Abstract
The function of the retinitis pigmentosa GTPase regulator interacting protein 1 (RPGRIP1) gene is currently not known. However, mutations within the gene lead to Leber Congenital Amaurosis and autosomal recessive retinitis pigmentosa in human patients. In a previously described knockout mouse model of the long splice variant of Rpgrip1, herein referred to as Rpgrip1tm1Tili mice, mislocalization of key outer segment proteins and dysmorphogenesis of outer segment discs preceded subsequent photoreceptor degeneration. In this report, we describe a new mouse model carrying a splice acceptor site mutation in Rpgrip1, herein referred to as Rpgrip1nmf247 that is phenotypically distinct from Rpgrip1tm1Tili mice. Photoreceptor degeneration in homozygous Rpgrip1nmf247 mice is earlier in onset and more severe when compared with Rpgrip1tm1Tili mice. Also, ultrastructural studies reveal that whereas Rpgrip1nmf247 mutants have a normal structure and number of connecting cilia, unlike Rpgrip1tm1Tili mice, they do not elaborate rod outer segments (OS). Therefore, in addition to its role in OS disc morphogenesis, RPGRIP1 is essential for rod OS formation. Our study indicates the absence of multiple Rpgrip1 isoforms in Rpgrip1nmf247 mice, suggesting different isoforms may play different roles in photoreceptors and underscores the importance of considering splice variants when generating targeted null mutations.
INTRODUCTION
Rpgrip1, which encodes the retinitis pigmentosa GTPase regulator interacting protein 1, was identified in a yeast two-hybrid screen as a RPGR interacting protein (1–3). It contains a coiled-coil domain that is normally found in proteins involved in vesicular trafficking (2) and a C-terminal RPGR interacting domain (RID) (1–3). Within the eye it is mainly expressed in amacrine cells and photoreceptors (1–5). Sub-cellular localization of RPGRIP1 appears to be species specific. Within photoreceptors, RPGRIP1 localizes predominantly to outer segments (OS) of rods and cones in humans, whereas in bovine eyes, it is found only in rod OS. Immunohistochemical staining within OS appear as distinctly spaced foci (4), suggesting an association with a structural component. In contrast, in mice, RPGRIP1 is found primarily in connecting cilia (CC), where it localizes between the ciliary axoneme and the plasma membrane (4,6). This has led to the proposal that RPGRIP1 is a structural component of the ciliary axoneme, acting as a scaffold for other proteins necessary for the transport of molecules through the CC from the inner segments (IS) to the OS of photoreceptors in mice (6).
Despite the differences in localization, mutations in both human and murine Rpgrip1 lead to a rapid, early onset retinal degeneration. In humans, mutations within RPGRIP1 are responsible for 4.5–6% of Leber Congenital Amaurosis (LCA) (summarized in Ref. 7) and for some cases of juvenile RP (8). Although LCA is a relatively rare autosomal recessive disorder with an incidence of 2–3 per 100 000 live births, it accounts for 5% of all retinal dystrophies (9,10) and is a severe and devastating disease. LCA is characterized by retinal degeneration at or within 6 months of birth, extinguished or severely reduced scotopic and photopic electroretinograms by 1 year of age with absent or abnormal visual evoked potentials, sluggish pupillary reaction, nystagmus and oculo-digital behavior such as eye poking, eye rubbing and eye pressing (9,11,12). Several nonsense mutations in RPGRIP1 that are predicted to lead to an early termination of the gene product have been described in LCA6 patients. These occur throughout the coding region from exons 3 through 23 (13,14).
In order to study the function of RPGRIP1, Zhao et al. (6) generated a mouse model with an insertional mutation into exon 14 of the Rpgrip1 gene, herein referred to as Rpgrip1tm1Tili mice. Rpgrip1tm1Tili mice initially develop a full complement of photoreceptors, but their OS are disorganized with grossly oversized discs that stack vertically, indicating the importance of RPGRIP1 in proper disc morphogenesis. A progressive photoreceptor degeneration was observed with a complete loss of the outer nuclear layer (ONL) by 3 months of age.
Here, we report a new mutant allele of Rpgrip1, nmf247, identified in a recessive ethyl nitrosourea (ENU) chemical mutagenesis screen at The Jackson Laboratory (JAX). Mice homozygous for the Rpgrip1nmf247 mutation have a more severe retinal phenotype than the purported null Rpgrip1tm1Tili mutant. OS rarely form, and the ONL loss that initiates between postnatal day (P) 12 and P14 is nearly complete by P28. Our results suggest that RPGRIP1 is important not only for OS disc morphogenesis as reported previously, but also for the formation of OS, particularly rod OS, in mice. It also underscores the power of studying allelic series of mutations to determine the function of domains and splice variants within genes.
RESULTS
Identification of the nmf247 allele as a point mutation within the splice acceptor site for intron 6 of the Rpgrip1 gene
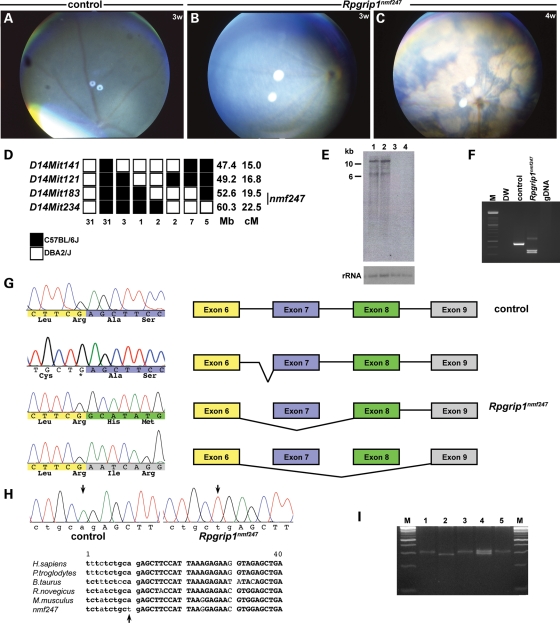
nmf247 mice were first identified by the Neuromutagenesis Facility (NMF) program at JAX. At 3 weeks of age, mice homozygous for the nmf247 mutation were identified by a granular fundus appearance (Fig. 1A and B). By 4 weeks of age, classic signs of retinal degeneration including attenuation of retinal blood vessels, focal and diffuse depigmentation of the retinal pigment epithelium (RPE) and patches of pigment deposits were apparent by indirect ophthalmoscopy (Fig. 1C).
Figure 1.
