5B). Since the ionic contribution difference between these concentrations of sodium citrate and NaCl would only be 6-fold, we suggest a Hofmeister effect, although this is not as pronounced as for deSUMOylating enzymes where enhancements at high sodium citrate are above 50-fold [13]. The details are not precisely defined, but Hofmeister salts tend to order flexible components of protein structures [32]. A more extended analysis will be required to fully explain the catalytic enhancement by high concentrations of sodium citrate, but we raise the following hypothesis. All three enzymes above consist essentially of a single catalytic domain, and their activation by sodium citrate is consistent with a loop ordering typical of the Hofmeister effect. Indeed, it is likely the loop that is ordered corresponds to UCH-L3 residues 147–166, which are disordered in the structure of the free enzyme, but ordered and in contact with portions of the substrate in the ubiquitin-bound form of the enzyme (Fig.3A)[29, 30]. This hypothesis is supported by our finding that placing an Arg at P5 of a peptidyl substrate overcomes the salt-dependent enhancement of catalysis. This Arg is in contact with residues 156 and 157 and may help to lock down the mobile loop, thereby enhancing alignment of the peptide substrate. This scenario for the Hofmeister effect explains the data, and is supported by the well documented mobility and re-ordering of the active-site loop in UCH-L3 and closely related DUBs[29, 30]. But of course structural investigations would be required to further test the hypothesis.
In conclusion, we have designed and tested for the first time a combinatorial fluorogenic substrate library to define the specificity of the catalytic cleft of DUBs. Our results show that the four enzymes tested have evolved a preference for binding the ubiquitin C-terminal tail. Catalytic rates are low, and as previously demonstrated for Iso-T, the natural substrate is cleaved several orders more efficiently. Consequently, specificity for ubiquitin has been greatly enhanced by exosite interactions that consist of epitopes on the surface of ubiquitin, tuning the enzymes for their specific function. Different DUBs have tolerance for different residues in their active site clefts. For example, UCH-L3 has a previously unexpected tolerance for Val in P2, which clearly distinguishes it from the other DUBs tested here. This provides proof of concept that small molecules that target the cleft could, in principle, deliver inhibition selectively, and the positional libraries used here could be applied to other DUBS in guiding drug development according to these specificities.
Acknowledgment
We thank Scott Snipas for outstanding technical assistance and Adam Richardson for the help with the Cluster software analysis.
Abbreviations used
- Ac
acetyl
- ACC
7-amino-4-carbamoylmethylcoumarin
- AFC
7-amino-4-trifluoromethylcoumarin
- AMC
7-amino-4-methylcoumarin
- Boc
tert-butoxycarbonyl
- DMF
N,N-dimethyl formamide
- DTT
dithiothreitol
- DUB
deubiquitinating enzyme
- ESI
electrospray ionization
- PS-SCL
positional scanning substrate combinatorial library
- SUMO
small ubiquitin-related modifier
- SENP
Sentrin/SUMO-specific protease
- TFA
trifluoroacetate
References
- 1.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opinion Cell. Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson KD, Ventii KH, Friedrich KL, Mullally JE. The ubiquitin signal: assembly, recognition and termination. Symposium on ubiquitin and signaling. EMBO Reports. 2005;6:815–820. doi: 10.1038/sj.embor.7400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 7.Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikko E, Andre B. Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic. 2007;8:566–581. doi: 10.1111/j.1600-0854.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- 9.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. tNature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 10.Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, Kolli N, Gan-Erdene T, Wilkinson KD, Gill G, Lima CD, Ploegh HL, Ovaa H. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borodovsky A, Ovaa H, Meester WJ, Venanzi ES, Bogyo MS, Hekking BG, Ploegh HL, Kessler BM, Overkleeft HS. Small-molecule inhibitors and probes for ubiquitin-and ubiquitin-like-specific proteases. Chembiochem. 2005;6:287–291. doi: 10.1002/cbic.200400236. [DOI] [PubMed] [Google Scholar]
- 12.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. 10. [DOI] [PubMed] [Google Scholar]
- 13.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 14.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 15.Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 17.Stein RL, Chen Z, Melandri F. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry. 1995;34:12616–12623. doi: 10.1021/bi00039a017. [DOI] [PubMed] [Google Scholar]
- 18.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nature Chem. Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 19.Russell NS, Wilkinson KD. Deubiquitinating enzyme purification, assay inhibitors, and characterization. Meth. Mol. Biol. 2005;301:207–219. doi: 10.1385/1-59259-895-1:207. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z, Salvesen GS. Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem. J. 2008;409:461–469. doi: 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- 22.Alves LC, Almeida PC, Franzoni L, Juliano L, Juliano MA. Synthesis of N alpha-protected aminoacyl 7-amino-4-methyl-coumarin amide by phosphorous oxychloride and preparation of specific fluorogenic substrates for papain. Peptide Res. 1996;9:92–96. [PubMed] [Google Scholar]
- 23.Backes BJ, Ellman JA. An Alkanesulfonamide “Safety-Catch” Linker for Solid- Phase Synthesis. J Org. Chem. 1999;64:2322–2330. [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS, Ellman JA. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino- 4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem. 2002;67:910–915. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- 27.Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 28.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. USA. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 30.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Curr. Opinion Chem. Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Ziebuhr J, Schelle B, Karl N, Minskaia E, Bayer S, Siddell SG, Gorbalenya AE, Thiel V. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007;81:3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUM02/3-conjugated species. J. Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]