SUMMARY
The Ski-interacting protein, SKIP/SNW1, associates with the P-TEFb/CDK9 elongation factor and coactivates inducible genes, including HIV-1. We show here that SKIP also associates with c-Myc and Menin, a subunit of the MLL1 histone methyltransferase (H3K4me3) complex, and that HIV-1 Tat transactivation requires c-Myc and Menin, but not MLL1 or H3K4me3. RNAi-ChIP experiments reveal that SKIP acts downstream of Tat:P-TEFb to recruit c-Myc and its partner TRRAP, a scaffold for histone acetyltransferases, to the HIV-1 promoter. By contrast, SKIP is recruited by the RNF20 H2B ubiquitin ligase to the basal HIV-1 promoter, in a step that is bypassed by Tat and down-regulated by c-Myc. Interestingly, we find that SKIP and P-TEFb are dispensable for UV stress-induced HIV-1 transcription, which is strongly up-regulated by treating cells with the CDK9 inhibitor, flavopiridol. Thus SKIP acts with c-Myc and Menin to promote HIV-1 Tat:P-TEFb transcription at an elongation step that is bypassed under stress.
Keywords: SKIP/SNW1, c-Myc, TRRAP, Menin, MLL1, P-TEFb, HIV-1 Tat
INTRODUCTION
Highly-expressed regions of the genome are enriched for specific histone modifications that are established during transcription by RNA Polymerase II (RNAPII; Li et al., 2007; Shilatifard, 2008). The chromatin modification state of active genes is linked to the C-terminal domain (CTD) of the RNAPII Rpb1 subunit, a unique structure of 52 heptapeptide repeats related to the consensus sequence, Y1S2P3T4S5P6S7 (Egloff and Murphy, 2008). Phosphorylation of the CTD at the Ser5 position is associated with promoter clearance and recruitment of Setd1/MLL-type histone methyltransferase (HMT) complexes, which modify histone H3 at the Lys-4 position (H3K4me3) at promoter-proximal nucleosomes. The Ser5P RNAPII is induced to pause after initiation by the negative elongation factor complex, NELF, and the DRB-sensitivity inducing factor, DSIF/Spt4:Spt5, which are counteracted by the positive-acting transcription elongation factor, P-TEFb (Cyclin T1:CDK9; reviewed by Brès et al., 2008; Core and Lis, 2008; Price, 2008). P-TEFb/CDK9 phosphorylates the RNAPII CTD repeats at the Ser2 position, which triggers the binding of the Setd2 HMT complex, which mediates H3K36 methylation during elongation.
Mammalian cells contain multiple Setd1-type H3K4me3 complexes, which share a set of common subunits (Wdr5, Ash2L, RbBP5, and hDPY-30), but also contain unique components. For example, only Setd1A,B complexes contain Wdr82, whereas MLL3,4 complexes uniquely contain UTX, and MLL1,2 complexes selectively incorporate the tumor suppressor, Menin (Cho et al., 2007; Shilatifard, 2008). H3K4me3 has been linked to transcription initiation (Vermeulen et al., 2007) and assembly of pre-mRNA splicing complexes (Sims et al., 2007). H3K4me3 by Setd1A,B complexes requires mono-ubiquitination of H2B by the RNF20 E3 ubiquitin ligase (Weake and Workman, 2008), which promotes binding of certain Setd1-specific subunits to chromatin (Lee et al, 2007) and directly enhances histone methylation by the Setd1 HMT (Kim et al., 2009). H2Bub also facilitates elongation and nucleosome reassembly by the FACT complex (Fleming et al., 2008; Pavri et al., 2006), and is required for transcription at a subset of cellular genes in vivo (Shema et al., 2008).
In mammalian cells, the order in which factors are recruited to specific promoters varies, depending upon the activator. The HIV-1 Tat protein is a unique activator that directly up-regulates transcription elongation through its ability to bind CycT1 and recruit P-TEFb to the TAR RNA element at the viral promoter (reviewed by Brès et al., 2008). The S. cerevisiae CTD kinase Ctk1, a homologue of P-TEFb, is recruited to yeast genes downstream of the BRE1/hRNF20 H2B ubiquitin ligase, at a step that requires the subsequent removal of the ubiquitin moiety from H2B (Wyce et al., 2007). At mammalian genes, P-TEFb recruitment is often mediated by the bromodomain protein, Brd4 (Jang et al., 2005; Mochizuki et al., 2008; Yang et al., 2007; Yang et al., 2005). At the HIV-1 promoter, the Tat:P-TEFb complex also stimulates pre-mRNA 5′-end capping as well as histone acetylation and H3K4me3 (Zhou et al., 2004), which may reflect the fact that Tat induces P-TEFb to phosphorylate the CTD at both Ser2 and Ser5 positions in vitro (reviewed by Brès et al., 2008).
Beyond its unique mode of recruitment, relatively little is known about the steps downstream of P-TEFb at the Tat-induced HIV-1 promoter. We previously reported that P-TEFb associates with the Ski-interacting protein, SKIP/SNW1 (also termed NCoA62; Prp45 in S. cerevisiae; BX42 in D. melanogaster), which is required for Tat:P-TEFb transcription elongation in vivo and in vitro (Brès et al., 2005). SKIP is an important co-activator of induced nuclear receptor, Notch, and TGFß/SMAD2,3-regulated genes, and functions as a corepressor under basal conditions (Folk et al., 2004; MacDonald et al., 2004). SKIP is also a required pre-mRNA splicing factor, and has been reported to associate with the SNIP1 complex, which controls Cyclin D1 mRNA stability (Bracken et al., 2008). P-TEFb has also been found to interact with the c-Myc oncoprotein, and is required for c-Myc-dependent transcription and transformation (Eberhardy and Farnham, 2002; Gargano et al., 2007; Kanazawa et al., 2003). Ectopic expression of the c-Myc activation domain elevates global Ser2P-RNAPII levels in vivo (Cowling and Cole, 2007), indicating that c-Myc can also stimulate P-TEFb activity. In addition, c-Myc up-regulates H3K4me3 in vivo through its ability to bind and inactivate the JARID1A/PLU-1/LID H3K4me3-specific demethylase (Secombe et al., 2007). The c-Myc protein can function either as a DNA-binding activator or transcription coactivator/corepressor, and its genomewide distribution on chromatin correlates with high levels of promoter histone acetylation and methylation (Guccione et al., 2006; Martinato et al., 2008). As a coactivator, c-Myc up-regulates histone acetylation through direct binding to the transformation-transactivation domain-associated protein, TRRAP, which interacts with and recruits several different histone acetyltransferase complexes to responsive promoters (Murr et al., 2007). Previous studies identified c-Myc as a transcriptional corepressor for latent integrated HIV-1 proviruses, acting together with histone deacetylases at the core promoter (Jiang et al., 2007).