Fundus photographs of (A) WT control at 3 weeks, and (B, C) Rpgrip1nmf247 mutants at 3 weeks (B) and at 4 weeks of age (C), indicating a rapid progression of the retinal degenerative phenotype. (D) Haplotype analysis. F2 mice from a (B6D2F2-nmf247/+) F1 intercross were phenotyped for the retinal spotting and genotyped for the indicated microsatellite markers. Black boxes represent the B6-derived allele, and white boxes represent the DBA/2J-derived allele. The number of chromosomes sharing the corresponding haplotype is indicated below each column of squares. The order of marker loci was determined by minimizing the numbers of crossovers. The genomic coordinates based on NCBI build 37 in mega base (Mb) and centimorgan (cM) are indicated next to each row of squares. The genotype for nmf247 was inferred from the phenotype or the results from progeny testing of non-informative recombinants. (E) Northern analysis of poly A+ RNA isolated from P14 mutant (lane 1, 3) and WT control (lane 2, 4) whole eyes (lane 1, 2) and brains (lane 3, 4). A 844 bp probe spanning nucleotides 3230–4074 was used for hybridization. The shorter band, ∼6 kb in size was not observed in both mutant and WT control in Northern analysis of total retinal RNA isolated from P15 eyes (data not shown) using the same probe, indicating either extraretinal splicing variants or non-specific cross-action of the probe. The 18S ribosomal RNA (rRNA) is shown as a loading control. (F) Amplification of cDNA synthesized from RNA isolated from eyes of mutant and wild-type mice with primers 2F in exon4 (5′GCACAGCCTCGAGTCCAC3′) and 2R in exon 9 (5′TTGGCTGAGGAAGGCATTAT3′) led to a smaller product in Rpgrip1nmf247 mutants. Distilled water (DW) and genomic DNA (gDNA) from tail were used as negative controls. (G) A deletion of exon 7 of Rpgrip1 from Rpgrip1nmf247 mice compared with WT was observed. The color-coded sequences indicate each exon; yellow, blue, green and grey indicate exon 6, 7, 8 and 9, respectively. The non-color coded sequences are from the intron. Skipping of exon 7 or both exon 7 and 8 results in frame-shift that is predicted to lead to a premature termination in exon 9 in mutant mice. Alternatively, the mutation causes an abnormal splicing into intron 6 generating a 96 bp insertion. The inserted sequences code for 32 amino acids, but the mutant protein is predicted to terminate at the mutation site, which generates a stop codon. (H) Genomic analyses of Rpgrip1 using primers 13F in intron 6 (5′GGTCTTCAGGTCTCGCTGTC3′) and 13R in exon 7 (5′ACTCGTGTCAGTTGGGCTTC3′) revealed a point mutation (A to T) that alters the splice acceptor site in intron 6 (arrows). Sequence comparison shows this nucleotide is highly conserved across species. Lower case letters indicates nucleotides in the intron, upper case letters indicate exon, and the bold face indicates conserved sequence. (I) An allele specific PCR assay (49) was designed using primers mRpgrip1-Ra1 (5′CTCCACGTTCTCCTTAATGGAAGCTTT3′), mRpgrip1-Ra2 (5′GCTTCTTAAGGCGAATCAGACCCACGTTCTCCTTAATGGAAGTTCA3′), and mRpgrip1-Fa1 (5′GCACATATTTAGTTGGGTTCTGTAAG3′). M; DNA ladder marker, lane 1, 3, 5: Rpgrip1nmf247, lane 2: WT, lane 4: het.
To chromosomally localize the mutation, affected nmf247 mice, on a C57BL/6J (B6) background were outcrossed to strain DBA/2J to generate F1 carriers, which were intercrossed to produce F2 progeny. Using SSLP markers distributed throughout the genome, a scan was carried out on DNA pools from affected and unaffected F2 mice. Significant skewing of B6 alleles in the affected DNA pool was observed with markers on proximal chromosome 14. Testing individual DNA samples confirmed that nmf247 mapped to chromosome 14 between markers, D14Mit141 and D14Mit234. Subsequently, 280 meioses were tested to narrow the region containing the mutant gene between the flanking SSLP markers, D14Mit183 (52.6 Mb)-nmf247-D14Mit234 (60.3 Mb). Contained within this region is Rpgrip1 at 52.7 Mb, a gene known to lead to retinal degeneration when mutated (Fig. 1D). Northern analysis of Rpgrip1 using polyA+ RNA isolated from whole eyes of mutant and wild-type mice showed bands of equal intensity and size (Fig. 1E), indicating that if a mutation within the gene was responsible for the disease phenotype, the expression level and size of the transcripts, at least within the resolution of the assay, were not altered. Amplification of cDNA isolated from retina of mutant and wild-type mice using primers from exons 4 and 9 of Rpgrip1 produced three amplicons in nmf247 mutants in comparison to a single amplicon observed in wild-type mice (Fig. 1F). The largest Rpgrip1 amplicon observed in nmf247 mutants is caused by abnormal splicing, resulting in incomplete removal of intron 6 (Fig. 1G). Sequencing of the two smaller amplified products showed that exon 7 (longer product) and exons 7 and 8 (shorter product) were deleted in nmf247 mutants (Fig. 1G). Comparison of mutant and B6 sequences from genomic DNA using primers spanning intron 6 and exon 7, revealed a mutation in the splice acceptor site (A to T) in intron 6. The point mutation in the splice acceptor site of intron 6 within Rpgrip1 is predicted to lead to frame shifts in the two smaller aberrant transcripts and an in-frame 32 amino acid insertion and generation of a premature termination codon in the largest aberrant transcript, and if translated, an early termination of RPGRIP1. The mutated adenine residue is highly conserved in all splice acceptor sites and across species (Fig. 1H). In order to genotype nmf247 mutants more efficiently, an allele-specific PCR assay was designed for use on genomic DNA (Fig. 1I).
Photoreceptor degeneration is more rapid in Rpgrip1nmf247 than in Rpgrip1tm1tili mice
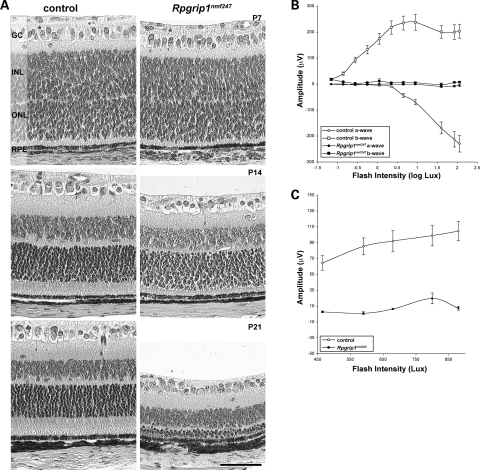
The photoreceptors of Rpgrip1nmf247 mutants show rapid and progressive degeneration (Fig. 2A). At P7, whereas apparent in wild-type mice, OS were not observable by light microscopy in mutant animals. At P14, OS were not observable in mutant mice and IS were slightly shorter than those of controls. However, no difference was observed in the number of photoreceptor cell bodies in the ONL of control and mutant retina. By P21, the ONL was reduced to a 3–4 nuclei thickness in mutant retina. In addition, IS of Rpgrip1nmf247 mutants were shorter than controls. Functional assessment of photoreceptors corresponded to the histological findings, with absence of rod and cone ERG responses by P21, the earliest time point examined (Fig. 2B and C).
