Abstract
As an adhesion receptor, the β2 integrin lymphocyte function-associated antigen-1 (LFA-1) contributes a strong adhesive force to promote T lymphocyte recirculation and interaction with antigen-presenting cells. As a signaling molecule, LFA-1-mediates transmembrane signaling, which leads to the generation of second messengers and costimulation resulting in T cell activation. We recently have demonstrated that, in costimulatory fashion, LFA-1 activation promotes the induction of T cell membrane urokinase plasminogen activator receptor (uPAR) and that this induced uPAR is functional. To investigate the mechanism(s) of this induction, we used the RNA polymerase II inhibitor 5,6-dichloro-1-β-d-ribobenzimidazole and determined that uPAR mRNA degradation is delayed by LFA-1 activation. Cloning of the wild-type, deleted and mutated 3′-untranslated region of the uPAR cDNA into a serum-inducible rabbit β-globin cDNA reporter construct revealed that the AU-rich elements and, in particular the nonameric UUAUUUAUU sequence, are crucial cis-acting elements in uPAR mRNA degradation. Experiments in which Jurkat T cells were transfected with reporter constructs demonstrated that LFA-1 engagement was able to stabilize the unstable reporter mRNA containing the uPAR 3′-untranslated region. Our study reveals a consequence of adhesion receptor-mediated signaling in T cells, which is potentially important in the regulation of T cell activation, including production of cytokines and expression of proto-oncogenes, many of which are controlled through 3′ AU-rich elements.
The integrin family of cell surface receptors mediates attachment to the extracellular matrix, as well as cell–cell adhesive interactions. Numerous consequences of integrin-mediated adhesion have been described, including cell growth, differentiation, apoptosis, cell shape change and cytoskeletal reorganization, leukocyte homing and activation, and gene expression (reviewed in ref. 1). The leukocyte integrin lymphocyte function-associated antigen-1 (LFA-1) (αLβ2, CD11a/CD18) is thought to play a major role in T lymphocyte function. In addition to providing part of the multi-step foundation for lymphocyte recirculation and transendothelial migration, LFA-1 binding to its antigen presenting cell-bound ligands intercellular adhesion molecule-1,-2, and sometimes -3 is required to strengthen the weak T cell receptor recognition of the MHC-peptide complex. This adhesion receptor engagement, in turn, contributes transmembrane signaling to the T cell activation process. Signaling through the T cell receptor-CD3 complex is essential to initiate T cell responses but is usually weak and transient, requiring costimulatory signals. The classical costimulator molecule, CD28, provides important synergistic signals to T cells, via its interaction with the costimulatory ligands B7–1 and -2. Functional responses of this costimulation include augmentation of T cell cytokine production and proliferation (2). Similarly, costimulation of OKT3- or antigen-primed memory T cells with recombinant intercellular adhesion molecule-1 (i.e., engagement of LFA-1) can lead to vigorous interleukin 2 production and proliferation (3), as well as induction of other T cell membrane molecules, such as CTLA-4 (4). Costimulation via LFA-1 can lead to expression of the activation antigens CD25 and CD69 (5, 6) as well as the biosynthesis and expression of tumor necrosis factor (3). Cooperativity between integrins and growth promoting pathways has been demonstrated in a number of experimental settings (7). Whether such cooperativity involves truly independent pathways, mutual permissive actions, or synergism is presently unclear.
We have previously demonstrated that T cell LFA-1 engagement results in phosphoinositide hydrolysis and a rise in [Ca2+]i (8). Others have shown that LFA-1 mediates a tyrosine kinase-dependent activation of phospholipase Cγ1 (9) with consequent activation of protein kinase C. To address other functional consequences of LFA-1-mediated transmembrane signaling in T cells, we screened T cells, stimulated by anti-CD3 plus anti-LFA-1 antibodies, for activation phenotypes by flow cytometry. We recently described the induction of urokinase plasminogen activator receptor (uPAR) mRNA and protein by antibody-mediated engagement of CD3 and LFA-1 (10). We demonstrated that this form of lymphocyte activation, mediated in part via LFA-1, enhances T cell invasion of Matrigel-based matrices, specifically through use of induced uPAR.
Prior reports have demonstrated that CD28-mediated costimulation in T cells leads to a stabilization of cytokine mRNAs (11). To address whether transmembrane signaling through LFA-1 could result in a similar effect on uPAR expression, we activated the human T cell leukemia line Jurkat with the aforementioned costimulation protocol. As with primary T cells, Jurkat uPAR is inducible at the mRNA and protein level. Transcription inhibitor experiments demonstrated that LFA-1 has a major uPAR mRNA-stabilizing effect. A serum-inducible cDNA construct encoding the stable rabbit β-globin reporter mRNA was used in a series of Jurkat transfection experiments. Modification of this construct to include the 3′-untranslated region (3′ UTR) of uPAR, and its mutated variants, demonstrated that the classical nonameric UUAUUUAUU sequence (12, 13) plays an important role in uPAR mRNA degradation. Engagement of LFA-1 on these transfected cells conferred stability to the 3′ UTR-containing, unstable reporter mRNA. We discuss the costimulatory role of adhesion receptor engagement in T cell activation, through marked effects on mRNA degradation.
MATERIALS AND METHODS
Cell Lines.
The Jurkat T cell leukemia line was obtained from the American Type Culture Collection. For cytokine assays, human peripheral T cells were freshly isloated, after leukopheresis of normal blood donors, as described (10). After monocyte, B cell, and natural killer cell depletion via adherence, nylon wool columns and panning, the purified population was >97% CD3+.
Antibodies and Chemicals.