In this study, we examine the role of SKIP in HIV-1 Tat transactivation. Interestingly, SKIP acts downstream of Tat:P-TEFb to recruit both c-Myc and TRRAP to the integrated HIV-1 promoter, and promotes H3K4me3 by the MLL1 HMT complex. We find that both SKIP and c-Myc interact directly with Menin, a subunit of MLL1,2 complexes, and that Tat transactivation requires c-Myc, TRRAP, and Menin, but not MLL1 or Ash2L, and is therefore independent of H3K4me3. Moreover, we find that Tat-induced transcription does not require the RNF20 H2B ubiquitin ligase. By contrast, transcription from the basal HIV-1 promoter depends upon RNF20, which functions upstream of SKIP and other factors, and is down-regulated by c-Myc. We also investigated whether these factors are involved in stress-induced transcriptional up-regulation of the integrated HIV-1 provirus. Unexpectedly, neither SKIP nor P-TEFb are required for HIV-1 transcription induction upon UV stress, and indeed HIV-1 mRNA levels increase synergistically upon exposure of UV-treated cells to flavopiridol, a chemical inhibitor of CDK9. These findings suggest that elongation controls are absent in cells exposed to these agents, and that the HIV-1 core promoter is regulated by a distinct set of factors under stress. Together, these data indicate that Tat bypasses the requirement for RNF20 through its ability to recruit P-TEFb and SKIP, which function together with c-Myc, TRRAP, and Menin in a step that is linked to transcription elongation and bypassed upon cellular stress.
RESULTS
SKIP recruits c-Myc and TRRAP to the Tat-activated HIV-1 promoter
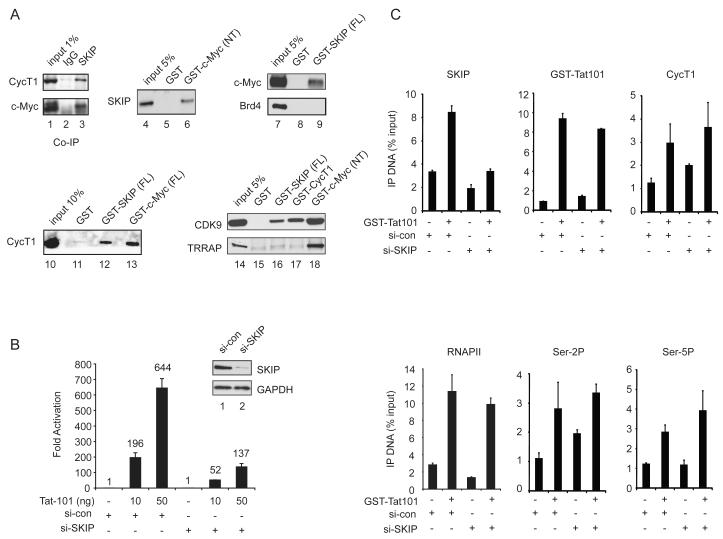
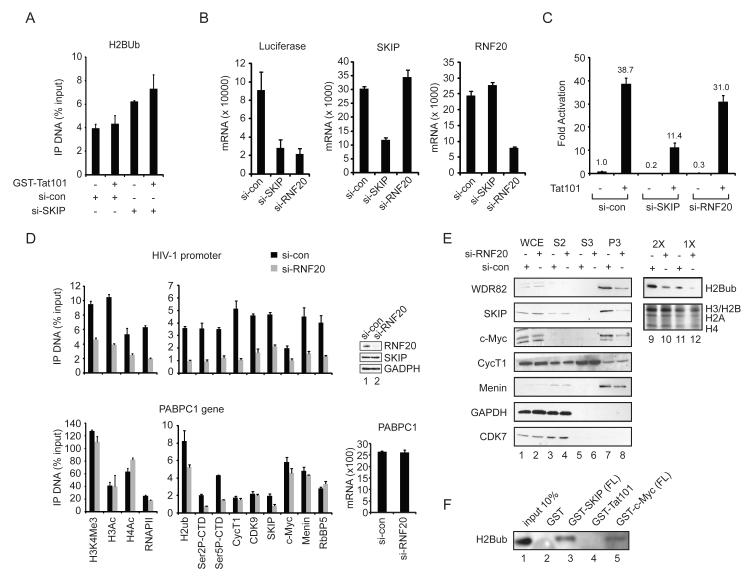
In this study, we used RNAi-ChIP and biochemical protein interaction experiments to assess the role of SKIP in Tat:P-TEFb transactivation at the integrated HIV-1 promoter in HeLa cells. Because c-Myc was shown previously to interact with P-TEFb (Eberhardy and Farnham, 2002; Gargano et al., 2007; Kanazawa et al., 2003), we first asked whether SKIP also associates with c-Myc in nuclear extracts. As shown in Fig. 1A, both CycT1 and c-Myc were detected in SKIP immunoprecipitates from a HeLa nuclear extract using a polyclonal SKIP antibody (lane 3), and were not detected with control anti-IgG immunosera (lane 2). In addition, HeLa nuclear SKIP also bound to the recombinant GST-Myc (NT) activation domain (lane 6) in GST pulldown experiments, and conversely, endogenous c-Myc was recovered through binding to GST-SKIP-coupled beads (lane 9). By contrast, the Brd4 bromodomain protein, which is also known to interact with P-TEFb, did not bind to the GST-SKIP beads (lane 9), indicating that these associations are not mediated via Brd4. Endogenous CycT1 and CDK9 also bound to the GST-SKIP (lanes 12 and 16) and GST-c-Myc beads (lanes 13 and 18). Although the c-Myc partner protein, TRRAP, bound avidly to the GST-c-Myc (NT) activation domain (lane 18, bottom panel), it did not interact efficiently with GST-SKIP (lane 16, bottom panel), indicating that SKIP does not recognize c-Myc indirectly, through TRRAP. As expected, the P-TEFb CDK9 subunit bound avidly to GST-CycT1(aa1-303) beads (lane 17), and none of these factors recognized the control GST protein-coupled beads (lanes 5, 8, 11, 15).
Figure 1.
SKIP associates with c-Myc and acts downstream of Tat:P-TEFb at the HIV-1 promoter.
(A) Co-immunoprecipitation of HeLa SKIP with CycT1 and c-Myc, as shown by immunoblot (lanes 1-3). GST pulldown fractions from HeLa nuclear extract were assessed by immunoblot, using antisera indicated to the left of each panel (lanes 4-18). (B) Analysis of Tat activity in HeLa cells treated with SKIP-siRNA or a control (si-con) siRNA. Inset shows immunoblot analysis of knockdown efficiency. (C) ChIP analysis of the integrated HIV-1 LTR:Luc reporter gene promoter in the presence (+) or absence (−) of transduced recombinant GST-Tat101 protein. Where indicated, the cells were transfected with si-SKIP or si-con RNAs prior to analysis by ChIP. All graphs represent mean and standard error obtained from three independent experiments.