Figure 2.
Early onset, progressive photoreceptor degeneration observed in Rpgrip1nmf247 mutants. (A) Retinal sections were obtained from control and Rpgrip1nmf247 at P7, P14 and P21 and stained by hematoxilin and eosin (H&E) and visualized by light microscopy. GC, ganglion cell; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. Bar indicates 50 µm. ERG analysis of (B) rod and (C) cone photoreceptors in 3-week-old Rpgrip1nmf247 mutants and controls.
The slower photoreceptor loss in the purported null mutant, Rpgrip1tm1Tili, could conceivably be the result of genetic background influences as the original characterization was done in a mixed B6 and 129X1/SvJ (129X1) background. In order to test this possibility, we outcrossed Rpgrip1nmf247 mutants to 129X1 mice and intercrossed the resulting F1s to examine mutants that were homozygous for the Rpgrip1nmf247 mutation. OS development was not observed in B6x129X1-Rpgrip1nmf247 mutants and their disease progression was earlier in onset than reported for Rpgrip1tm1Tili mutants (data not shown). This suggests that the phenotypic dissimilarity observed between Rpgrip1nmf247 and Rpgrip1tm1Tili mutant mice is unlikely to be due to the influences of genetic modifiers.
Mislocalization of OS proteins is observed in Rpgrip1nmf247 mutants
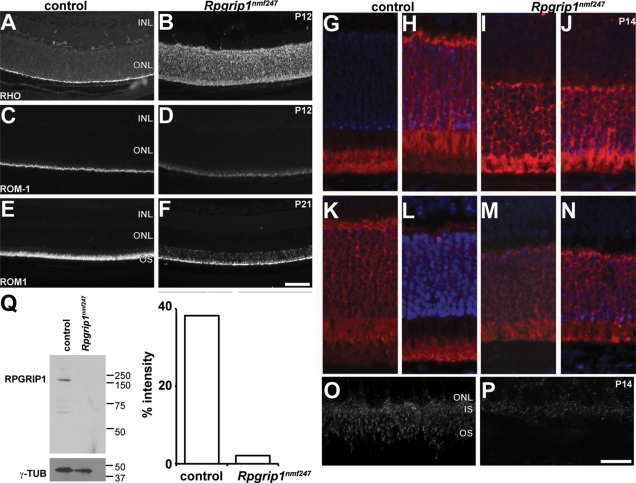
Due to the localization of RPGRIP1 to the CC and the presence of a coil-coiled domain within RPGRIP1, which is often observed in gene products involved in vesicular transport (2), it has been hypothesized that RPGRIP1 may be important for movement of proteins from the IS to the OS (2,3). Previous reports have shown that rod opsin is mislocalized in Rpgrip1tm1Tili mutants at P20 (6). Similarly in Rpgrip1nmf247 mutants, rhodopsin (Fig. 3A and B) was mislocalized to the ONL at P12, the earliest time point examined. However, in contrast to Rpgrip1tm1Tili mutants, rod outer membrane protein-1 (ROM1), a molecule that is transported via the CC by independent mechanisms than rhodopsin, was also mislocalized in Rpgrip1nmf247 mutants (Fig. 3C–F). The ROM1 mislocalization is more pronounced at P21. Other molecules that traverse the CC and that were normally localized in Rpgrip1tm1Tili mutants were mislocalized in Rpgrip1nmf247 mutants, such as transducin (Fig. 3G–J) and arrestin (Fig. 3K–N). It is likely that the combined effects of the lack of OS development and of aberrant transport across the CC leads to the more severe phenotype in Rpgrip1nmf247 mutants.
Figure 3.
Mislocalization of proteins in Rpgrip1nmf247 mutant retinas. Retinal sections were obtained from control and Rpgrip1nmf247 at P12 (A–D) and at P21 (E, F), and stained with (RHO; A, B) anti-rhodopsin, anti-ROM1 (ROM1; C–F). To examine the ability of transducin and arrestin to translocate in rod photoreceptor cells, (G–J) anti-transducin and (K–N) anti-arrestin were used to stain dark adapted (G, I, K, M) and light stimulated (H, J, L, N) retinas. RPGRIP1 localization was examined using a polyclonal anti-RPGRIP1 antibody (O, P). INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segment. Bars indicate 50 µm (A–F), 25 µm (G–N), and 10 µm (O, P). (Q) Western analysis indicates the absence of RPGRIP1 in Rpgrip1nmf247 mutants. γ-tubulin (γ-TUB) serves as a loading control (left). The ratio of the intensity of staining of RPGRIP1 to γ-tubulin was 38.2% in WT control but 2.1% in Rpgrip1nmf247 mutants.
To examine the effect of the mutation on RPGRIP1 localization, a polyclonal rabbit antibody to the C-terminal end of RPRGIP1 (3) was used on unfixed, frozen retinal preparations. Staining of the CC in Rpgrip1nmf247 mutants was absent (Fig. 3O and P). To determine if RPGRIP1 was mislocalized but still synthesized, western analysis of retinal lysates from mutant and control animals with antibody to the C-terminal end was carried out and showed that RPGRIP1 was not detected in mutant mice (Fig. 3Q).
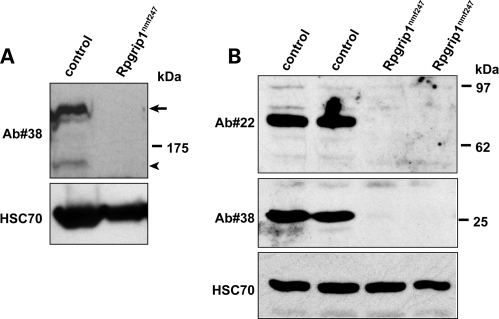
To examine the effect of the mutation on isoforms of RPGRIP1 (15), western blot analyses were performed with two additional antibodies against the N-terminal coiled-coil (Ab#22) and C-terminal RID (Ab#38) domains of RPGRIP1 (Fig. 4). In wild-type mice, Ab#22 detects a unique ∼70 kDa RPGRIP1 isoform (Fig. 4B), whereas Ab#38 recognizes a major ∼200 kDa, a minor 150 kDa (Fig. 4A) and a 25 kDa RPGRIP1 isoform (Fig. 4B). The Rpgrip1nmf247 mutant lacks all RPGRIP1 isoforms detected by Ab#22 and Ab#38 (Fig. 4, Supplementary Material, Fig. S1). In contrast, the Rpgrip1tm1Tili mutant lacks both the ∼200 and ∼150 kDa bands detected by Ab#38, but strongly expresses a unique transgenic derived 80 kDa RPGRIP1 variant, which was absent in wild-type and mutant Rpgrip1nmf247 mice (Supplementary Fig. S1).
Figure 4.
Immunoblot of retinal homogenates of wild-type, and Rpgrip1nmf247mice (P16), with antibodies against the N-terminal coiled-coil (Ab#22) and C-terminal RID (Ab#38) domains of RPGRIP1. The absence of the ∼200 kDa full length (arrow, A) and smaller ∼150kD (arrowhead, A) and other smaller (70 and 25 kDa, B) RPGRIP1 isoforms is observed in Rpgrip1nmf247 mice. HSC70 serves as a loading control in both panels.
