Abstract
Expression quantitative trait loci (eQTL) mapping is a powerful tool for identifying genetic regulatory variation. However, at present, most eQTLs in humans were identified using gene expression data from cell lines, and it remains unknown whether these eQTLs also have a regulatory function in other expression contexts, such as human primary tissues. Here we investigate this question using a targeted strategy. Specifically, we selected a subset of large-effect eQTLs identified in the HapMap lymphoblastoid cell lines, and examined the association of these eQTLs with gene expression levels across individuals in five human primary tissues (heart, kidney, liver, lung and testes). We show that genotypes at the eQTLs we selected are often predictive of variation in gene expression levels in one or more of the five primary tissues. The genotype effects in the primary tissues are consistently in the same direction as the effects inferred in the cell lines. Additionally, a number of the eQTLs we tested are found in more than one of the tissues. Our results indicate that functional studies in cell lines may uncover a substantial amount of genetic variation that affects gene expression levels in human primary tissues.
INTRODUCTION
Expression quantitative trait loci (eQTL) mapping studies in humans are widely used to identify genetic variation that affects gene regulation—specifically, transcript levels (reviewed in 1). The long-term goal of these studies is to elucidate how functional regulatory variation at the DNA level underlies morphological or physiological variation by using expression levels as intermediate molecular phenotypes.
To date, many of the surveys of putatively functional regulatory variation in humans have been conducted in lymphoblastoid cell lines (LCLs), rather than primary tissues, mostly by using the HapMap cell lines (2–6). Cell lines offer convenience and replicability, and the HapMap cell lines, in particular, represent the most complete catalog of human variation (7), a resource that will become even more useful as the genomes of many of these cell lines are sequenced as part of the 1000 Genomes project (8).
Thus, cell lines are expected to continue to be integral to improving our understanding of human regulatory variation. However, cell lines often carry chromosomal abnormalities (9), altered methylation patterns (10), may have pronounced batch effects related to preparation and/or growth rates (11,12) and the Epstein–Barr virus (EBV) transformation itself can alter the expression levels of a subset of genes (13). Furthermore, the modular nature of gene regulation may allow the genetic architecture of gene expression to vary substantially over tissues (14,15). For these reasons, work with cell lines is often criticized as being potentially uninformative regarding regulatory interactions in primary tissues. Yet, functional assays in LCLs may often be the only feasible approach (e.g. 16) to further study the potential function of large number of non-coding loci identified in recent genetic association studies of complex human diseases (17–24).
Indeed, collections of large numbers of primary tissues are not very common. To date, only a few eQTL studies in humans have been carried out in primary tissues (including studies in liver, blood, adipose tissue and brain (25–28)). These studies, however, provided little information regarding the overlap of eQTLs found in LCLs and primary tissues. For one, differences in study designs, platforms, sampling schemes, and the specific definitions of different classes of eQTLs (such as how one defines a significant eQTL, or how one defines a cis eQTL) made it difficult to interpret comparisons of eQTL mapping results across studies (1). Perhaps due to these reasons, only two studies of eQTLs in primary tissues have thus far compared eQTLs across pairs of tissues (25,28), and only one study (27) has compared their findings with eQTL results from the HapMap cell lines, and found only minimal overlap. We therefore set out to test, using a dedicated study design, whether SNPs identified as eQTLs in HapMap LCLs also contribute to functional regulatory variation in human primary tissues.
When we designed the study, we considered the two primary strategies that can be used to confirm cis regulatory variation (29,30): (i) identifying differences in expression levels between alleles of the same gene (using allele-specific expression measurements (31,32)), or (ii) identifying associations between SNP genotypes and the total expression (i.e. the overall expression across both alleles) of the gene (2–6,25–28). The allele-specific approach is attractive because it allows one to easily control for environmental and trans effects within individuals. However, this approach requires one to develop allele-specific assays, and is limited to individuals heterozygous at a SNP within the transcript. Moreover, the assayed SNP must be in linkage disequilibrium with the causal regulatory variant (often outside the transcript). In contrast, the eQTL mapping approach allows one to use all available samples (thus increasing power) and does not require one to identify additional SNPs within the expressed transcripts of interest. The eQTL mapping approach mitigates trans and environmental effects by averaging the genotypic affect across individuals.