The role of SKIP in HIV-1 Tat transactivation was assessed using a stable HeLa cell line that carries a single integrated HIV-1:Luciferase reporter gene (Tréand et al., 2006). In these experiments, HIV-1 Tat (101aa) was introduced to the cells via transient transfection or chloroquine-mediated protein transduction. Depletion of SKIP by siRNA transfection reduced HIV-1 luciferase activity by 3.7- and 4.7-fold at 10 ng and 50 ng of Tat, respectively, relative to cells treated with a control siRNA (Fig. 1B). Chromatin immunoprecipitation (ChIP) experiments revealed an increase in SKIP levels at the Tat-activated HIV-1 promoter in cells treated with the si-control, but not si-SKIP, RNAs (Fig. 1C). Interestingly, knockdown of SKIP had no effect on the recruitment of Tat, CycT1, RNAPII, or Ser2P and Ser5P levels at the HIV-1 promoter (Fig. 1C). Thus, SKIP functions downstream of Tat:P-TEFb recruitment and RNAPII phosphorylation.
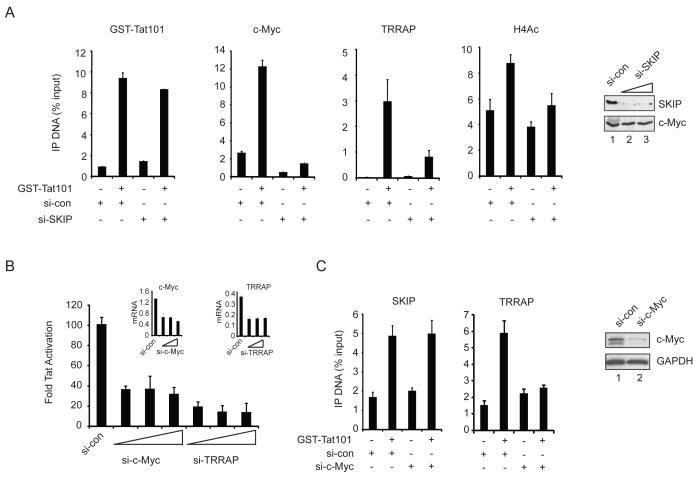
We next examined the binding of c-Myc and TRRAP to the HIV-1 promoter by ChIP. Although c-Myc is a known repressor of HIV-1 transcription (Jiang et al., 2007), we found that its occupancy increased at the HIV-1 promoter in the presence of Tat (Fig. 2A). While depletion of SKIP did not affect binding of GST-Tat to the HIV-1 LTR, we noted that recruitment of c-Myc was strongly reduced. Immunoblot experiments established that global c-Myc protein levels were unaffected in cells transfected with the SKIP-siRNA (lanes 1-3), indicating that c-Myc is present, but not recruited to the viral LTR. Similarly, we found that TRRAP occupancy increased at HIV-1 LTR upon Tat transactivation in normal, but not SKIP knockdown, cells (Fig. 2A). The Tat-dependent increase in histone H4 acetylation also required SKIP (Fig. 2A). Importantly, Tat transactivation was strongly reduced in c-Myc- and TRRAP knockdown cells (Fig. 2B). Quantitative RT-PCR analysis established that the c-Myc and TRRAP siRNAs selectively depleted their mRNA targets in these cells (Fig. 2B, inset). Moreover, HIV-1 Tat transactivation was strongly enhanced by ectopic expression of either c-Myc or TRRAP in the HeLa LTR:Luc cells (Suppl. Fig. 1). Additional analysis by RNAi-ChIP revealed that knockdown of c-Myc reduces binding of TRRAP, but not SKIP, to the HIV-1 promoter (Fig. 2C). Thus, c-Myc functions downstream of SKIP to recruit TRRAP, and both proteins are important coactivators for Tat in vivo.
Figure 2.
SKIP recruits c-Myc:TRRAP to the Tat-activated HIV-1 promoter.
(A) ChIP analysis of the basal and Tat-induced HIV-1 promoter in control or SKIP knockdown HeLa cells. Inset, immunoblot analysis of knockdown efficiency. (B) HeLa HIV-1 LTR:Luc (luciferase) activity induced with 5 ng of transfected Tat101 plasmid in si-control, si-c-Myc, or si-TRRAP transfected cells. Inset, analysis of knockdown efficiency by RT-PCR. (C) RNAi-ChIP analysis of the indicated proteins at the HIV-1 promoter in control or c-Myc knockdown cells. Inset, assessment of knockdown efficiency by immunoblot. All graphs represent mean and standard error obtained from three independent experiments.
SKIP is also required for H3K4me3 at the Tat-induced HIV-1 promoter
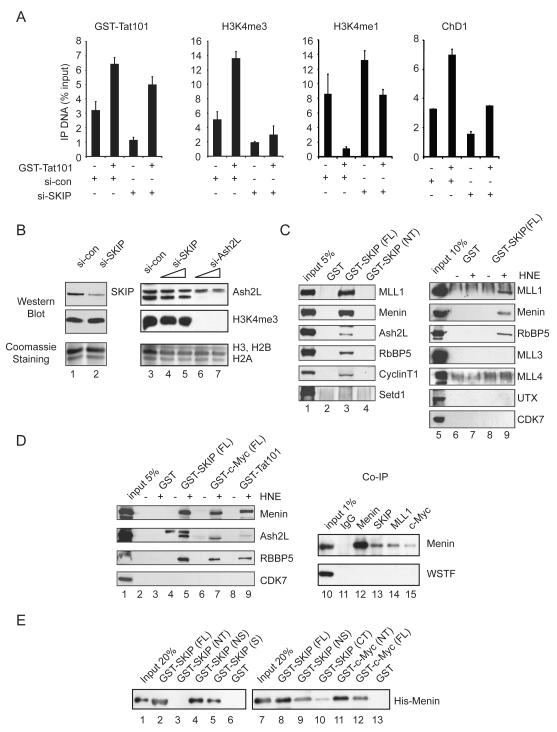
Binding of c-Myc to active genes often correlates with promoter H3K4me3, which is induced at the Tat-activated HIV-1 promoter (Zhou et al., 2004). Interestingly, both basal and Tat-induced H3K4me3 levels at the HIV-1 promoter declined significantly in the SKIP knockdown cells by ChIP (Fig 3A, second panel). The induction of H3K4me3 by Tat was accompanied by a drop in the level of monomethylated H3K4 (H3K4me1), which was not observed in the SKIP-depleted cells. In addition, we observed lower levels of promoter-bound ChD1, a chromatin remodeling protein that recognizes H3K4me3, in cells treated with the SKIP-siRNA as compared to the control siRNA (si-con; Fig. 3A). Thus SKIP is required for H3K4me3 at the basal and Tat-induced HIV-1 promoter in these cells.
Figure 3.
SKIP is required for gene-specific H3K4me3, and both SKIP and c-Myc interact with nuclear MLL1:Menin complexes and recombinant Menin in vitro.