Ultrastructural analysis reveals that OS are rarely observed in Rpgrip1nmf247
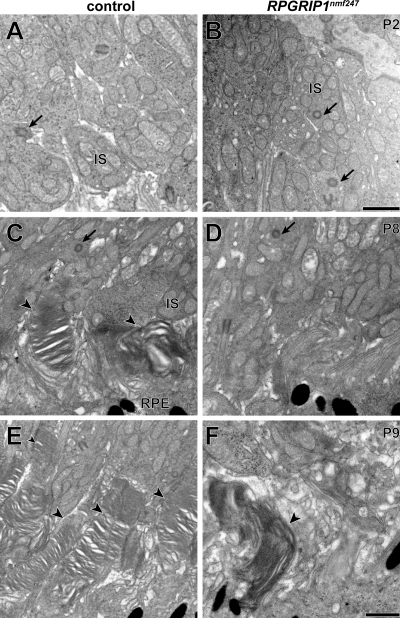
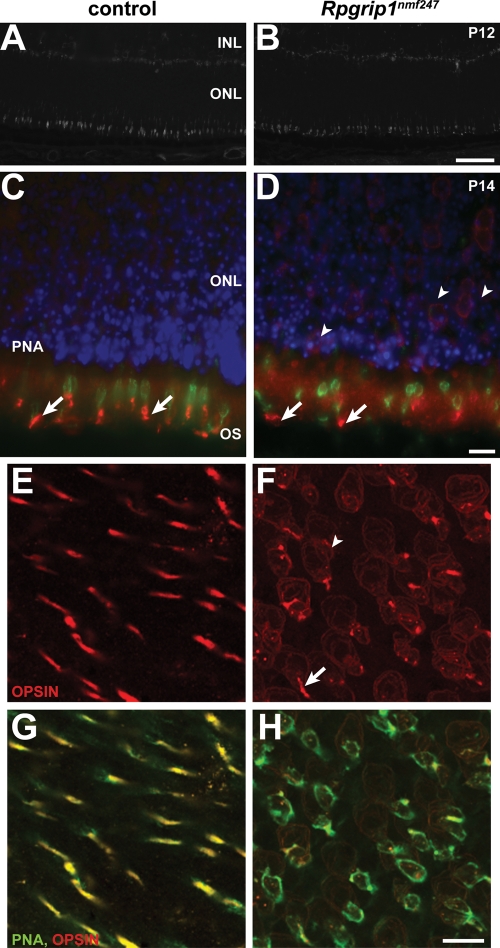
Because multiple proteins were mislocalized in Rpgrip1nmf247 mutants, we examined the CC and OS by transmission electron microscopy to determine if they were structurally compromised. As in the Rpgrip1tm1Tili mutants, the CC appeared to be structurally intact with the normal 9+0 doublet configuration (Fig. 5A and B). However, unlike the Rpgrip1tm1Tili mutants, the Rpgrip1nmf247 mutants rarely developed OS; at P8 no OS were observed (Fig. 5C and D). To ascertain whether OS formation was simply delayed in mutants, we examined retinal sections at P9 as well. The very rare OS that were formed appeared elongated and showed a vertical orientation, similar to those found in Rpgrip1tm1Tili mutants (Fig. 5E and F). It is likely that the abnormal OS are cone OS as Rpgrip1nmf247 mutant retinas show linear peanut agglutinin (PNA) staining, shorter but comparable in density to controls at P12 (Fig. 6A and B). As observed in Rpgrip1tm1Tili mutants, both blue and red/green opsin were partially mislocalized to the ONL of Rpgrip1nmf247 mutants (Fig. 6C–H).
Figure 5.
Ultrastructural transmission electron microscopy analysis suggests normal CC formation and aberrant OS development. The CC (arrows) in (A) wild-type controls and (B) Rpgrip1nmf247 mutants did not differ (P2). At P8 OS discs are observed (arrow heads) in (C) wild-type controls but not in (D) Rpgrip1nmf247 mutants. At P9, OS are lengthened in (E) wild-type controls but (F) in the rare instance in which OS are observed in Rpgrip1nmf247 mutants, OS discs are enlarged and have a vertical orientation. IS, inner segment; RPE, retinal pigment epithelium. Bars = 500 nm.
Figure 6.
Defective cone OS morphogenesis in Rpgrip1nmf247 mutant retina. Staining of (A) wild-type control and (B) Rpgrip1nmf247 mutant retinas with PNA. Double staining of blue cone opsin (red) with PNA (green) in control (C) and Rpgrip1nmf247 mutant retina (D) at P12. Retinas were obtained at P14, flat mounted, and stained with red/green opsin (red, E–H) and double stained with PNA (green, G–H). Arrows indicate OS and arrowheads indicate mislocalized opsin in nuclear bodies. INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segment. Bars; 50 µm (A, B), 10 µm (C–H).
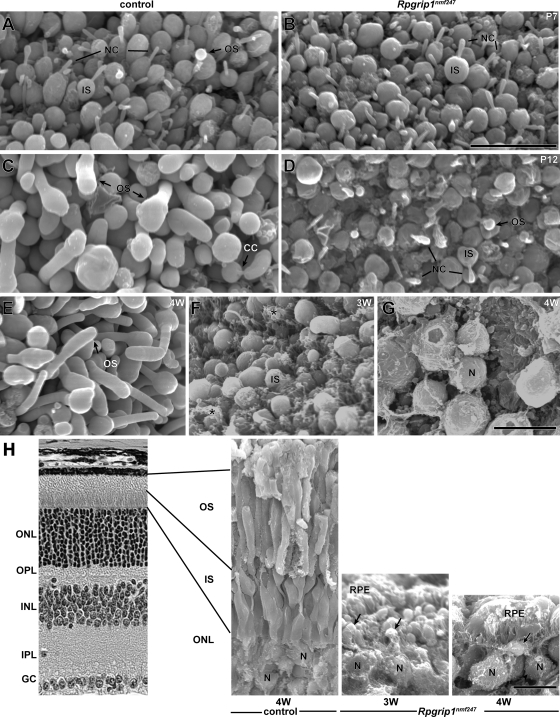
OS development and morphology was examined by scanning electron microscopy (SEM) at P7 and P12 (Fig. 7A–D). At P7, no IS abnormalities were observed in Rpgrip1nmf247 mutants, confirming our observations by light microscopy (Fig. 2A). Also, the number and the length of cilia in mutant mice were comparable with WT control (Fig. 7A and B). However, although rudimentary OS emerging from the CC were observed in WT control, they were rarely found in Rpgrip1nmf247 mutants at P7. By P12, OS are elongated in WT controls (Fig. 7C) and as previously reported, paddle-like structures were observed at the apex of some OS (16). In contrast, in Rpgrip1nmf247 mutant retinas, OS formation was virtually absent at the same age (Fig. 7D). In the few cases in which OS were observable, they were rudimentary and misshapen.