We thus followed Veyrieras et al. (6) and others and chose to design an eQTL association study. As we were limited by the small numbers of samples per primary tissue (10–23 per tissue), we chose to focus on a subset of previously identified large-effect HapMap cis-eQTLs (6) for which we estimate to have reasonable power to detect an effect on gene expression levels in the primary tissues, given the genotypes of our samples (see Materials and Methods for details). We thus selected 21 cis-eQTLs for investigation, and examined whether these eQTLs are also associated with variation in gene expression in one or more of the primary tissues.
RESULTS
In order to test whether eQTLs found in cell lines are also functional in other contexts, we asked whether strong candidate cis-eQTLs found in the CEU (European-ancestry from Utah, USA; Centre d'Etude du Polymorphisme Humain collection) HapMap cell lines can also explain inter-individual variation in gene expression data from panels of multiple human primary tissues (13 hearts, 23 kidneys, 18 livers, 20 lungs and 10 testes; in total comprising 84 tissues sampled from 63 individuals identified as Caucasian). As the number of primary tissue samples available to us is small, we focused on a subset of eQTLs for which we have reasonable power to detect a regulatory effect in the primary tissues. We chose this subset of eQTLs using the following approach (see Fig. 1 for an illustration of our SNP selection procedure): We first focused on cis-eQTLs that were previously identified using microarray expression data from HapMap LCLs (6,33), ignoring trans-acting variants as their effect sizes tend to be small (26). We then excluded SNPs with minor allele frequencies ≤0.2 in the CEU population (as our primary tissues samples were collected from Caucasians), and excluded all but the one largest-effect eQTL for each gene (in the original analysis more than one significant cis-eQTL per gene may have been identified). We further excluded eQTLs that were not detected in all the HapMap populations in which the SNP was typed, as eQTLs identified in multiple populations are more likely to be true positives, as well as less likely to result from an artifact particular to a single preparation of cell lines. Finally, we ranked the remaining eQTLs by their effect sizes, and arbitrarily chose to focus on those eQTLs for which the genotype explains ≥19% of the variation in expression in the CEU population. This series of exclusion steps resulted in a list of 206 eQTLs.
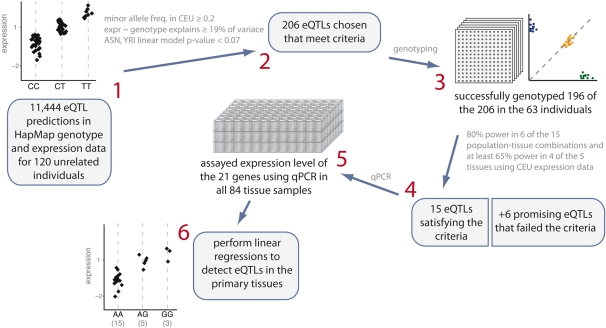
Figure 1.
Illustration of candidate eQTL selection strategy. (1) In selecting SNPs to test for an association with expression in primary human tissues, we began with a set of predictions based on a Bayesian analysis of HapMap genotypes and expression data. (2) We narrowed these 11 444 eQTL predictions down to 206 based on several criteria, as detailed in the Materials and Methods section. (3) We successfully genotyped 196 of the 206 in the 63 individuals from which we have tissue samples. (4) For each eQTL in each of the five tissues, we conducted simulations to estimate power conditional on the actual genotypes of our samples, using expression data from one of the three HapMap populations. On the basis of these simulations, we chose 15 eQTLs that we estimated to have 80% power in at least six of the 15 population-tissue combinations and at least 65% power in four of the five tissues using data from CEU. To these 15, we added six more eQTLs that fell slightly below our cutoffs, but looked promising. (5) We then assayed expression level of the 21 genes using qPCR in all 84 samples. (6) We used linear regressions to determine significance of each of the eQTLs in each of the five human primary tissues.
The next step was to examine whether the specific genotypes in our tissue collection would allow us to test the function of the 206 eQTLs. To determine that, we designed multiplex Sequenom Mass-Spec genotyping assays for 196 of the 206 SNPs (we were not able to include 10 SNPs in the multiplex design), and genotyped these 196 SNPs in all our samples. We then performed resampling simulations to assess the power for detection of each of the 196 eQTLs. Specifically, we sampled gene expression levels from the HapMap microarray data from a collection of individuals with genotypes that match the genotypes of our actual primary tissues and asked what is the proportion of such replicate simulations (for each tissue-eQTL combination) in which we detect the eQTL. In other words, we estimated the power to detect the HapMap eQTL if the only available data was from HapMap cell lines with the same genotypes as those we have in our primary tissues.