(A) RNAi-ChIP analysis of the basal and Tat-activated HIV-1 promoter in control or SKIP knockdown cells. All graphs represent mean and standard error obtained from three independent experiments. (B) Immunoblot analysis of global H3K4me3 levels in SKIP or Ash2L knockdown HeLa cells. Total acid-extracted histones were visualized by Coomassie staining. (C) Immunoblot analysis of the binding of MLL complex subunits (input, lanes 1, 5) to GST (lane 2, 6 and 7), GST-SKIP (full-length, FL; lane 3, 8 and 9) or GST-SKIP (N-terminal fragment, NT; lane 4) beads in pull-down experiments from a HeLa nuclear extract. (D) Binding of nuclear MLL1 complex factors (input, lane 1) to GST (lanes 2,3), GST-SKIP (FL; lanes 4,5), GST-c-Myc (FL; lanes 6,7) or GST-Tat101 (lanes 8,9) beads in the presence (+) or absence (−) or HeLa nuclear extract (HNE), as visualized by immunoblot. Analysis of the association of endogenous HeLa SKIP, MLL1, or c-Myc with Menin by co-immunoprecipitation with anti-Menin antisera, as visualized by immunoblot (lanes 10-15). (E) Analysis of the binding of wild-type or truncated GST-SKIP and GST-c-Myc proteins to purified recombinant baculovirus-expressed His-tagged Menin was assessed by immunoblot (lanes 1-13) with an anti-Menin antibody.
Immunoblot analysis of total acid-extracted histones revealed that global levels of H3K4me3 are unaffected by depletion of SKIP (Fig. 3B, lanes 4,5), but were strongly reduced upon depletion of Ash2L, a conserved and critical Setd1/MLL complex subunit (lanes 6,7). Thus, SKIP promotes gene-specific, but not global, histone H3K4 methylation. In mammalian cells global H3K4me3 is mediated predominantly by the Setd1 HMTs (Wu et al., 2008), suggesting that SKIP may function selectively with the gene-specific MLL-type HMTs.
SKIP and c-Myc interact with Menin and the MLL1 complex
To assess whether SKIP associates with human Setd1/MLL complexes, GST pull-down experiments were carried out in HeLa nuclear extracts using the full-length or truncated GST-SKIP proteins. Interestingly, the endogenous MLL1 and Menin proteins bound avidly to GST-SKIP(FL), but did not interact with the SKIP N-terminal domain, GST-SKIP(NT), or with GST alone (Fig. 3C, lane 3). GST-SKIP pulldown fractions also contained low levels of Ash2L and RbBP5, but lacked Setd1 (lane 3), MLL3, or UTX, which is found in MLL3,4 complexes, and only nonspecific binding was observed for MLL4. In a separate experiment, Menin efficiently bound to GST-SKIP (Fig. 3D, lane 5), GST-c-Myc (lane 7), and GST-Tat101 (lane 9) beads. Endogenous SKIP, MLL1, and c-Myc proteins were also detected in immunoprecipitates of anti-Menin (Fig. 3D, lanes 13-15), but not control IgG- (lane 11) or anti-WSTF (lanes 10-15) antisera. In addition, we found that affinity-purified baculovirus-expressed His-Menin protein bound directly to GST-SKIP(FL) beads (Fig. 3E, lanes 2 and 8), as well as to GST-SKIP(NS) (lanes 4 and 9) and GST-SKIP(S) (lane 5) SNW domain-containing proteins in vitro. By contrast, His-Menin did not bind to GST alone (lanes 6, 13), or to GST-SKIP (NT) (lane 3) or GST-SKIP(CT) (lane 10), indicating that Menin interacts directly with the SKIP SNW domain. His-Menin also bound to full-length GST-c-Myc(FL) (Fig. 3E, lane 11) and to the GST-c-Myc(NT) activation domain (lane 12). We conclude that SKIP associates selectively with MLL1 complexes, at least in part through direct binding to Menin.
HIV-1 Tat transactivation requires Menin, but not MLL1 or Ash2L
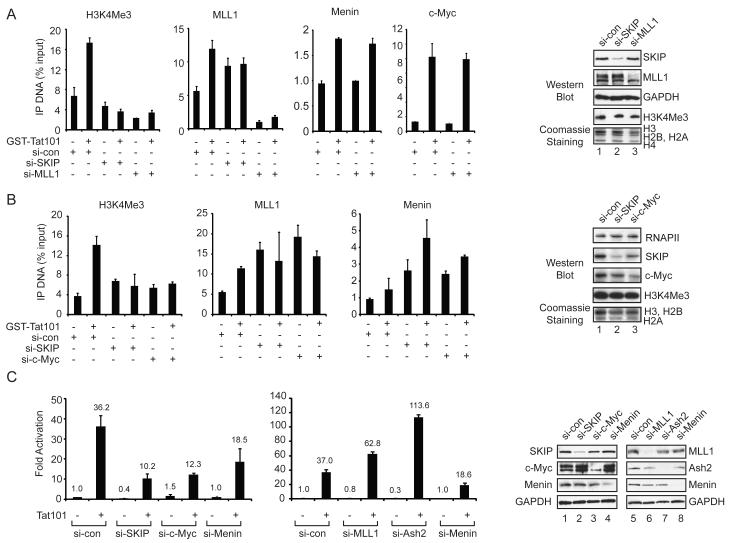
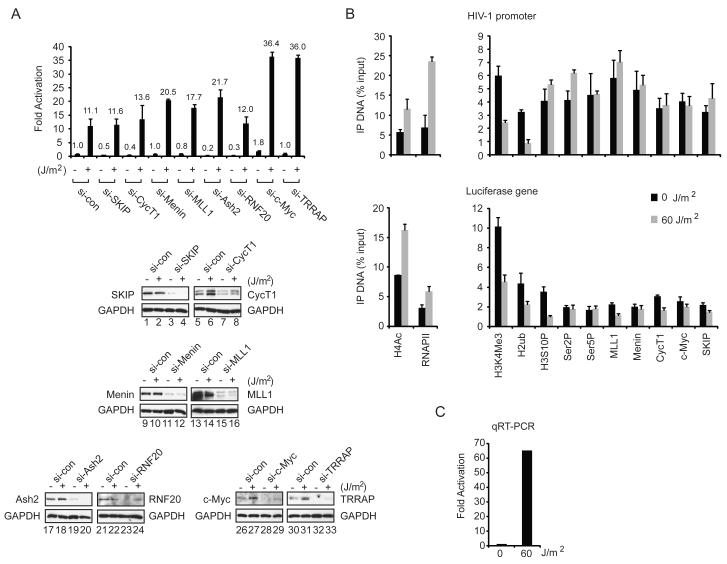
We next used RNAi-ChIP experiments to assess whether MLL1 is responsible for H3K4me3 at the Tat-activated HIV-1 promoter. As shown in Fig. 4A, both basal and Tat-induced H3K4me3 levels were strongly reduced in either MLL1-siRNA or SKIP-siRNA treated cells, as compared with cells transfected with the control siRNA. ChIP experiments confirmed the loss of MLL1 protein at the HIV-1 promoter and the knockdown of MLL1 was confirmed by immunoblot (Fig. 4A, lane 3). Significantly, knockdown of MLL1 did not reduce binding of Menin or c-Myc to the Tat-activated HIV-1 promoter. Further analysis showed that knockdown of either c-Myc or SKIP was sufficient to reduce Tat-induced H3K4me3 (Fig. 4B), despite the successful recruitment MLL1, Menin, RbBP5, and Ash2L to the HIV-1 promoter (Fig. 4B and Suppl. Fig. 2). Indeed, for reasons that are not clear, the binding of these MLL1 complex proteins to the basal HIV-1 promoter was enhanced in either SKIP or c-Myc knockdown cells (Fig. 4A, B). Consistent with a role for SKIP and c-Myc in gene-specific methylation through MLL1 complexes, global Setd1-dependent H3K4me3 was unaffected in HeLa cells depleted of SKIP or c-Myc (Fig. 4A,B, lanes 1-3).