Figure 7.
OS morphogenesis and degeneration were observed by scanning electron microscopy of control and Rpgrip1nmf247 mutant retina at P7 (A, B), P12 (C, D), 3 weeks (F, H) and 4 weeks (E, G, H). Specimens were viewed from an RPE orientation (A–G) or fractured and viewed from a neuroretinal orientation (H). In control retinas, at P7, developing OS were observed (OS, A) and at P12, elongation of OS was observed (C). In contrast, in Rpgrip1nmf247 mutant retinas no OS were observed at P7 (B) and rare rudimentary OS budding was observed at P12 (OS, D). Asterisks indicate RPE adherence to the IS (F) in Rpgrip1nmf247 mutants. The photoreceptor nuclei (N) are noted (G, H). The degeneration is rapid and progressive with CC no longer observable on IS at 3 weeks (F and arrows, H) and with very rare IS at 4 weeks (G and arrow, H). OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GC, ganglion cells; RPE, retinal pigment epithelium; CC, connecting cilium; NC, nascent cilia. Bars = 10 µm.
Abnormalities and loss of photoreceptors was also observed by SEM at 3 and 4 weeks of age (Fig. 7E–H). By 3 weeks of age, the IS in Rpgrip1nmf247 mutant retina appeared irregular in both shape and size (Fig. 7F). RPE microvilli, juxtaposed to the IS in mutant retinas, were found to adhere avidly to IS (Fig. 7H, arrows). Nascent cilia were rarely detected in the degenerating photoreceptors at 3 weeks and photoreceptor nuclei were reduced to three layers (data not shown). By 4 weeks of age, the reduction in IS length was readily apparent (Fig. 7H), and photoreceptor cell bodies remaining, now reduced to one layer, bear IS (Fig. 7G and H).
DISCUSSION
In this study, we examined a new mutant allele of Rpgrip1, nmf247, identified in an ENU-induced mutagenesis screen. Careful examination of the mutant eye revealed that RPGRIP1 is not only important in OS disc morphogenesis, as previously reported (6), but also OS formation. Rpgrip1nmf247 mutants do not develop rod OS, however, cone OS are formed initially and degenerate rapidly, indicating that the mechanism for OS formation in respect to RPGRIP1 is likely to be different between rods and cones. Although an early time course for the miniature longhaired dachshunds carrying a nonsense mutation in exon 2 of Rpgrip1 is not available, PNA staining of cone OS and mislocalization of rhodopsin to the ONL was observed in 6-week-old mutant dogs (17,18). This suggests that like Rpgrip1nmf247 mutant mice, dogs form cone photoreceptor OS.
Rho−/− (19), Prph2Rd2 (20) and Crxtm1Clc (16) mutant mice also do not form OS. Rhodopsin, the visual pigment that makes up ∼90% of protein contained within the OS discs (21), and peripherin/RDS, a membrane glycoprotein that localizes to the rims of the OS discs (22,23), appear to be necessary structural components for OS elaboration. CRX, a transcription factor, regulates the expression of many retina-related proteins including rhodopsin and peripherin/RDS (24). Both of these proteins are significantly down-regulated in the Crxtm1Clc mice (25,26), and this may contribute to the phenotypic similarities, namely lack of OS disc formation, in these mutants (16).
A coiled-coil domain normally found in proteins involved in vesicular trafficking and its localization to the CC suggests that RPGRIP1 may function in transport of proteins from the IS to the OS. Because protein synthesis occurs in the IS, all proteins necessary for outer segment structure and function must be transported through the CC. The outer segment is in fact considered to be a modified cilium, and basal bodies are located at the base of the CC. Transport of proteins occurs via intraflagellar transport (IFT) particles (27). Outer segment proteins, like opsin, are packaged in vesicles that bud off the endoplasmic reticulum and move toward the base of the CC where they fuse with the cell membrane and the protein cargo is tethered to the IFT particle. The membrane bound cargo protein and the IFT particles are then moved by a kinesin motor along the microtubule axoneme of the CC to the OS. The IFT is returned to the inner segment via dynamin-mediated transport. Mutations in a number of proteins thought to function in vesicular transport (e.g. TULP, ALMS1, BBS proteins) lead to accumulation of vesicles within the IS (28–31). Prph2Rd2 and Crxtm1Clc also accumulate vesicles at the base of the IS (16,32). A number of investigators have shown that these vesicles in Prph2Rd2 mutants are rhodopsin positive (32,33) indicating that the initial steps in outer segment development are occurring in this model. Likewise, in Rho−/− mice, peripherin/RDS and ROM1 are still incorporated into the OS membrane and accumulate at the distal end of the cilium (34). We did not observe the accumulation of vesicles near the base of the IS nor at the distal ends of the CC in Rpgrip1nmf247 mutants. However, all of the OS proteins that we examined in the Rpgrip1nmf247 mutants were mislocalized to some degree. This suggests that RPGRIP1 functions prior to the assembly of vesicles, docking of vesicles to the plasma membrane or attachment of cargo to the IFT machinery.
As noted above, how RPGRIP1 mediates its function in ciliary transport or initiation of OS formation is not known. However, it has been shown that RPGR, a binding partner of RPGRIP1 is synthesized but not localized to the CC in Rpgrip1tm1Tili mutants (6). A targeted null allele of Rpgrtm1Tili also shows mislocalization of visual pigments, develops OS as do Rpgrip1tm1Tili mice, but has a milder phenotype than Rpgrip1tm1Tili mice (6,35). This suggests that some of the pathological features, including early mislocalization of outer segment proteins observed in mutant Rpgrip1nmf247 mice may occur via its interaction with RPGR, but other phenotypes, including the actual formation of OS, may occur as a result of ancillary mechanisms dependent on the role of specific RPGRIP1 isoform(s), which is not contingent on RPGR function.
The Rpgrip1nmf247 model represents a second mutant mouse allele that has a more severe retinal phenotype than that of the originally described Rpgrip1tm1Tili null allele (6). It was originally reported that there were no murine alternative splice variants for Rpgrip1 (6). However, since the majority of genes in the genome have splice variants (36,37) and multiple splice variants for both human and bovine Rpgrip1 are known (2,4,38), it is reasonable to assume that alternative splice variants for murine Rpgrip1 do occur. Indeed, Lu and Ferreira (38) reported the presence of a short splice variant, named Rpgrip1b, encompassing exons 1 through 13 plus three additional novel amino acids. It was predicted that the Rpgrip1tm1Tili mutant gene, which carries an insertion in exon 14, may still produce a functional protein from the shorter splice variant, Rpgrip1b (15,38). This prediction is now validated in this work, which shows that Rpgrip1tm1Tili mice produce a unique 80 kDa RPGRIP1 isoform likely resulting from the translation of some inserted or mispliced transgenic exons (Supplement Fig. S1). In contrast, the Rpgrip1nmf247 mutant is null for both the full length and shorter variant forms (Fig. 4), since the point mutation in intron 6 leads to either a deletion of exon 7 or exons 7 and 8 and early termination. The difference in phenotypes between Rpgrip1tm1Tili and Rpgrip1nmf247 mutant mice has led us to hypothesize that whereas the full length Rpgrip1 variant is necessary for proper OS disc morphogenesis, the loss of other RPGRIP1 isoforms cause a more severe block in OS formation. In support of this hypothesis, Pawlyk et al. (39) showed that introduction of a recombinant AAV construct containing the full length RPGRIP1 cDNA driven by the murine opsin promoter was able to restore proper OS morphogenesis in Rpgrip1tm1Tili. The rescue observed from this latter experiment also indicates that the 80 kDa RPGRIP1 immunoreactive product still retains its biological function(s).