Using the results of the power simulations we selected 21 cis-eQTLs, which we expected to be able to detect in at least one or more of the primary tissues (see Fig. 1 and Supplementary Material, Table S1). We then proceeded to measure the expression levels of each of the 21 cis genes in all tissues using quantitative RT-PCR (see Materials and Methods). For a subset of these genes, we also confirmed the original inference from the microarray data using newly extracted RNA from 9 HapMap cell lines (see Supplementary Material, Fig. S1).
To test for an association between the genotypes of each putative eQTL (i.e. a particular SNP) and the expression level of the cis gene, we used an additive linear model to analyze the quantitative RT-PCR data from each tissue. We note that the association between genotype and expression level may not be additive (e.g. 34). However, in our case, we focus on eQTLs that showed strong additivity in the HapMap cell line expression data, and thus simple additive linear models may be expected to capture most of the association between genotypes and expression levels in these cases.
Given a prior expectation of the direction of the genotype effect based on the HapMap cell-line expression data, we chose to increase power by constructing the test to be one-sided in the direction of the predicted genotype effect. Therefore, we tested for an association of each putative eQTL with gene expression data from all five tissues, for a total of 105 tests. Using this additive association approach, we found that 19 of the 105 tests were significant (at a one-sided P-value < 0.05) and, as we assumed, results were highly similar when we included both additive and dominance terms in the linear model (Supplementary Material, Table S2). With our false positive rate set to 0.05, we have an estimated positive false discovery rate, or pFDR (35), of 14%, suggesting that about 16 of the eQTLs are expected to be true positives (and we note that based on permutation tests, our P-values appear well calibrated; see Materials and Methods).
We detected eQTLs in at least one primary tissue for 11 of the genes (52%, P-value = 0.003, binomial test, see Materials and Methods), while five genes had significant eQTLs in more than one primary tissue (see Fig. 2 and Supplementary Material, Fig. S4 and Fig. 3A for a specific example, in which we detect the eQTL associated with USMG5 in four of five tissues). If we relax the stringency of our test to a one-sided P-value < 0.1 (which may be a reasonable approach given that all these loci were previously detected as eQTLs in the LCLs), 67% of the eQTLs are present in at least one tissue. Moreover, 70 of the 105 tests were in the expected direction based on the HapMap gene expression data (significant by one-sided sign test; P = 0.0004, null expectation is 52.5). Thus, our results indicate that putative eQTLs found in cell lines can often be detected in one or more primary tissues.
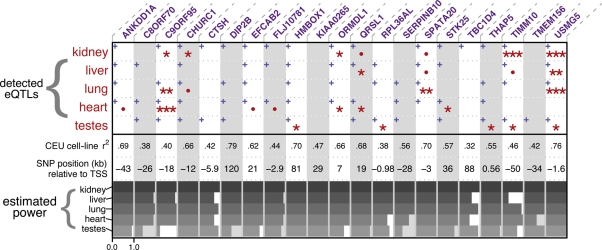
Figure 2.
Summary of detected eQTLs and our predicted power. Columns correspond to the 21 genes we tested for eQTLs. One-sided P-values from linear models testing the eQTL are summarized by either one (0.05 > P > 0.01), two (0.01 > P > 0.001), or three (0.001 > p) red asterisks; marginally significant tests (0.1 > P > 0.05) are marked with a red dot (see Supplementary Material, Table S1 for all P-values). Blue plus signs indicate the effect is in the same direction as the effect observed in the LCLs. CEU cell-line r2 is the proportion of variance explained by genotype in a linear model fit to the original CEU cell-line data. Estimated power is illustrated with horizontal gray bars for each eQTL-tissue combination using microarray expression data sampled from CEU HapMap LCLs with matching genotypes (see Materials and Methods), with the axis (ranging from 0 to 1) shown below only for the first column.
