Mass spectrometry-based proteomics has become an indispensable tool in system biology generating a need for accurate and precise quantitation of peptide standards. The presented method utilizes ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) to accurately quantify peptide standards at concentrations of 0.1-10 μM. The ability for accurate quantitation of micro-molar concentrations has the advantages that quantitation can be performed routinely with high precision and the high sensitivity of the method minimizes the amounts required.
Mass spectrometry-based proteomics has evolved into an indispensable tool for molecular and cellular biology that will significantly contribute to the emerging field of systems biology. Early work focused on qualitative identification of proteins and peptides. More recently, there has been increasing interest in quantitative proteomics [1]. Progress in the area of mass-spectrometry-based proteomics has been made using stable isotopes. Stable isotopes can be introduced chemically by reacting peptide digests with an isotope tag (ICAT) [2], or the iTRAQ method [3;4] which currently allows the quantitative comparison of up to 8 samples in a single analysis. Whole proteomes can be labeled metabolically during cell culture (SILAC) by providing stable isotope precursors using either stable-isotope-labeled amino acids [1;5-8] or stable-isotope-labeled salts [9]. Although these isotopic labeling techniques are very powerful for biomarker discovery studies, their expense precludes their use in studies involving large numbers of samples, such as biomarker verification or validation. Moreover, SILAC can only be used in cell culture, and not for clinical studies.
Recently, there has been interest in label-free techniques, including GE Healthcare's DeCyder MS where two chromatograms are compared, the Empai scoring technique [10], ion accounting [11] and spectral counting [12;13], where the relative abundances of peptides are used to give the relative abundances of proteins. Without stable-isotope-labeled standards, however, the effect of other components in the samples can cause suppression effects, as has been noted in our FIA-based metabolomics studies using high resolution FTICR [14], and even in other studies utilizing LC-MS/MS separation [15]. This will probably mean that label-free methods will be limited to comparison of samples where the majority of the components do not change – i.e., a set of samples where only the concentrations of a few components change, and the matrix is effectively the same for all of the samples in the set.
A successful absolute quantitation of proteins can be achieved using double exact matching isotope dilution mass spectrometry (IDMS) as shown in the excellent paper of Burkitt et.al [16]. Alternatively, this can be applied using custom-synthesized stable-isotope labeled standard peptides [17-20]. These labeled standards can include peptides which contain post-translational modifications (for example, phosphorylation and chemical alkylation) [21]. We have studied globin alkylation by chemical carcinogens for several years, and have established assays for specific quantitation of alkylated peptides which can be used as biomarkers for exposure assessment [22;23]. These assays are based on proteolysis with trypsin, immunoaffinity enrichment, and LC-MS/MS. Recently, we used a similar approach to develop an immunoaffinity MALDI-MS/MS (iMALDI) assay for targeted protein quantitation [24].
During these studies, however, it became obvious that accurate quantitation of the standard peptides was challenging. The reason for this is that the amounts of standard peptides available are usually so small that accurate weights cannot be obtained. Peptides are hygroscopic and easily absorb moisture from the air, and the presence of water and salts leads to overestimation of peptide amounts. We and other researchers have been forced to use a combination of weight, HPLC-UV, and LC-MS/MS for quantitation of peptide standards [25]. However, this method is labor-intensive and is therefore not suitable for routine quantitation of peptides for multiplex proteomic applications, which may require thousands of standard peptides.
To overcome these limitations, we have developed a new approach for the peptide quantitation based on the analysis of their component amino acids after acid hydrolysis, using ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). The high specificity of MS/MS combined with fast chromatography makes this a suitable approach for high-throughput analyses.
Single standard amino acids were purchased from Sigma-Aldrich (St. Louis, MO), concentrated hydrochloric acid (12 N) from Fisher Scientific (Pittsburgh, PA), and all other reagents and solvents were ACS grade or higher. Amino acid standards for instrument calibration and the internal standard amino acid, norleucine, were prepared from single amino acid standards which are stable in 100 mM HCl solution. Stock solutions at 10 mM were made in water with 0.1 mM HCl. Norleucine was added to each calibration standard and samples at a final concentration of 2.5 mM.