Retinitis pigmentosa and LCA are genetically and phenotypically heterogeneous diseases (RetNet website: http://www.sph.uth.tmc.edu/RetNet/disease.htm). Although phenotypic heterogeneity can result from mutations in multiple loci, the observed variability may also be due to allelic variants within different protein domains of the same gene. Through genetic efforts, many loci have been identified (40–42) and as a result, we are beginning to achieve a greater understanding of what is necessary for normal retinal development, function and maintenance. Also, as allelic series of mutations within genes become available in model systems, such as the allelic series of Pde6b (43) and of Pde6a (44) mutants, and now for Rpgrip1, we can begin to elucidate the unique functions of protein domains and their alternative splice variant forms.
MATERIAL AND METHODS
Origin of the nmf247 mutant and husbandry
nmf247 mice were identified in a B6 G3 ENU mutagenesis screen carried out by the NMF at JAX. Once heritability of the observed retinal degeneration was established as a recessive trait, mutants were bred and maintained in the Research Animal Facility at JAX. Mice were provided with NIH 6% fat chow diet and acidified water, with 12:12 h dark:light cycle in pressurized individual ventilation caging which were monitored regularly to maintain a pathogen free environment. Procedures involving the use of mice were approved by the Institutional Animal Care and Use Committee.
Clinical evaluation and electroretinography
Mice, dark-adapted for a minimum of 1 h, were treated with atropine prior to examination by indirect ophthalmoscopy with a 60 or 78 diopter aspheric lens. Fundus photographs were taken with a Kowa small animal fundus camera using a Volk superfield lens held 2 inch. from the eye as previously described (45).
For functional electroretinographic evaluation of Rpgrip1nmf247 mutants, following an overnight dark adaptation, mice were anesthetized with an intraperitoneal injection of xylazine (0.5 mg/cc) and ketamine (1 mg/cc) in normal saline. In adult mice, a dose of 0.1 cc/g was administered. Additional anesthetic was given if akinesia was inadequate. The equipment and protocol used here were those described in reference (46).
Genetic mapping
Genomic DNA was isolated from tail tips using a PCR buffer with nonionic detergents (PBND) preparation, which was adapted from a protocol from Perkin Elmer Cetus (47). Tail tips were digested in PBND buffer + Proteinase K overnight at 55°C. Samples were heated to 95°C for 10 min and 1 µl of the DNA preparation was used in a 12 µl PCR reaction. Amplicons were visualized with ethidium bromide after electrophoretic separation on a 4% agarose gel.
For mapping purposes, phentoypically affected mice, presumed to be homozygous for the nmf247 mutation, were mated with DBA/2J mice. The resulting F1 offspring were intercrossed and F2 offspring were phenotyped by indirect ophthalmoscopy. DNA isolated from tail tips of 13 affected and 13 unaffected mice were pooled and subjected to a genome wide scan using 92 SSLP markers distributed throughout the genome. Samples used in the DNA pools were tested individually to confirm the map location. One hundred and forty F2 mice were produced to generate a fine structure map of the region.
For testing genetic background influences, affected Rpgrip1nmf247 mice were outcrossed to 129X1 mice and the resulting F1 progeny were intercrossed. Mice homozygous for the nmf247 mutation were examined by indirect ophthalmoscopy and light microscopy to determine the onset and severity of the retinal disease.
Preparation of RNA samples and subsequent analysis
Total RNA was isolated from whole eyes and brains of affected nmf247 and B6 mice using TRIzol Reagent (Life Technologies) per manufacturer's protocol. Total RNA was treated with RNase-free DNaseI (Ambion) and quantity determined by NanoDrop (ND-1000 spectrophotometer). RNA quality was evaluated with an Agilent Technologies 2100 Bioanalyzer. cDNA was generated using the Retroscript kit (Ambion).
Primers to sequence the coding region of Rpgrip1 was designed from exon sequences obtained from the Ensembl Database. RT–PCR was done using eye cDNA in a 24 µl PCR reaction containing 1×PCR buffer (10 mm Tris–HCl pH 8.3, 50 mm KCl), 250 µm of each dATP, dCTP, dGTP, dTTP, 0.2 µm of each forward and reverse primer, 1.5 mm MgCl2, and 0.6 U Taq polymerase. The following PCR program was used: 94°C for 1 min 30 s followed by 35 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s and a final extension of 72°C for 2 min. PCR products were electrophoresed on a 1% agarose gel and visualized by ethidium bromide staining. DNA fragments were sequenced on an Applied Biosystems 3730XL (using a 50 cm array and POP7 polymer).
For northern blot analysis, total RNA was isolated from whole eyes and brain of P14 C57BL/6J mice and affected nmf247 mice. Poly(A)+ RNA was purified from total RNA using a Dynabeads mRNA Purification kit (Dynal Biotech). Poly(A)+ RNA (1.8 µg eye and 1.4 µg brain) was fractionated on 1% agarose-formaldehyde gels and transferred to Hybond-N+ nylon membranes (Amersham Biosciences). 32P-random-labeled probes were made using a ∼840 bp cDNA fragment corresponding to the C-terminus of the Rpgrip1 transcript. Whole eye cDNA was amplified with Rpgrip1 primers 9F and rpY2h1R (9F: 5′ AACCAGGTGTCCAAGACAAGA 3′, rpY2h1R: 5′ TTCATGAAAACAGATCTTCAG 3′). Hybridization was carried out at 42°C in ULTRAhyb solution (Ambion NorthernMax) overnight. Blots were washed 2× at room temperature in 2×SSC/0.1%SDS for 5 min, followed by 2× at 42°C in 0.1XSSC/0.1% SDS for 15 min.
Histological and immunological analysis
Mice were asphyxiated by carbon dioxide inhalation and enucleated eyes were fixed overnight in cold methanol/acetic acid solution (3:1, v/v). The paraffin embedded eyes were cut into 6 µm sections, stained by H&E and examined by light microscopy. Alternately, fresh eyes were immersed in optimal cutting temperature solution and frozen for sectioning. Sections were incubated with anti-rhodopsin (Leico Technologies), anti-red/green opsin (Chemicon), anti-ROM1, (a kind gift from R. Mclnnes), anti-guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 (gnat1, Santa Cruz), anti-arrestin (ABR) and anti-RPGRIP1 (3). Localization of the specific proteins was visualized by fluorescence microscopy or confocal microscopy following cy-3 conjugated secondary antibody (Jax Immunoresearch) labeling.