Figure 4.
SKIP, c-Myc and Menin are required for Tat transactivation, but act independently of MLL1-mediated H3K4me3.
(A) ChIP analysis of the integrated HIV-1 LTR:Luc promoter in control, SKIP- or MLL1-knockdown cells, as indicated below each panel. Right panel, immunoblot analysis of total cell extracts to assess knockdown efficiency (lanes 1-3). Global H3K4me3 levels were assessed by immunoblot and Coomassie staining from cells transfected with each si-RNA. (B) ChIP analysis at the HeLa HIV-1 LTR:Luc promoter in cells treated with control, SKIP- or c-Myc-specific siRNAs, as indicated below each panel. Right panel, immunoblot analysis of knockdown efficiency (lanes 1-3). (C) Luciferase analysis of the HIV-1 Tat transactivation efficiency in cells transfected either with 2 ng of an empty or a Tat expression vector and treated with control or factor-specific siRNAs, as indicated below each panel. Right panel, immunoblot analysis of total cell extracts to assess the efficiency of knockdown for each factor (lanes 1-8). All graphs represent mean and standard error from three independent experiments.
Importantly, Tat transactivation of the HIV-1 LTR:Luc reporter gene was significantly impaired in cells depleted of SKIP, c-Myc or Menin (Fig. 4C and Suppl. Fig. 3). By contrast, knockdown of MLL1 or Ash2L elevated Tat transactivation (Fig. 4C), indicating that these proteins interfere with Tat activity in vivo. Interestingly, we noted that in these conditions, global c-Myc protein levels increased in the SKIP or Menin knockdown cells (Fig. 4C, lanes 1-4), although c-Myc occupancy at the HIV-1 promoter declined in SKIP-depleted cells, and was unaffected in Menin-depleted cells (Fig. 4A). In the absence of Tat, basal transcription was down-regulated by c-Myc, but required SKIP and Ash2L. Thus Tat transactivation requires Menin, but not MLL1 or Ash2L, and is therefore is independent of H3K4me3.
SKIP functions downstream of RNF20 at the basal HIV-1 promoter
These data indicate that SKIP binds to Menin and recruits c-Myc:TRRAP to the HIV-1 promoter, stimulating Tat transactivation and H3K4 methylation. We next asked whether SKIP also affects histone H2B ubiquitylation. ChIP experiments revealed no change in H2Bub levels upon Tat transactivation. Indeed, H2Bub levels were slightly increased in SKIP-depleted cells. Nevertheless, basal HIV-1 transcription was significantly impaired by knockdown of either SKIP or the H2B ubiquitin ligase, RNF20, and RT-PCR experiments confirmed that silencing of SKIP did not affect expression of RNF20, and visa-versa (Fig. 5B). By contrast, HIV-1 Tat:P-TEFb transactivation was only modestly affected in RNF20 knockdown cells (Fig. 5C), suggesting that Tat bypasses the need for RNF20 and H2Bub. ChIP analysis of the basal HIV-1 promoter confirmed that H2Bub levels dropped sharply in RNF20 knockdown cells (Fig. 5D). Interestingly, the silencing of RNF20 reduced the occupancy of RNAPII, P-TEFb, SKIP and the other factors we examined, indicating that RNF20 acts at a very early step in basal HIV-1 transcription initiation. By contrast, transcription of a P-TEFb-independent housekeeping gene, PABPC1, was unaffected by knockdown of RNF20 (Fig. 5D). RNAi-ChIP experiments revealed a modest decline in H2Bub at the PABPC1 gene in RNF20-depleted cells, and lower levels of RNAPII Ser2P, Ser5P, and SKIP (Fig. 5D), with no loss of RNAPII or other factors. We conclude that RNF20 regulates an early step at the basal HIV-1 promoter, which is effectively bypassed by Tat.
Figure 5.
RNF20 is required for basal HIV-1 transcription and to load transcription factors to the LTR:luc promoter.
(A) ChIP analysis of H2Bub levels at the HeLa HIV-1 LTR:Luc promoter in si-con and si-SKIP transfected cells. (B) qRT-PCR analysis of basal HIV-1 LTR:Luc mRNA levels, normalized to beta-actin mRNA, in si-control, si-SKIP, or si-RNF20 silenced cells (left panel). The efficiency of SKIP or RNF20 knockdown was assessed by qRT-PCR analysis of the endogenous genes in the middle and right panels, respectively. (C) Analysis of basal and Tat-activated transcription from the integrated HIV-1 LTR:Luc reporter gene in HeLa cells transfected with control, SKIP-, or RNF20-specific siRNAs. (D) ChIP analysis of the integrated HIV-1 promoter (top panel) and PABPC1 gene (bottom panel) in si-con (black) and si-RNF20 (gray) transfected HeLa cells using the factor-specific antisera listed below the bottom panel. Top right panel shows the efficiency of the RNF20 knockdown by immunoblot (lanes 1, 2). Bottom right panel, qRT-PCR analysis of PABPC1 mRNA levels, normalized to beta-actin mRNA, in control and RNF20 knockdown cells. All graphs represent mean and standard error obtained from three independent experiments. (E) Immunoblot analysis of the binding of indicated HeLa proteins to chromatin fractions derived from control or RNF20 knockdown cells (lanes 1-8). Fractions tested were: WCE, whole cell extract; S2, cytoplasmic fraction; S3, soluble nuclear fraction; and P3, chromatin-associated protein fraction. Global H2Bub levels in total acid-extracted histones were assessed by immunoblot and Coomassie staining (lanes 9-12). (F) Immunoblot analysis of H2Bub levels in GST-SKIP (FL), Gst-Tat101, or GST-c-Myc (FL) protein pulldown fractions from a HeLa core histone preparation (lanes 1-5), using an anti-H2Bub antibody.