DISCUSSION
Overall, our results show that a large number of eQTLs originally identified in the HapMap LCLs can also be detected in primary tissues, suggesting that studies in cell lines are helpful for identifying functional regulatory variation. However, it should be noted that, perhaps not surprisingly, there are a number of assays for which we expected to have good power to detect eQTLs, but did not detect one in any of the tissues that we tested (i.e. Fig. 3B). For example, under the (admittedly) extreme scenario that all 21 cis-eQTLs are truly affecting gene regulation in all five tissues with the same effect size as seen in the HapMap LCLs, then given our power calculations, we would expect 96/105 significant tests (at one-sided P ≤ 0.05). One possible explanation for the apparent discrepancy between this expectation and our observations is that eQTLs observed in the LCLs have a systematically smaller effect sizes in the primary tissues. Indeed, we estimated that a two-third reduction in effect size (from those observed in HapMap) is consistent with our observed number of significant tests (see Materials and Methods). A number of factors may underlie such lower effect sizes. First, the effect sizes can be over-estimated when the original study is underpowered, due to the so-called Winner's Curse (e.g. 36), leading to smaller observed effect sizes when considering an entirely different sample of individuals. Second, the true effect size of an eQTL in a different tissue may tend to be systematically smaller because we selected the very strongest eQTLs in the first tissue, a biological (rather than statistical) winner's curse.
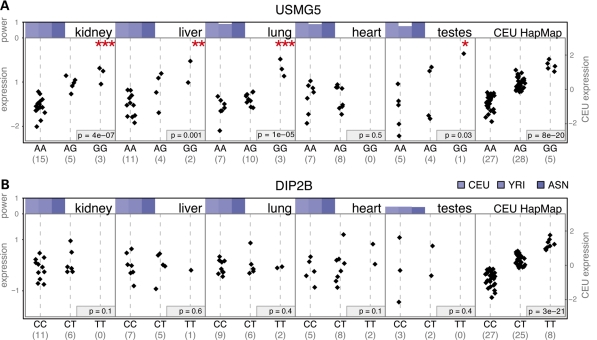
Figure 3.
Results for two illustrative genes. (A) We detect the cis-eQTL for the gene USMG5 in four of five tissues while in (B) we do not confirm the eQTL for the gene DIP2B, despite estimating that we have good power to detect it. The first five panels in parts (A) and (B) correspond to qPCR expression data in the five primary tissues (vertical axis on left) while the last panel shows microarray expression data from the original CEU HapMap LCLs (vertical axis on right); note that we show the CEU cell-line data for comparison purposes—there is no overlap in individuals between cell lines and primary tissues. Black diamonds correspond to normalized expression levels for the genotypes indicated below (number of observations in parenthesis). P-values for the five tissues are one-sided (*0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001); P-values for the cell-line data are two-sided tests. Power is shown above each plot (vertical axis ranging between 0 and 1 as indicated) and was estimated by resampling cell-line expression values from each HapMap population in turn (color indicates HapMap population), conditioning on the genotypes observed for the tissue samples we assayed.
That said, perhaps a more obvious explanation for the overall relatively low eQTL detection rate in our primary tissue study is biological, namely that certain genetic variants affect expression levels only in a subset of tissues. Indeed, genes are regulated in a tissue- and timing-specific manner, at least in part, due to the modular nature of cis-regulatory elements (15). Thus, although the expression of all assayed genes was detected in all five tissues, we do not know which cis-regulatory elements are employed in each tissue. For example, under an alternative scenario, if we assume that each cis-eQTL were present in exactly one primary tissue with an effect size similar to that observed in the cell lines, then given our power calculations, we would expect 23/105 significant tests (including false positives, at one-sided P ≤ 0.05). Our confirmation rate (19/105) is only somewhat lower than this estimate.
Of course, it remains possible—likely perhaps—that some eQTLs detected in the LCLs are artifacts, and would never be seen in primary tissues. We expect that, in actuality, our observations are accounted for by an unknown combination of all three explanations, as often is the case in biology. However, our results and others (37,38) indicate that a non-negligible fraction of cis variation affects multiple tissues. To investigate and characterize further how shared is the genetic basis of variation in gene expression, large-scale, multiple-tissue studies will be best suited and are imminently feasible. Importantly, our observations (albeit using limited sets of genes and samples) are encouraging, as they indicate that investigations of genetic and regulatory variation in cell lines can certainly inform our understanding of regulatory variation in diverse primary tissues.