For translocation studies, mice were dark adapted in a darkened room for 16 h. For light stimulation, their eyes were dilated and the mice were transferred to a mirror box in which they were exposed to 1200 lux of light for 30 min. Eyes were collected immediately after dark adaptation or after light stimulation as described above.
Ultrastructural analysis
Eyes were removed immediately after carbon dioxide euthanasia and fixed for 3 h in a cold, phosphate-buffered, glutaraldehyde–paraformaldehyde mixture. After 3 h, the anterior segment was removed and the posterior segment cut into 1 × 2 mm blocks of retina, choroid and sclera. The additional fixation of the whole eye before dissection improved adhesion of the retina to the RPE and did not alter the quality of preservation. The dissected tissue was placed in fresh fixative for an additional 2–8 h and was post-fixed in 1% osmium tetroxide, dehydrated and embedded in plastic. Thick sections were cut for orientation and thin sections cut and stained with uranyl acetate and lead citrate and examined using a transmission electron microscope.
Scanning electron microscopy
Neural retinas were dissected from the RPE and immersion fixed in 3% glutaraldehyde and 1% paraformaldehyde solution overnight. Tissues were treated with 1% osmium tetroxide and 1% thiocarbohydrazide (TCH, Electron Microscopy Science, Hatfield, PA). Tissues were washed six times with water, dehydrated with an ascending grade of ethanol, dried with hexamethyldisilazane and sputter coated. Specimens were mounted and examined by SEM.
Western analysis
Total protein from wild-type and Rpgrip1nmf247 eyes was obtained by standard methods in RIPA buffer (1% NP-40, 0.1% SDS and 0.5% sodium deoxycholate in PBS) containing a protease inhibitor cocktail (Roche). Hundred microgram of total protein was separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was stained with anti-RPGRIP1 (rabbit polyclonal used in immunohistochemical analysis) and anti-γ tubulin (Sigma) for loading control. All immunoblots were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies followed by the ECL detection system (Perkin-Elmer). The intensity of bands was determined using ImageJ software (NIH).
For the western analysis with antibodies Ab# 22 and 38 (2,5), retinal homogenates were prepared as previously described (5) and protein concentration was determined by the BCA method. Approximately 200 µg of homogenates were loaded and resolved by SDS-PAGE, transferred to PVDF membranes (Millipore) and western blot analyses were performed as described elsewhere (48). Affinity purified rabbit polyclonal antibodies, Ab#22 and 38, against RPGRIP1, were used at 125 ng/ml. Hsc70 antibody (loading control, 1:2000) was from StressGen (Ann Arbor, MI, USA).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This study was supported by National Institutes of Health grant, EY016501 (P.M.N.), National Institutes of Health core grant, 2P30-EY005722-21 (P.A.F.), a TJL institutional core grant, CA34196, and the Jules & Doris Stein Research to Prevent Blindness Professor (P.A.F.).
ACKNOWLEDGEMENTS
We would like to thank Drs Kenneth Johnson, Pete Fuerst and Neal Peachey for careful review of the manuscript, Pete Finger for assistance with the SEM analysis and Douglas Howell for the ERG analysis. An anti-ROM1 antibody was a kind gift from Dr R. McInnes.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Boylan J.P., Wright A.F. Identification of a novel protein interacting with RPGR. Hum. Mol. Genet. 2000;9:2085–2093. doi: 10.1093/hmg/9.14.2085. [DOI] [PubMed] [Google Scholar]
- 2.Roepman R., Bernoud-Hubac N., Schick D.E., Maugeri A., Berger W., Ropers H.H., Cremers F.P., Ferreira P.A. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 3.Hong D.H., Yue G., Adamian M., Li T. Retinitis pigmentosa GTPase regulator (RPGR)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J. Biol. Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 4.Mavlyutov T.A., Zhao H., Ferreira P.A. Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species. Hum. Mol. Genet. 2002;11:1899–1907. doi: 10.1093/hmg/11.16.1899. [DOI] [PubMed] [Google Scholar]
- 5.Castagnet P., Mavlyutov T., Cai Y., Zhong F., Ferreira P. RPGRIP1s with distinct neuronal localization and biochemical properties associate selectively with RanBP2 in amacrine neurons. Hum. Mol. Genet. 2003;12:1847–1863. doi: 10.1093/hmg/ddg202. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y., Hong D.H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:3965–3970. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzynski E., Lee Y., Sakamoto K., Naggert J.K., Nishina P.M. From vivarium to bedside: lessons learned from animal models. Ophthalmic. Genet. 2006;27:123–137. doi: 10.1080/13816810600977192. [DOI] [PubMed] [Google Scholar]
- 8.Booij J.C., Florijn R.J., ten Brink J.B., Loves W., Meire F., van Schooneveld M.J., de Jong P.T., Bergen A.A. Identification of mutations in the AIPL1, CRB1, GUCY2D, RPE65 and RPGRIP1 genes in patients with juvenile retinitis pigmentosa. J. Med. Genet. 2005;42:e67. doi: 10.1136/jmg.2005.035121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan J., Bonneau D., Frezal J., Munnich A., Dufier J.L. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum. Genet. 1990;85:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- 10.Perrault I., Rozet J.M., Gerber S., Ghazi I., Leowski C., Ducroq D., Souied E., Dufier J.L., Munnich A., Kaplan J. Leber congenital amaurosis. Mol. Genet. Metab. 1999;68:200–208. doi: 10.1006/mgme.1999.2906. [DOI] [PubMed] [Google Scholar]
- 11.Foxman S.G., Heckenlively J.R., Bateman J.B., Wirtschafter J.D. Classification of congenital and early onset retinitis pigmentosa. Arch. Ophthalmol. 1985;103:1502–1506. doi: 10.1001/archopht.1985.01050100078023. [DOI] [PubMed] [Google Scholar]
- 12.De Laey J.J. Leber's congenital amaurosis. Bull. Soc. Belge. Ophtalmol. 1991;241:41–50. [PubMed] [Google Scholar]