Because RNF20 regulates SKIP occupancy at these genes, without affecting SKIP protein stability (Fig. 5D), we asked whether it also facilitates binding of SKIP to cellular chromatin. For these experiments, HeLa whole cell extracts were fractionated into cytoplasmic, soluble nuclear, and nuclear pellet (chromatin) fractions and probed by immunoblot. In extracts from cells treated with a control siRNA, the endogenous Wdr82, SKIP, c-Myc and Menin proteins were highly enriched in the chromatin fraction (Fig 5E, lanes 7, 8), whereas CDK7 and GAPDH were recovered predominantly in the cytoplasmic fraction (lanes 3, 4) and the P-TEFb CycT1 subunit was widely dispersed, with a minor component in the chromatin (P3) fraction (lanes 5, 6). Interestingly, knockdown of RNF20 reduced the amount of Wdr82, SKIP and c-Myc in the chromatin fraction, whereas CycT1 and Menin were unaffected. Immunoblot analysis of GST-SKIP and GST-c-Myc pulldown fractions revealed the presence of H2Bub (Fig. 5E), indicating that these factors may associate with complexes on H2Bub-modified chromatin (Fig. 5F). Thus SKIP regulates transcription downstream of RNF20 at the basal HIV-1 promoter, and binds to cellular chromatin in an H2Bub-sensitive manner.
P-TEFb, SKIP and c-Myc are dispensable for UV stress-induced HIV-1 transcription
UV and other forms of genotoxic stress strongly induce HIV-1 transcription in HeLa and Jurkat cells (Valerie et al., 1988). This increase in proviral transcription correlates with enhanced P-TEFb activity in UV-treated cells that accompanies the release of active P-TEFb from an inhibitory complex with 7SKRNA (Zhou and Yik, 2006). Consequently, we asked whether SKIP, c-Myc and Menin are also needed for HIV-1 LTR:Luc transcription in UV-treated cells. As shown in Fig. 6A, basal HIV-1 transcription increased 11-fold in UV-treated cells. Remarkably, RNAi-mediated knockdown of SKIP, CycT1, Menin, MLL1, Ash2L or RNF20 either had no effect on transcription or modestly elevated the activity of the HIV-1 luciferase reporter gene in vivo (Fig. 6A). Moreover, HIV-1 LTR:Luc reporter gene expression increased 3-4-fold in c-Myc and TRRAP knockdown cells, indicating that the c-Myc:TRRAP complex is repressive to HIV-1 transcription under UV stress. The selective knockdown of each factor was confirmed by immunoblot, which also revealed that CycT1, and to a lesser extent, c-Myc and TRRAP, protein levels increase in UV-treated HeLa cells (Fig. 6A, lanes 1-33). ChIP analysis of the HIV-1 promoter and luciferase reporter gene coding region revealed that H3K4me3, H2Bub, and H3S10P levels decline sharply upon induction of transcription by UV, and that transcription proceeds without an increase in Ser2P or Ser5P (Fig. 6B). By contrast, RNAPII occupancy increased at the HIV-1 promoter and coding region in UV-treated cells, concomitant with an increase in histone H4 acetylation. The strong induction of HIV-1 transcription was confirmed by RT-PCR (Fig. 6C). ChIP analysis of the PABPC1 housekeeping gene in these cells revealed no effect on H3K4me3 levels, although a drop was observed for H2Bub and H3S10P (Suppl. Fig. 4). We conclude that SKIP co-operates with c-Myc and TRRAP to promote transcription downstream of Tat:P-TEFb, in a step that is bypassed in UV-stressed cells.
Figure 6.
UV-induced basal HIV-1 LTR:Luc transcription does not require SKIP, CycT1 (P-TEFb), Menin, or RNF20, and is repressed by c-Myc and TRRAP.
(A) Analysis of basal and UV-induced HIV-1 LTR:Luc transcription in cells treated with control or various factor-specific siRNAs, as indicated. Bottom panels, immunoblot analysis of knockdown efficiency in this experiment (lanes 1 to 33). (B) ChIP analysis of the HIV-1 LTR:Luc promoter (top panel) or coding region (bottom panel) in control (black) or UV-induced (gray) cells. (C) qRT-PCR analysis of HIV-1 luciferase mRNA levels in control or UV-treated cells, normalized to beta-actin mRNA, for this experiment. All graphs represent mean and standard error obtained from three independent experiments.
The P-TEFb inhibitor flavopiridol synergistically increases HIV-1 mRNA levels in UV-induced cells
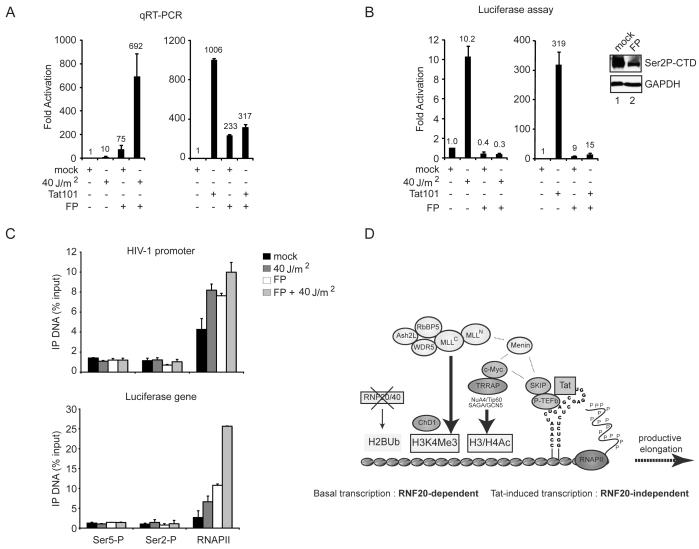
These observations predicted that UV-induced HIV-1 transcription should be resistant to the P-TEFb inhibitor, flavopiridol (FP). Indeed, previous studies have shown that mRNA elongation at some cellular genes is only transiently blocked in cells treated with FP (Ni et al., 2008). As shown in Fig. 7A, basal HIV-1 transcription was increased approximately 10-fold in UV-stressed cells, and even more strongly (75-fold) in FP-treated cells. Most interestingly, we found that the addition of FP synergistically up-regulated HIV-1 transcription in UV-treated cells (Fig. 7A). The net 692-fold increase in HIV-1 mRNA levels is comparable to that observed upon Tat transactivation in unstressed cells (Fig. 7A). As expected, Tat transactivation was strongly inhibited in FP-treated cells, both in transient transfections as well as in cells transduced with recombinant GST-Tat protein, and FP reduced global Ser2P in these cells (Fig. 7B lanes 1,2). Thus FP further increases basal HIV-1 transcription in UV-treated cells, opposite to its effects on Tat:P-TEFb-regulated transcription. In addition UV- and Tat-induced HIV-1 LTR:Luc reporter gene activity, as measured in luciferase assays, was potently blocked by FP in these cells (Fig. 7B), and control experiments further established that FP does not interfere with luciferase activity in vitro (Suppl. Fig. 5). These results indicate that P-TEFb remains critical for luciferase gene expression in UV-treated cells, perhaps reflecting its requirement for mRNA capping, export, or translation (Ni et al. 2004, 2008; Hargreaves et al. 2009). ChIP analyses revealed that total RNAPII levels increase at the HIV-1 promoter and coding region upon induction by UV and FP, without a corresponding increase in Ser2P or Ser5P (Fig. 7C). Thus transcription induction in UV and FP-induced cells does not depend upon SKIP, P-TEFb or RNAPII phosphorylation, indicating that the events that pause transcription and confer a requirement for P-TEFb may be lost in stressed cells.
Figure 7.