The Peptides were synthesized at a 5 μmol scale using Fmoc chemistry on a Prelude peptide synthesizer (Protein Technology, Tucson, AZ). The C-terminal amino acids were conjugated to TentaGel R resin (Rapp Polymere, Tübingen, Germany) and subsequent residues, at a concentration of 100 mM, were double coupled using 20% piperidine as the deprotector and 1H-Benzotriazolium 1-[bis(dimethylamino)methylene]-5chloro-,hexafluorophosphate (1),3-oxide (HCTU) as the activator. Cleavage and deprotection was performed with 95:2.5:2.5 TFA:water: triisopropylsilane. The peptides were resolubilized in 0.1% TFA and purified by reverse-phase HPLC Ultimate 3000 (Dionex, Sunnyvale, CA) monitoring peptide elution at 230 nm using a Vydac C18 column (10 × 250 mm, 10μm resin) with a linear gradient of 0.1% TFA in water (v/v) and 0.85% TFA in 50% acetonitrile (v/v) at a flow rate of 4mL/minute over 60 minutes. The fractions of interest were spotted onto stainless steel MALDI plates and measured by MALDI-TOF (Applied Biosystems/MDS SCIEX, Foster City, CA) mass spectrometry. Fractions containing greater than 80% of the target peptide were pooled and lyophilized.
For HCl hydrolysis, peptide solutions were mixed with 10 μL of 20 μM norleucine, as internal standard, in a small culture vial and lyophilized in a vacuum centrifuge. The vials were placed inside a 10 mL glass container, 500 μL of 6N HCl were added to the bottom of the container, and the container was closed with an air tight PTFE lined cap. The vials were placed in an oven at 150°C for 4 hr, then cooled to room temperature for 30 minutes in a water bath to condense the acid vapor on the inside of the sample vials. Prior to analysis the hydrolysates were dried in a vacuum centrifuge for 15 min to remove traces of acid.
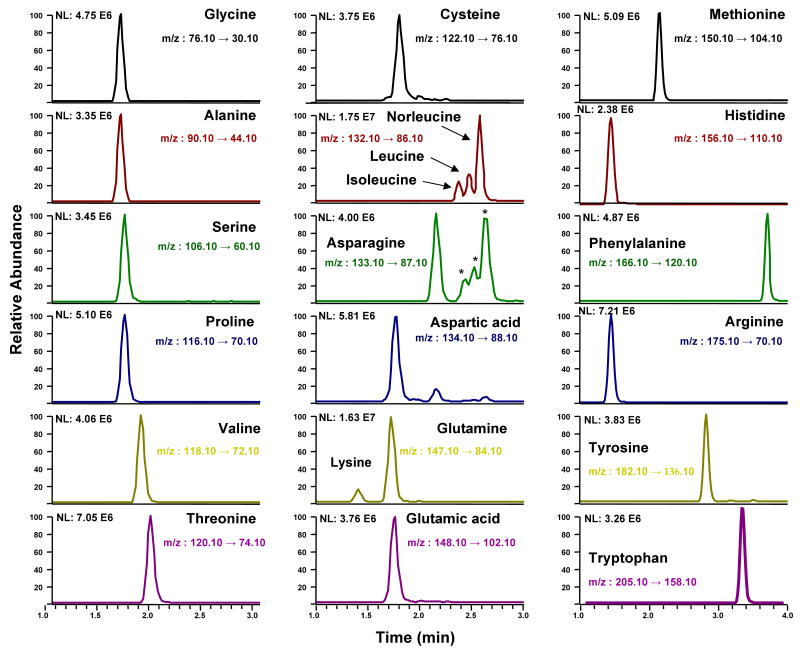
The quantitative analysis of hydrolyzed peptide was performed with an UPLC (Waters, Milford, MA) coupled to a TSQ-Quantum ultra triple quadrupole mass analyzer (ThermoFinnigan, San Jose, CA). A 2.1 mm × 150 mm UPLC™ BEH SHIELD RP18 1.7 μm column was operated with a linear gradient from 20% methanol-0.1% formic acid to 80% methanol-0.1% formic acid for 7 minutes at a flow rate of 200 μL/min. The individual amino acids were monitored in selected reaction monitoring mode (SRM) (Figure 1 and Supplemental Table S1). Amounts of amino acids were calculated from the area ratio of the corresponding ion transitions of amino acids to ion transition of the internal standard, norleucine. Calibration curves of the area of amino acid over area of norleucine were constructed daily, to guarantee linearity. Area ratios of analyte over internal standard were multiplied by the amount internal standard added and adjusted by a corresponding response factor to correct to difference in MS response. Calibration solution repeatedly showed linear response from 0.1 pmol up to 200 pmol/injection (high amount injected) representing 4 orders of magnitude.
Figure 1.
Extracted ion chromatograms for common amino acid standards and norleucine used as internal standard. * Indicates 13C-isotopes of isoleucine, leucine and norleucine.