MATERIALS AND METHODS
Tissue samples and DNA/RNA extraction
We genotyped and assayed gene expression in 84 samples from five human tissues: 13 hearts, 23 kidneys, 18 livers, 20 lungs and 10 testes. We had multiple tissues from 11 of the 63 individuals (kidney and liver from two individuals, heart and lung from three, heart, liver and lung from one, heart liver and testes from one and all but testes from four individuals). All samples are from self-identified Caucasian individuals. We note that tissues were sampled from largely non-overlapping sets of individuals for each tissue type. Given we are primarily interested in the effect of cis regulatory variation, and not the genetic basis of expression covariation among tissues, having largely non-overlapping sets of individuals is preferable to having all tissues from each individual because non-overlapping sets will reduce the impact of individual-specific effects, such as environmental effects, which otherwise could confound the analysis. Thus, although we desired independent sets of individuals we included all tissues available to us in order to increase power, as the trade-off is likely to be minimal. The human adult tissue samples were collected for us by the National Disease Research Interchange (NDRI). We extracted DNA from each tissue using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD). RNA was extracted from the tissues using Trizol (Invitrogen, Carisbad, CA) and first strand cDNA was synthesized using a poly-T oligo and the superscript kit (Invitrogen, Carisbad, CA).
SNP selection and genotyping
To select which SNPs to genotype in our samples (see Fig. 1), we used a heuristic that prioritized SNPs based on two criteria: (i) The ability of the genotype at a SNP to significantly explain expression in at least two of the three HapMap populations using simple additive linear models and (ii) a minor allele frequency ≥0.2 in the CEU HapMap population. The motivation for the latter criterion is that our samples are from individuals identified as Caucasian and by conditioning on a minor allele frequency above 0.2 we increase the odds that the SNPs chosen will segregate among our samples in as many tissues as possible. A more precise statement of criterion (i) is that we required genotype to explain more than 19% of the variance in expression in the linear model for the CEU population, and for the P-value for the slope coefficient to be <0.07 in each of the YRI (Yoruba in Ibadan, Nigeria) and ASN (pooled Han Chinese from Beijing, China and Japanese from Tokyo, Japan) populations for those populations for which the SNP in question has been genotyped. We chose these thresholds to optimize what we considered the important factors affecting our power to detect eQTLs and because we desired a list of approximately 200 eQTLs to pick from. The criteria outlined above resulted in a SNP associated with each of 206 genes, of which 196 SNPs were successfully genotyped (Supplementary Material, Table S1). Genotyping was performed on a Sequenom Mass Array Genotyping System using methods previously described (39). Average call rate was 91% (range 3–100%).
Power simulations
Following the genotyping of the 196 eQTLs in our samples, we performed power simulations using published HapMap expression data (33) as normalized by (and provided by) (6). For each candidate SNP, we calculated the number of samples from our tissue collection possessing each genotype at that SNP. Then for each genotype, we sampled (without replacement) an equivalent number of cell-line gene expression levels from HapMap individuals with matching genotypes for that SNP. We only considered SNPs with at least two genotypes, each of which is found in at least two individuals. For each combination of HapMap population (from which we sampled expression levels) and SNP, we generated 1000 replicate samplings. We then estimated power as the fraction of replicate samplings for which the slope coefficient in a linear model predicting expression with genotype was significant (at the 0.05 level).
Our goal in selecting SNPs was simply to prioritize eQTL predictions that are likely to be real in the cell lines and for which we should have decent power to detect in our samples of primary tissues. We believe that eQTLs detected separately in multiple HapMap populations are more likely to reflect true functional variation. We hold this view because we are concerned about pursuing falsely inferred eQTLs that are really attributable to non-genetic batch effects (11). We acknowledge that the possible trade-off of our approach is to miss population-specific cis effects (40). Moreover, one important drawback of a strategy that focuses on eQTLs that are shared across populations is that we might enrich for effects attributable to the EBV transformation itself (i.e. artifacts, which are expected to be shared across populations). However a substantial enrichment of this sort requires that a large fraction of real cis eQTLs are population specific, which appears unlikely given that only a small proportion of the variation in gene expression can be attributed to population-specific differences (41). Thus we chose to use both the ASN and YRI populations when we selected the SNPs (above) as well as in resampling the expression data in our power simulations. Specifically, for each gene we estimated power conditioning on genotypes from each of the five tissues among our samples and each of the three HapMap populations (CEU, YRI, ASN), resulting in 15 power estimates per gene. We chose the 15 genes with more than 80% power in at least six population-tissue combinations and at least 65% power in at least four of the five CEU tests. A number of genes fell slightly below one or both of these cutoffs, but still met our subjective judgment of good candidate genes based on the power analyses and the linear models. So rather than relax the criteria, we manually added the six genes that looked similarly promising.