The CDK9 inhibitor, flavopiridol, strongly increases basal HIV-1 mRNA levels in UV-treated cells, but inhibits Tat-induced HIV-1 transcription.
(A) qRT-PCR analysis of HIV-1 LTR:Luc (luciferase) mRNA levels, normalized to beta-actin mRNA, in control cells or cells exposed to UV, Tat101 (20 ng), or flavopiridol (FP – 500 nM). (B) Analysis of HeLa HIV-1 LTR:Luc (luciferase) activity in the presence of UV, Tat101 (20 ng), or FP (500 nM), as indicated. Inset, immunoblot analysis of the effect of FP on RNAPII Ser2P levels. (C) ChIP analysis of the HIV-1 LTR:Luc promoter and coding region in control, UV-induced and FP (300 nM)-treated cells. All graphs represent mean and standard error obtained from three independent experiments. (D) Model diagram depicting the role of SKIP, c-Myc and Menin in HIV-1 Tat transactivation and induction of MLL1-mediated H3K4me3 at the integrated HIV-1 promoter. See text for details.
DISCUSSION
SKIP is a unique protein that can activate or repress transcription of induced genes, depending upon the cellular context, and also functions in splicing through mechanisms that are not well understood. We previously showed that SKIP associates with the active P-TEFb complex and is required for Tat transactivation in vivo and in vitro (Brès et al., 2005). Here we examine the role of SKIP in basal and Tat transactivation at the integrated HIV-1 promoter in HeLa cells. Our findings highlight a role for SKIP in recruiting the c-Myc:TRRAP complex to the viral promoter, which stimulates H3K4me3 by the MLL1 HMT complex. In vitro, SKIP and c-Myc interact directly with the MLL1 subunit Menin, and all three factors are required for Tat transactivation in vivo. However, Tat transactivation does not depend upon MLL1, Ash2L or H3K4me3. Interestingly, Tat:P-TEFb activity is also independent of histone H2B ubiquitination through RNF20. By contrast, the basal HIV-1 promoter requires RNF20, which promotes the loading of SKIP, RNAPII and other factors, and is down-regulated by c-Myc. Our studies also indicate that a different strategy is responsible for UV stress-induced HIV-1 transcription, which is accompanied by increased histone acetylation and a loss of H2B ubiquitination and H3K4me3. Surprisingly, P-TEFb and SKIP are no longer required for elongation in UV-treated cells, and transcription increases synergistically upon addition of the CDK9 inhibitor, flavopiridol. Thus the mechanisms that confer a requirement for P-TEFb and SKIP are lost under conditions of stress.
A role for SKIP and c-Myc:TRRAP in Tat transactivation
These data suggest a model in which SKIP is recruited to the Tat:P-TEFb complex upon binding to TAR RNA at the paused RNAPII complex at the HIV-1 promoter (Fig. 7D). Although P-TEFb interacts strongly with c-Myc (Eberhardy and Farnham, 2002; Kanazawa et al., 2003), it is unable to recruit c-Myc to the viral promoter without SKIP. In turn, c-Myc directly recruits TRRAP, a component of SAGA/GCN5- and NuA4/Tip60-type histone acetyltransferases (Murr et al., 2007), and we find that both c-Myc and TRRAP are required for Tat transactivation in HeLa cells. Thus SKIP can regulate Tat transactivation and histone acetylation through recruitment of the c-Myc:TRRAP complex. Because TRRAP/GCN5 complexes cooperate with other promoter-bound factors to promote phosphoacetylation of histone H3, which is a preferred substrate for H3K4 methylation (Meyer et al., 2008), these findings could explain how SKIP and c-Myc:TRRAP promote H3K4me3 (Fig. 7D).
However the underlying mechanism is likely to be more complicated, because we also find that SKIP and c-Myc selectively associate with MLL1, and not Setd1, complexes in nuclear extracts, and promote gene-specific H3K4me3 by MLL1 without affecting Setd1-dependent global H3K4me3 (Wu et al., 2008). This specificity can be attributed in part to direct binding of SKIP and c-Myc to the Menin tumor suppressor, which is a dedicated subunit of MLL1,2 complexes, and helps to recruit MLL1 to cellular genes (Milne et al., 2005). Although we find that SKIP and c-Myc do not regulate the binding of Menin and the MLL1 HMT subunits to the HIV-1 promoter, these factors may stimulate MLL1 HMT activity on chromatin. Indeed, previous studies have shown that Drosophila and mammalian c-Myc proteins can regulate H3K4me3 levels through inactivation of the H3K4me3-specific histone demethylase, Jarid1A/LID/PLU-1 (Secombe et al., 2007). This mechanism might also be operative at the HIV-1 promoter, and could help stabilize de novo H3K4 methylation at induced promoters.
The observation that Menin, but not MLL1 or Ash2L, is required for Tat activity in vivo, indicates that H3K4me3 is dispensable for transcription elongation, and that Menin can influence transcription independently of the MLL1 complex. Consistent with a possible role in transcription elongation, Menin localizes to both the promoter and coding regions of target genes (Milne et al., 2005). Although H3K4me3 is not required for Tat transactivation, it has also been shown to promote spliceosome complex assembly (Sims et al., 2007), and therefore may play a role in SKIP-dependent splicing events, and SKIP may transfer to the spliceosome at this step. Given the critical role for Menin in transformation by translocated MLL fusion proteins in acute leukemias, and as a tumor suppressor in endocrine tissues (Yang and Hua, 2007), it will also be interesting to assess whether SKIP contributes to Menin or c-Myc-dependent cancer promotion or suppression pathways.
Mechanistic differences between basal and Tat-activated transcription
SKIP is required for H3K4me3 but not H2Bub, and functions downstream of RNF20 at the basal, but not Tat-activated, promoter. RNF20 appears to regulate HIV-1 transcription initiation at an early step, and has been shown to function as a gene-specific coactivator and corepressor in HeLa cells (Shema et al., 2008). We find that SKIP associates with bulk chromatin in an RNF20-dependent manner, and therefore it may function with P-TEFb downstream of H2B ubiquitination at cellular genes. By contrast, SKIP, c-Myc and associated factors are recruited to the HIV-1 promoter through the Tat:P-TEFb complex, and RNF20 is no longer required.
Our findings confirm earlier reports that basal HIV-1 transcription is down-regulated by c-Myc (Jiang et al., 2007), but surprisingly show that c-Myc and TRRAP are required coactivators for Tat. One reason for this discrepancy may be the ability of c-Myc to ‘squelch’ transcription when expressed ectopically at high levels, although we find that c-Myc and TRRAP stimulate Tat transactivation when expressed at low levels (Suppl. Fig. 1). Interestingly, repression of the HIV-1 promoter in latently-infected T cells was recently shown to be regulated by the CBF-1 DNA-binding protein and the CIR-1 corepressor (Tyagi and Karn, 2007), both of which have been shown to interact directly with SKIP to repress Notch target genes. Thus, in addition to its role in Tat transactivation, SKIP may interact with CBF-1:CIR-1 to repress the latent HIV-1 provirus in resting T cells. Thus both SKIP and c-Myc may serve dual roles as corepressors and coactivators of HIV-1 transcription in vivo.