We have shown that all 20 common amino acids can be detected by UPLC-MS/MS analysis without any derivatization step (Figure 1). It is also shown that while accurate quantitation of all amino acids is desirable, using 1 to 3, 5 or 6 amino acids for quantitation produced CVs of <17% and, the mean did not significantly change by increasing the number of amino acids used for quantitation. Thus we concluded that three AAs were sufficient for accurate quantitation of peptide solutions.
Method validation was assessed by analyzing the NIST standard peptide reference material 8327 three times on the same day and on three different days to obtain intraday and interday results (Supplemental Table S2). Therefore, hydrolyzed peptide solutions were initially quantified by the amounts of alanine, serine, proline, threonine, valine, leucine or isoleucine, if present. The mean relative standard deviations (RSD) for intraday and interday assay reproducibility (n=3) were 8% and 5%, respectively. Accuracies of the validation samples ranged from 95% to 110%.
To illustrate that the complete hydrolysis of the standard peptide had been acheived, the release of the free amino acids from the peptide was monitored over time. The ratio of amino acid that was released from the peptide to the internal standard amino acid (norleucine) that was added prior to hydrolysis was measured at several time points (1 hr, 2 hrs, 4 hrs, 8 hrs, 16 hrs, and 24 hrs) after the start of the acid hydrolysis. Duplicate samples were analyzed for each time point and the release of six amino acids was monitored. Results for alanine, serine, proline, valine, threonine, and leucine are shown in supplement Figure S1. The result of this time course experiment indicated that hydrolysis need to be performed at 4 hr since this time gave high yield of the six amino acids studied. Longer hydrolysis time (8-24 hrs) showed no difference in the yield.
UPLC-MS/MS was chosen for the analysis of peptide hydrolysates because of its high speed and resolution and the advantage of automation of sample preparation and processing. Initially, multiple stable isotope labeled amino acids were used as internal standards, but the addition of 20 extra ion transitions significantly reduces the number of points over the peaks. Comparison of results between multiple stable isotope label amino acid and single amino acid (Norleucine) as the internal standard was performed. The result of peptide quantitation using 4 amino acids, their isotope-labeled counterparts and norleucine are shown in supplement figures S2 & S3 and table S3. No difference was found in the value measured for the concentration of peptide. Therefore, we decided to use a single internal standard for all amino acids and to correct for different MS response using a response factor obtained from calibration solution made from single amino acid standards since this has been shown to be sufficient using the NIST standard peptide reference material (Table S3).
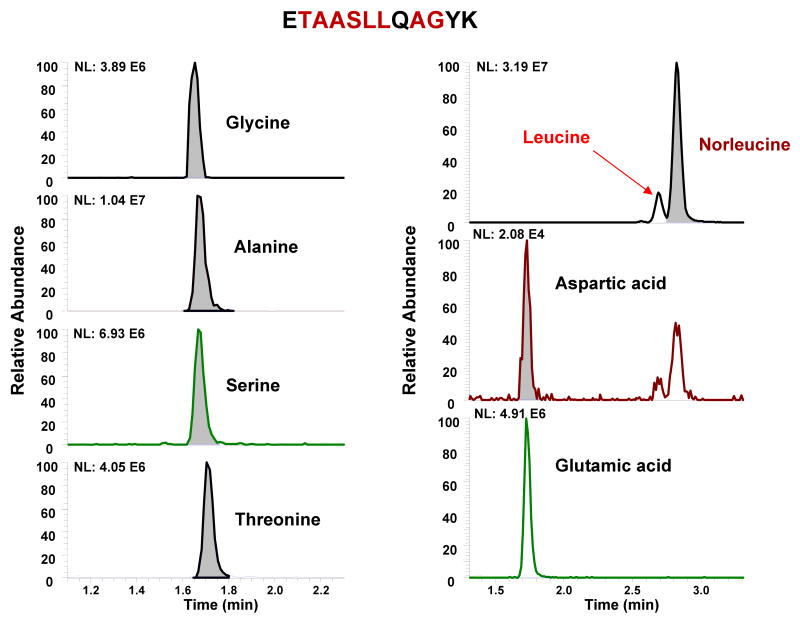
The study was extended to the analysis of standard peptides used for quantitative proteomic applications (Figure 2, and Supplemental Table S4). These peptides were synthesized for projects that required absolute quantitation of proteins in biological samples. Usually, the peptides are quantified by micro-weight and a minimum amount of 100 μg is required for these measurements. Quantitation by UPLC-MS/MS produces similar results to those previously determined by weight, but required less than 10 ng for accurate analysis. This represents a 10,000 fold increase in sensitivity without sacrifice of accuracy and specificity. Furthermore, ∼30% of the peptide concentrations were not accurate based on weight quantitation. Thus, there is an unacceptable and unavoidable uncertainty using weight for standardization.