We note that our power simulations gave us an estimate of power assuming that expression in our samples can be approximated by sampling from the HapMap expression levels. This approach will tend to overestimate power because the effect sizes of eQTLs sampled from the tail of an empirical distribution will tend to be overstated, resulting in a classic ‘Winner's Curse’ problem (e.g. 36). It is unclear how to do power simulations that avoid this feature, as the underlying effect sizes are unknown.
We calculated the expected number of significant tests, based on our power simulations under two scenarios. First, we assumed that all eQTLs would be functional in all tissues with the same effect size as originally seen in the cell lines. For this we simply summed over tissues our expected power estimated conditioning on the sample genotypes, to approximate the total number of eQTLs we would detect. In the second, equally arbitrary, but perhaps informative scenario, we assumed that each gene would have a real eQTL in exactly one of the five tissues. We estimate the number of eQTLs we would expect to detect under this scenario by performing the following simulation. For each gene we chose a tissue to contain the hypothesized real eQTL. We then sampled HapMap microarray expression data from samples with matching genotypes at the SNP under consideration, and used a linear model to test for an association (significant at 0.05 level) between these expression values and the SNP genotype. For all tests assessing eQTLs that are absent by hypothesis, we called them significant at the false positive rate of 0.05 (i.e. the probability under the null). We repeated this procedure for all genes 1000 times, observing the average number of detected eQTLs.
Assessing effect-size reduction
We estimated how much smaller, on average, the effect sizes must have been than what we observed in the cell-line data if we were to only find the observed number of eQTLs (19), assuming all eQTLs were real in all tissues. In brief, for each eQTL, we use a linear model to partially regress out the effect of genotype from the HapMap LCL expression data and determine by how much we would need to reduce the effect so that on average only 19 of the 105 tests are significant. More specifically, we conducted new power analyses similar to those above, but instead of resampling the original microarray expression data from the HapMap LCLs, we resampled from the residuals of a linear model that entirely regresses out the effect of genotype from these HapMap expression data, but then adds back the mean effect and only a fraction of the genotype effect. In our original power simulations, the linear model was: yi∼ μ+ βg+ εi and thus for these power simulations to assess the reduction in effect size, we used as expression values: εi+ μ+ βfg where yi is the expression level from the microarray data for individual i for the gene, and f is the factor by which we reduce the effect size. We then determine which f in the range of [0.1, 0.9] would result in 19 expected detected eQTLs.
Expression assay and analysis
We used quantitative PCR (qPCR) to measure gene expression levels. We designed qPCR primers for gene regions within 1 kb upstream of the predicted 3′-UTR—within the region that was originally probed by the microarray (6). Primer sequences are available upon request. As templates, we used first strand cDNA samples from each tissue. Quantitative RT-PCR was performed in a 25 µL reaction containing 2X SYBR master mix (Sigma), 0.2 pM each primer and 1 µL cDNA template. PCR was performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems, Inc.). We assayed each sample in triplicate on the same plate, with three 96-well plates required to assay all the samples for each gene. Although on each plate we included a dilution series over four concentrations (1X, 1/5X, 1/25X and 1/125X) to allow for a plate-specific estimate of the rate at which the PCR product accumulates (log r), we found these estimates to be rather noisy and instead assumed the doubling rate was identical for all plates (see below). We note that incorporating the plate-specific dilution curves in our analyses did not yield markedly different results.