- 13.Dryja T.P., Adams S.M., Grimsby J.L., McGee T.L., Hong D.H., Li T., Andréasson S., Berson E.L. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am. J. Hum. Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber S., Perrault I., Hanein S., Barbet F., Ducroq D., Ghazi I., Martin-Coignard D., Leowski C., Homfray T., Dufier J.L., et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur. J. Hum. Genet. 2001;9:561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- 15.Ferreira P.A. Insights into X-linked RP3, allied diseases and underlying pathomechanisms. Hum. Mol. Genet. 2005;14:R259–R267. doi: 10.1093/hmg/ddi272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrow E.M., Furukawa T., Raviola E., Cepko C.L. Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC Neurosci. 2005;6:5. doi: 10.1186/1471-2202-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellersh C.S., Boursnell M.E., Pettitt L., Ryder E.J., Holmes N.G., Grafham D., Forman O.P., Sampson J., Barnett K.C., Blanton S., et al. Canine RPGRIP1 mutation establishes cone-rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics. 2006;88:293–301. doi: 10.1016/j.ygeno.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Turney C., Chong N.H., Alexander R.A., Hogg C.R., Fleming L., Flack D., Barnett K.C., Bird A.C., Holder G.E., Luthert P.J. Pathological and electrophysiological features of a canine cone-rod dystrophy in the miniature longhaired dachshund. Invest. Ophthalmol. Vis. Sci. 2007;48:4240–4249. doi: 10.1167/iovs.04-0737. [DOI] [PubMed] [Google Scholar]
- 19.Humphries M.M., Rancourt D., Farrar G.J., Kenna P., Hazel M., Bush R.A., Sieving P.A., Sheils D.M., McNally N., Creighton P., et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- 20.Jansen H.G., Sanyal S. Development and degeneration of retina in rds mutant mice: electron microscopy. J. Comp. Neurol. 1984;224:71–84. doi: 10.1002/cne.902240107. [DOI] [PubMed] [Google Scholar]
- 21.Palczewski K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farjo R., Naash M.I. The role of Rds in outer segment morphogenesis and human retinal disease. Ophthalmic. Genet. 2006;27:117–122. doi: 10.1080/13816810600976806. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg A.F. Role of peripherin/rds in vertebrate photoreceptor architecture and inherited retinal degenerations. Int. Rev. Cytol. 2006;253:131–175. doi: 10.1016/S0074-7696(06)53004-9. [DOI] [PubMed] [Google Scholar]
- 24.Morrow E.M., Furukawa T., Cepko C.L. Vertebrate photoreceptor cell development and disease. Trends Cell Biol. 1998;8:353–358. doi: 10.1016/s0962-8924(98)01341-5. [DOI] [PubMed] [Google Scholar]
- 25.Livesey F.J., Furukawa T., Steffen M.A., Church G.M., Cepko C.L. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr. Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 26.Blackshaw S., Fraioli R.E., Furukawa T., Cepko C.L. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 27.Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 28.Heckenlively J.R., Chang B., Erway L.C., Peng C., Hawes N.L., Hageman G.S., Roderick T.H. Mouse model for Usher syndrome: linkage mapping suggests homology to Usher type I reported at human chromosome 11p15. Proc. Natl. Acad. Sci. USA. 1995;92:11100–11104. doi: 10.1073/pnas.92.24.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J.Y. Rabphilin-3A is transported with fast anterograde axonal transport and associated with synaptic vesicles. Synapse. 1996;23:79–88. doi: 10.1002/(SICI)1098-2396(199606)23:2<79::AID-SYN3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 30.Hagstrom S.A., Duyao M., North M.A., Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 31.Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen H.G., Sanyal S., De Grip W.J., Schalken J.J. Development and degeneration of retina in rds mutant mice: ultraimmunohistochemical localization of opsin. Exp. Eye Res. 1987;44:347–361. doi: 10.1016/s0014-4835(87)80170-7. [DOI] [PubMed] [Google Scholar]
- 33.Nir I., Agarwal N., Papermaster D.S. Opsin gene expression during early and late phases of retinal degeneration in rds mice. Exp. Eye Res. 1990;51:257–267. doi: 10.1016/0014-4835(90)90022-m. [DOI] [PubMed] [Google Scholar]
- 34.Lee E.S., Burnside B., Flannery J.G. Characterization of peripherin/rds and Rom-1 transport in rod photoreceptors of transgenic and knockout animals. Invest. Ophthalmol. Vis. Sci. 2006;47:2150–2160. doi: 10.1167/iovs.05-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong D.H., Pawlyk B.S., Shang J., Sandberg M.A., Berson E.L., Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3) Proc. Natl. Acad. Sci. USA. 2000;97:3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lareau L.F., Green R.E., Bhatnagar R.S., Brenner S.E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Zavolan M., van Nimwegen E. The types and prevalence of alternative splice forms. Curr. Opin. Struct. Biol. 2006;16:362–367. doi: 10.1016/j.sbi.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Lu X., Ferreira P.A. Identification of novel murine- and human-specific RPGRIP1 splice variants with distinct expression profiles and subcellular localization. Invest. Ophthalmol. Vis. Sci. 2005;46:1882–1890. doi: 10.1167/iovs.04-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlyk B.S., Smith A.J., Buch P.K., Adamian M., Hong D.H., Sandberg M.A., Ali R.R., Li T. Gene replacement therapy rescues photoreceptor degeneration in a murine model of Leber congenital amaurosis lacking RPGRIP. Invest. Ophthalmol. Vis. Sci. 2005;46:3039–3045. doi: 10.1167/iovs.05-0371. [DOI] [PubMed] [Google Scholar]
- 40.Dryja T.P., Li T. Molecular genetics of retinitis pigmentosa. Hum. Mol. Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 41.Daiger S.P., Bowne S.J., Sullivan L.S. Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 2007;125:151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKusick V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thaung C., West K., Clark B.J., McKie L., Morgan J.E., Arnold K., Nolan P.M., Peters J., Hunter A.J., Brown S.D., et al. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum. Mol. Genet. 2002;11:755–767. doi: 10.1093/hmg/11.7.755. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto K., McCluskey M., Wensel T.G., Naggert J.K., Nishina P.M. New mouse models for recessive retinitis pigmentosa caused by mutations in the Pde6a gene. Hum. Mol. Genet. 2009;18:178–192. doi: 10.1093/hmg/ddn327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawes N.L., Smith R.S., Chang B., Davisson M., Heckenlively J.R., John S.W. Mouse fundus photography and angiography: a catalogue of normal and mutant phenotypes. Mol. Vis. 1999;5:22. [PubMed] [Google Scholar]
- 46.Hawes N.L., Chang B., Hageman G.S., Nusinowitz S., Nishina P.M., Schneider B.S., Smith R.S., Roderick T.H., Davisson M.T., Heckenlively J.R. Retinal degeneration 6 (rd6): a new mouse model for human retinitis punctata albescens. Invest. Ophthalmol. Vis. Sci. 2000;41:3149–3157. [PubMed] [Google Scholar]
- 47.Higuchi R. Rapid, efficient DNA extraction for PCR from cells or blood. Amplifications (Perkin Elmer Cetus) 1989;2:1–3. [Google Scholar]
- 48.Ferreira P.A. Characterization of RanBP2-associated molecular components in neuroretina. Methods Enzymol. 2000;315:455–468. doi: 10.1016/s0076-6879(00)15861-6. [DOI] [PubMed] [Google Scholar]
- 49.Rust S., Funke H., Assmann G. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 1993;21:3623–3629. doi: 10.1093/nar/21.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.