Figure 2.
Representative extracted ion chromatograms of peptide hydrolysate determined by UPLC-MS/MS.
The present method is about 5-fold more sensitive than a previously reported LC-MS/MS method for underivatized amino acids [14]. Alternatively, Amino acids analysis can be performed accurately at the femtomole level by a method employing fluorescence detection after derivatization with o-phthalaldehyde (OPA). However, inter-laboratories comparisons have shown high variability in the results due to inconsistency in derivatization conditions [15].
These discrepancies between the methods and laboratories are probably caused by impurities (such as TFA salts) of the peptides and absorption of moisture during peptide handling. Precautions can be implemented to better assure purity of the standard peptides; however, as the number of peptide standards increases, it will be difficult to maintain high purity and accuracy due to the chemical nature of peptides. Therefore, the ability of accurate quantitation of peptide solutions at micromolar concentrations, instead of dry material has the advantage of avoiding inaccuracy due to weighing, chemical derivatization and errors during preparation of dilutions. In addition, the increased sensitivity reduces the amounts needed for analysis and reduces costs, especially if chemically unique peptides are needed.
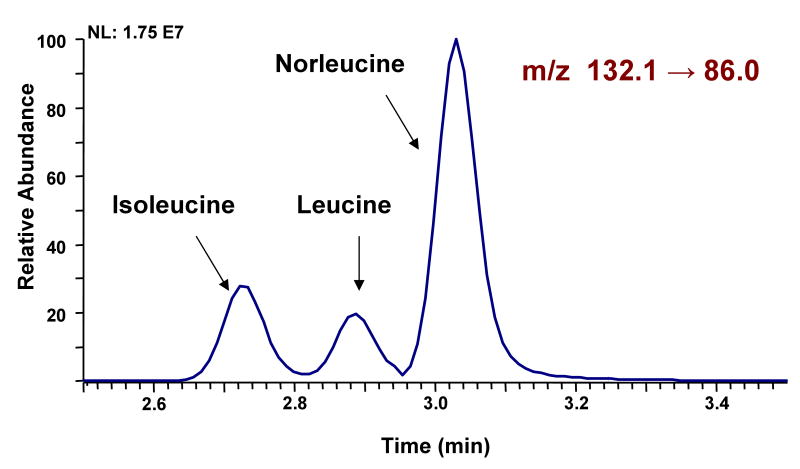
One of the problems with the MS/MS methodology is the fact that isobaric amino acids, such as leucine, isoleucine, and norleucine often fragment very similarly. This limitation was overcome by utilization of UPLC which achieves baseline separation for these amino acids (Figure 3). In addition asparagine and aspartic acid were separated to avoid cross peaks form natural 13C isotopes (Figure 1). Unfortunately, the reported fast separation did not separate glutamine and glutamic acid and therefore glutamic acid cannot be used for quantitation if the peptides also contain glutamine. Further, the method was optimized for speed and analysis of known peptides. Therefore, in the current approach is unsuitable for analysis of amino acid content in unknown proteins or biological specimens. For such traditional amino acid analyses a modified UPLC gradient can be applied to separate all amino acids by time (Data not shown).
Figure 3.
Separation of isobaric amino acids leucine, isoleucine, and norleucine by UPLC-MS/MS.
Peptide concentrations of 0.1-10 μM could be analyzed with this method using as little as 10 μl. This new method is more sensitive than other LC-MS/MS [14] and common methods like ion chromatography (IC) using ninhydrin post column derivatization [26], and gas chromatography/mass spectrometry (GC/MS) [27]. Moreover, the specificity of single reaction monitoring mode allowed specific analysis of twenty amino acids in less than 10 minutes per sample, making it suitable for high through put analysis.
In conclusion, amino acid quantitation of peptide solutions by UPLC-MS/MS is a viable alternative to traditional amino acid analysis methods or quantitation by weight. The UPLC-MS/MS method utilizes a few simple steps that can be completed within 6 hr, which represents a great advantage over previous GC/MS and IC/ninhydrin methods requiring derivatization and numerous steps for sample preparation. The ability to accurately quantitate μM solutions of peptides significantly reduces the amounts needed and allows for high throughput quantitative analysis of peptide standards. This method is expected to become a standard repertoire in proteomic laboratories.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant P30 ES10126, P42-ES05948, and Genome Canada.