For normalization, we developed a linear model-based method that, in effect, normalized our data by all genes. This linear model estimates the unknown relative, sample-specific concentration of cDNA (fis) and the unknown, plate-specific amplification threshold (Tp) assuming qPCR amplification on plate p of cDNA from individual i from tissue t using primers for gene g reaches some concentration Tp at PCR cycle Citg according to the following equation: Tp= Iitg fit rCitg. Taking the log of both sides and rearranging gives us a linear model with a response variable proportional to the cycle number and factors specifying the plate (p) and sample (s) and residuals corresponding to our estimated log expression level before amplification (Iitg):
This linear model, which contains as predictors only two factors for sample and plate, allows us to remove these effects and use in all subsequent analysis the residuals, which represent the log expression level. To then assess the significance of a SNP in explaining variation in expression, we used a gene/tissue combination-specific additive linear model: log Ii ∼ μ+ βg+ ε, where g is the number of A1 alleles in each individual and errors are normally distributed with a gene-specific variance. Significant eQTLs are thus those that have a β coefficient that is significantly different than zero in the direction predicted (6) at the 0.05 test level. We also consider a linear model for each gene/tissue combination that includes a dominance coefficient: log Ii ∼ μ+ β1g+ β2I(g = 1) + ε, where g is as before, β2 is the dominance coefficient and I(g = 1) is the indicator variable that evaluates to 1 if the individual is heterozygous for the SNP and 0 if homozygous.
To ensure that the results are robust with respect to the methodology used, we also tried quantile-normalizing the negative raw qPCR cycle numbers of each plate, using these in the above gene-tissue-specific linear models, and we observed nearly equivalent results to the above first method (see Supplementary Material, Fig. S2). For quantile normalization we normalized to a normal distribution, as the qq-plots suggested a good fit to a normal.
False discovery rate
We calculated the positive false-discovery rate (35) at a false positive rate of 0.05 using the qvalue package (version 1.1) downloaded from CRAN (http://cran.r-project.org/) for the R programming language (42). We also tested whether our test is well calibrated by permuting the genotypes and assessing the number of significant tests under this non-parametric null distribution, and found it to be well calibrated, with, for example, 5% of the tests significant at the 5% test level.
Additionally we calculate the probability that we see 11 or more eQTLs detected in at least one tissue based on the assumption of a well-calibrated test, which we assessed for the 5% level using a permutation method. Thus, under the null hypothesis of no eQTL in any tissue, each gene has a 1 – 0.955= 0.226 probability of falsely showing one or more eQTL. Therefore we calculated the P-value for observing 11 or more genes with one or more eQTL using a binomial distribution with P = 0.226.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online. Raw data available at http://giladlab.uchicago.edu/data.html.
AUTHORS CONTRIBUTIONS
K.B., G.C. and Y.G. conceived and designed the experiments. C.I.C. performed the wet lab experiments. K.B. analyzed the data and K.B., G.C. and Y.G. interpreted the results and wrote the paper.
FUNDING
This work was supported by the National Institutes of Health (GM077959 to Y.G.); the Sloan Foundation (Y.G., G.C.); the National Science Foundation (NSF GRF to K.B.); the Packard Foundation (to Jonathan Pritchard supporting G.C.); and the University of California, Davis (funds to G.C.).
ACKNOWLEDGEMENTS
We would like to thank Jean-Baptiste Veyrieras for providing us access to his data and results and Jonathan Pritchard, Molly Przeworski, Joseph Pickrell and Jack Degner for helpful comments on the manuscript.
Conflict of Interest statement.None declared.