UV stress-induced HIV-1 transcription is independent of P-TEFb and SKIP
Global levels of RNAPII Ser2P are increased in cells exposed to genotoxic and UV stress, in conjunction with a release of active P-TEFb from an inhibitory complex that forms with 7SKRNA (Zhou and Yik, 2006). The increase in P-TEFb activity correlates with enhanced transcription from the HIV-1 promoter, yet surprisingly we find that P-TEFb and SKIP are dispensable for viral transcription in UV-treated cells. Moreover, HIV-1 mRNA levels in UV-stressed cells increase dramatically in cells treated with the P-TEFb inhibitor, flavopiridol (FP), indicating that the two agents affect viral transcription by different mechanisms. Interestingly, UV-induced HIV-1 transcription is accompanied by a drop in H3K4me3, H2Bub, and H3S10P levels at the HIV-1 promoter, whereas levels of acetylated histone H4 increase. Transcription proceeds without an increase in either Ser2P or Ser5P RNAPII in the coding region, although total RNAPII levels increase both at the promoter and transcribed region. Thus the mechanism of UV stress-induced HIV-1 transcription differs fundamentally from Tat transactivation, which is P-TEFb-dependent and inhibited by FP. Consequently, it will be important to learn which cellular factors drive viral transcription under conditions of DNA damage, and how the HIV-1 core promoter responds differently to stress induced by UV and FP.
Our findings raise the possibility that the negative controls on HIV-1 transcription elongation mediated by factors such as NELF and DSIF, which are normally counteracted by P-TEFb, may be inactivated in UV- or FP-treated cells. Consistent with this possibility, the Spt5 DSIF subunit, which functions both in transcriptional pausing and elongation, was previously found to be absent from the HIV-1 promoter in FP-treated cells (Zhou et al., 2004). Similarly, p53-dependent activation of the p21 gene in cells treated with the CDK inhibitor, DRB, was also found to be independent of P-TEFb (Gomes et al., 2006). Although HIV-1 transcription is up-regulated by UV and FP, our data show that expression of the HIV-1 LTR:Luc reporter gene is nevertheless potently blocked by FP in both resting and UV-treated cells. Consequently other steps in gene expression that lie downstream of transcription elongation, likely including the binding of pre-mRNA splicing, polyadenylation and export complexes to the RNAPII Ser2P CTD, remain dependent upon P-TEFb even under stress. Taken together, these findings strongly suggest that SKIP functions in concert with P-TEFb to overcome constraints to transcription elongation that are effectively bypassed in cells exposed to stress.
EXPERIMENTAL PROCEDURES
Plasmids, recombinant proteins and antibodies
pTat101, pRL-TK, pGEX-Tat101, pGEX-HA-Tat86, pGEX-SKIP (FL) and derived truncation mutants, pGEX-CycT1 (1-303) and pGEX-c-Myc (NT) were described previously (Brès et al., 2005). pGEX-c-Myc (FL) was generated by subcloning c-Myc (FL) cDNA into XbaI and XhoI sites of pGEX-KG (Pharmacia). Recombinant His-Menin protein was affinity-purified from baculovirus-infected Sf9 cell extracts using Ni-NTA superflow column (Qiagen). Sources for antisera are listed in Supplemental Methods.
Cell culture, Tat protein transduction, UV-induction, and siRNAs
HeLa HIV-1 LTR:Luc cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum. Transfection of pTat101 was carried out using Effectene (Qiagen), and Tat protein transduction was as described (Brès et al., 2005). UV-induction was carried out with a UV Stratalinker 2400 (Stratagene), and cells were incubated for a further 18 hr prior to harvesting. Where indicated, flavopiridol was added 1 to 4 hr prior to induction, and was maintained during the UV treatment. Luciferase and Renilla luciferase activities were assayed 24 to 48 hr after transfection, according to the manufacturer’s protocol (Promega). siRNAs were transfected using either RNAi MAX or Lipofectamine2000 (Invitrogen). The procedures used for total acid extraction of HeLa histones and small-scale chromatin fractionation are listed, along with the siRNA sequences used in this study, in Supplemental Methods.
Quantitative RT-PCR and Chromatin Immunoprecipitation analyses
Total RNAs were isolated using Trizol, and subjected to DNAseI treatment prior to reverse transcription using random primers and SuperScript II reverse transcriptase (Invitrogen). For ChIP experiments, HeLa LTR:Luc cells were cultured with 2 ug/ml of recombinant Tat protein for 4 hr as described in Supplemental Methods. Both RT and ChIP samples were analyzed by MX3005P q-PCR machine (Stratagene) using SYBR master mix (Applied Biosystems). PCR primer sequences are listed in the Supplemental Methods.
Protein GST pull-down interaction assays, direct binding and co-immunoprecipitation experiments
GST pull-down experiments were carried out as described previously (Brès et al., 2005), and direct binding and co-immunoprecipitation procedures are described in the Supplemental Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank RBIO-J lab members for helpful suggestions throughout this study, and Jean-Claude Farré (UCSD) for help with recombinant protein purification. We are also grateful to Drs. Robert N. Eisenman (University of Washington) for the c-Myc FL cDNA, Peggy Farnham (U.C. Davis) for the c-myc NT cDNA, Naganari Ohkura (NCCRI, Japan) for the Menin cDNA, Jose-Ramon Suarez (Aventis Pharmaceuticals inc.) for flavopiridol, David Skalnik (University of Indiana) for the WDR82 antibody, Keiko Ozato (NIH) for the Brd4 antibody, Kai Ge (NIH) for MLL3 and MLL4 antisera, and Robert Perry (Fox Chase Cancer Centre, PA) for the ChD1 antibody. This work was supported by NIH grant AI044615, and V.B was supported by The Auen Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Folk P, Puta F, Skruzny M. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell. Mol. Life Sci. 2004;61:629–640. doi: 10.1007/s00018-003-3215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano B, Amente S, Majello B, Lania L. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle. 2007;6:2031–2037. doi: 10.4161/cc.6.16.4554. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Hargreaves D, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 2007;81:10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- MacDonald PN, Dowd DR, Zhang C, Gu C. Emerging insights into the coactivator role of NCoA62/SKIP in Vitamin D-mediated transcription. J. Steroid Biochem. Mol. Biol. 2004;89-90:179–186. doi: 10.1016/j.jsbmb.2004.03.097. [DOI] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, Taatjes DJ. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. USA. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréand C, du Chéné I, Brès V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Delers A, Bruck C, Thiriart C, Rosenberg H, Debouck C, Rosenberg M. Activation of human immunodeficiency virus type 1 by DNA damage in human cells. Nature. 1988;333:78–81. doi: 10.1038/333078a0. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hua X. In search of tumor suppressing functions of menin. Mol. Cell. Endocrinol. 2007;265-266:34–41. doi: 10.1016/j.mce.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell Biol. 2007;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J. Virol. 2004;78:13522–13533. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.