References
- 1.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 2.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 5.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 6.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. PNAS. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong SE. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Robert FB, Jr, Yvan RP. Stable isotope metabolic labeling for analysis of biopolymers. PCT Int Appl. 2000 WO1999US19434. 2000. [Google Scholar]
- 9.Chait BT, Cowbrun D, Oda Y. Method for the comparative quantitative analysis of proteins and other biological material by isotopic labeling and mass spectroscopy. PCT Int Appl. 2000 WO0013025 (A1) [Google Scholar]
- 10.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Tha Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3:354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Danell RM, Patel JR, Gumerov DR, Scarlett CO, Speir JP, Parker CE, Rusyn I, Zeisel S, Borchers CH. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 2008;4:128–140. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alderman JM, Flurkey K, Brooks NL, Naik SB, Gutierrez JM, Srinivas U, Ziara KB, Jing L, Boysen G, Bronson R, Klebanov S, Chen X, Swenberg JA, Stridsberg M, Parker CE, Harrison DE, Combs TP. Neuroendocrine inhibition of glucose production and resistance to cancer in dwarf mice. Exp Gerontol. 2009;44:26–33. doi: 10.1016/j.exger.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkitt WI, Pritchard C, Arsene C, Henrion A, Bunk D, O'Connor G. Toward Systeme International d'Unite-traceable protein quantification: from amino acids to proteins. Anal Biochem. 2008;376:242–251. doi: 10.1016/j.ab.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 18.Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, Vandenesch F, Garin J. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007;6:2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Dupuis A, Hennekinne JA, Garin J, Brun V. Protein Standard Absolute Quantification (PSAQ) for improved investigation of staphylococcal food poisoning outbreaks. Proteomics. 2008;8:4633–4636. doi: 10.1002/pmic.200800326. [DOI] [PubMed] [Google Scholar]
- 20.Gerber SA, Kettenbach AN, Rush J, Gygi SP. The absolute quantification strategy: application to phosphorylation profiling of human separase serine 1126. Methods Mol Biol. 2007;359:71–86. doi: 10.1007/978-1-59745-255-7_5. [DOI] [PubMed] [Google Scholar]
- 21.Pflieger D, Junger MA, Muller M, Rinner O, Lee H, Gehrig PM, Gstaiger M, Aebersold R. Quantitative proteomic analysis of protein complexes: concurrent identification of interactors and their state of phosphorylation. Mol Cell Proteomics. 2008;7:326–346. doi: 10.1074/mcp.M700282-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Boysen G, Georgieva NI, Upton PB, Jayaraj K, Li Y, Walker VE, Swenberg JA. Analysis of diepoxide-specific cyclic N-terminal globin adducts in mice and rats after inhalation exposure to 1,3-butadiene. Cancer Research. 2004;64:8517–8520. doi: 10.1158/0008-5472.CAN-04-3184. [DOI] [PubMed] [Google Scholar]
- 23.Georgieva NI, Boysen G, Upton PB, Jayaraj K, Gold A, Swenberg JA. Quantitative analysis of N-terminal valine peptide adducts specific for 1,2-epoxy-3-butene. Chem Biol Interact. 2007;166:219–225. doi: 10.1016/j.cbi.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Parker CE, Fuller JR, Kawula TH, Borchers CH. An immunoaffinity tandem mass spectrometry (iMALDI) assay for detection of Francisella tularensis. Anal Chim Acta. 2007;605:70–79. doi: 10.1016/j.aca.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fred C, Kautiainen A, Athanassiadis I, Törnqvist M. Hemoglobin adduct levels in rat and mouse treated with 1,2:3,4-diepoxybutane. Chem Res Tox. 2004;17:785–794. doi: 10.1021/tx034214g. [DOI] [PubMed] [Google Scholar]
- 26.Macchi FD, Shen FJ, Keck RG, Harris RJ. Amino acid analysis, using postcolumn ninhydrin detection, in a biotechnology laboratory. Methods Mol Biol. 2000;159:9–30. doi: 10.1385/1-59259-047-0:009. [DOI] [PubMed] [Google Scholar]
- 27.Namera A, Yashiki M, Nishida M, Kojima T. Direct extract derivatization for determination of amino acids in human urine by gas chromatography and mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;776:49–55. doi: 10.1016/s1570-0232(02)00075-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.