REFERENCES
- 1.Gilad Y., Rifkin S.A., Pritchard J.K. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deutsch S., Lyle R., Dermitzakis E.T., Attar H., Subrahmanyan L., Gehrig C., Parand L., Gagnebin M., Rougemont J., Jongeneel C.V., et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 2005;14:3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 3.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 4.Duan S., Huang R.S., Zhang W., Bleibel W.K., Roe C.A., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang R.S., Duan S., Kistner E.O., Zhang W., Bleibel W.K., Cox N.J., Dolan M.E. Identification of genetic variants and gene expression relationships associated with pharmacogenes in humans. Pharmacogenet. Genomics. 2008;18:545–549. doi: 10.1097/FPC.0b013e3282fe1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veyrieras J.-B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden E.C. International genome project launched. Nature. 2008;451:378–379. doi: 10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 9.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannula K., Lipsanen-Nyman M., Scherer S.W., Holmberg C., Höglund P., Kere J. Maternal and paternal chromosomes 7 show differential methylation of many genes in lymphoblast DNA. Genomics. 2001;73:1–9. doi: 10.1006/geno.2001.6502. [DOI] [PubMed] [Google Scholar]
- 11.Akey J.M., Biswas S., Leek J.T., Storey J.D. On the design and analysis of gene expression studies in human populations. Nat. Genet. 2007;39:807–808. doi: 10.1038/ng0707-807. [DOI] [PubMed] [Google Scholar]
- 12.Choy E., Yelensky R., Bonakdar S., Plenge R.M., Saxena R., De Jager P.L., Shaw S.Y., Wolfish C.S., Slavik J.M., Cotsapas C., et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter K.L., Cahir-McFarland E., Kieff E. Epstein–Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 2002;76:10427–10436. doi: 10.1128/JVI.76.20.10427-10436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnone M.I., Davidson E.H. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 15.Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 16.Smirnov D.A., Cheung V.G. ATM gene mutations result in both recessive and dominant expression phenotypes of genes and micrornas. Am. J. Hum. Genet. 2008;83:243–253. doi: 10.1016/j.ajhg.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsche L.G., Loenhardt T., Janssen A., Fisher S.A., Rivera A., Keilhauer C.N., Weber B.H.F. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat. Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 18.Graham D.S.C., Graham R.R., Manku H., Wong A.K., Whittaker J.C., Gaffney P.M., Moser K.L., Rioux J.D., Altshuler D., Behrens T.W., et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat. Genet. 2008;40:83–89. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt K.A., Zhernakova A., Turner G., Heap G.A.R., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarroll S., Huett A., Kuballa P., Chilewski S., Landry A., Goyette P., Zody M., Hall J., Brant S., Cho J., et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyagawa T., Kawashima M., Nishida N., Ohashi J., Kimura R., Fujimoto A., Shimada M., Morishita S., Shigeta T., Lin L., et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat. Genet. 2008;40:1324–1328. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto Y., Mabuchi A., Shi D., Kubo T., Takatori Y., Saito S., Fujioka M., Sudo A., Uchida A., Yamamoto S., et al. A functional polymorphism in the 5′-UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki A., Yamada R., Kochi Y., Sawada T., Okada Y., Matsuda K., Kamatani Y., Mori M., Shimane K., Hirabayashi Y., et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat. Genet. 2008;40:1224–1229. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 25.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 26.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 28.Heinzen E.L., Ge D., Cronin K.D., Maia J.M., Shianna K.V., Gabriel W.N., Welsh-Bohmer K.A., Hulette C.M., Denny T.N., Goldstein D.B. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastinen T., Ge B., Hudson T.J. Influence of human genome polymorphism on gene expression. Hum. Mol. Genet. 2006;15(Spec No 1):R9–R16. doi: 10.1093/hmg/ddl044. [DOI] [PubMed] [Google Scholar]
- 30.Yan H., Zhou W. Allelic variations in gene expression. Curr. Opin. Oncol. 2004;16:39–43. doi: 10.1097/00001622-200401000-00008. [DOI] [PubMed] [Google Scholar]
- 31.Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verlaan D.J., Ge B., Grundberg E., Hoberman R., Lam K.C.L., Koka V., Dias J., Gurd S., Martin N.W., Mallmin H., et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–127. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson G., Riley-Berger R., Harshman L., Kopp A., Vacha S., Nuzhdin S., Wayne M. Extensive sex-specific nonadditivity of gene expression in drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storey J. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 2002;64:479–498. [Google Scholar]
- 36.Zollner S., Pritchard J.K. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hovatta I., Zapala M.A., Broide R.S., Schadt E.E., Libiger O., Schork N.J., Lockhart D.J., Barlow C. DNA variation and brain region-specific expression profiles exhibit different relationships between inbred mouse strains: implications for eQTL mapping studies. Genome Biol. 2007;8:R25. doi: 10.1186/gb-2007-8-2-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bento J.L., Bowden D.W., Mychaleckyj J.C., Hirakawa S., Rich S.S., Freedman B.I., Segade F. Genetic analysis of the GLUT10 glucose transporter (SLC2A10) polymorphisms in Caucasian American type 2 diabetes. BMC Med. Genet. 2005;6:42. doi: 10.1186/1471-2350-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., Cheung V.G. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price A.L., Patterson N., Hancks D.C., Myers S., Reich D., Cheung V.G., Spielman R.S. Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet. 2008;4:e1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.