Mouse anti-human CD18 (LFA-1 β subunit) mAb (clone TS1/18) was purchased from Endogen (Cambridge, MA), and mouse anti-human CD11a (LFA-1 α subunit) mAb (clone TS1/22) was purified from ascites generated in our laboratory. Mouse anti-human CD3 mAb (clone UCHT-1) was purchased from Immunotech (Westbrook, ME). Rabbit anti-human uPAR antibody was generated in our laboratory (F.B.). The 5,6-dichloro-1-β-d-ribobenzimidazole (DRB) was purchased from Sigma.
T Cells Activation.
Cells were incubated with murine anti-CD3 mAb (0.1 μg/107 cells) and/or anti-LFA-1α (1:40 dilution of ascites) plus anti-LFA-1β (1.5 μg/107 cells) mAbs on ice for 20 min. Excess antibodies were washed out, and cells were plated on goat anti-mouse-coated plates at 4°C for 60 min. The unbound cells were aspirated, and bound cells were cultured in medium at 37°C for the indicated times, after which bound cells were recovered by vigorous pipetting.
Plasmid DNA Constructs.
Plasmids pBBB, containing the rabbit β-globin cDNA under the control of the c-fos promoter, and pEF-BOS-CAT for transfection normalization, were kindly provided by J. Belasco (Harvard Medical School, Boston) and have been described (14, 15). All pBBB-derived constructs were made by PCR amplification of the uPAR 3′ UTR from cellular genomic DNA, digestion with restriction enzymes BamHI and BglII, and insertion into the BglII site of pBBB. All PCR products used a common 5′ primer corresponding to nucleotides 1040–1057 of human uPAR cDNA sequence, upstream of the BamHI site at position 1084. The complementary PCR primers were as follows: (i) For construct pBBB-3′ uPAR, a primer corresponding to the complement of nucleotides 1338–1363 was synthesized, which included a mutation at position 1351 to generate a BglII site at position 1347; (ii) for construct pBBB-DE, a primer corresponding to the complement of nucleotides 1289–1315, including mutations at positions 1300, 1302, and 1304, was designed to generate a BglII site at position 1304, and the amplified fragment did not contain the nonameric mRNA degradation sequence; and (iii) pBBB-MU was constructed using a primer corresponding to the complement of nucleotides 1307–1363, which contained a mutation at position 1351 to generate a BglII site at position 1347, and also mutations at positions 1324, 1325, 1331, and 1332, to mutate the dinucleotides at both ends of the nonameric mRNA degradation sequence to G, C, G, and C, respectively.
Transfections.
The 108 Jurkat T cells were incubated at 37°C with 20 μg of plasmid DNA in TS buffer (16) in the presence of 200 μg/ml DEAE-dextran for 24 h. For LFA-1 activation experiments, transfected cells were panned with mAb before resuspension in medium containing 20% fetal bovine serum.
Northern Blot Analysis.
Twenty-five micrograms of total RNA or 1 μg of poly(A)+ RNA was subjected to Northern analysis as described (17). Normalization was performed by densitometric analysis of the same filters hybridized with a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Ribonuclease Protection Assay (RPA).
Rabbit β-globin (224 nt) and chloramphenicol acetyltransferase (CAT) (150 nt) probes were synthesized by using a MAXIscript in vitro Transcription Kit (Ambion, Austin, TX) and have been described (14). Full length probes were gel-purified and hybridized with 30 μg of total RNA, according to the manufacturer’s protocol, by using a RPA II kit (Ambion). Protected fragments were separated on 5% acrylamide/8 M urea gels and quantitated as described for Northern analysis, using a probe specific for the mRNA generated from the cotransfected pEF-BOS-CAT plasmid.
RESULTS
Jurkat Costimulation via LFA-1 Results in uPAR Induction at the mRNA and Protein Levels.
As we have shown with primary T lymphocytes (10), there is no detectable uPAR expression in unstimulated Jurkat cells. Flow cytometry after our two-step activation protocol demonstrated that anti-CD3- plus anti-LFA-1-treated cells express membrane uPAR, whereas singly anti-CD3- or anti-LFA-1-activated cells remain uPAR-negative (not shown). Flow cytometric analysis in permeabilized Jurkat failed to detect uPAR in resting cells, demonstrating that the membrane expression represents a de novo induction and not a translocation of preformed protein. Similar results were obtained when using a natural β2 integrin ligand, human intercellular adhesion molecule-1, stably expressed in a murine fibroblast cell line, to engage LFA-1 (data not shown).
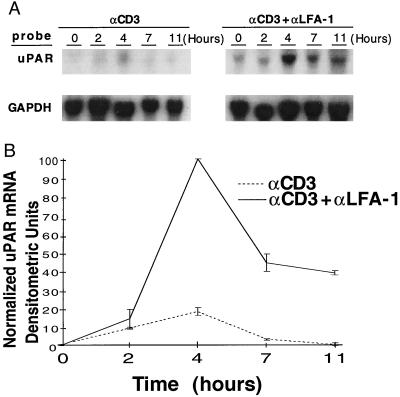
Northern blot analyses also were performed on antibody-activated cells to determine differences in mRNA levels and kinetics following Jurkat activation. CD3 stimulation alone induced a small rise in uPAR message, peaking at 4 h and rapidly returning to negligible levels by 7 h (Fig. 1A). In contrast, the costimulation through CD3 and LFA-1 resulted in a large increase (>10-fold) in uPAR mRNA, also peaking at 4 h. Of note is that substantially increased levels of message persist at 11 h in the costimulated cells, raising the possibility that LFA-1 engagement extends the half-life of the induced mRNA. These results are displayed graphically in Fig. 1B. LFA-1 engagement alone failed to increase uPAR mRNA levels (data not shown).
Figure 1.
LFA-1-mediated costimulatory effects on uPAR mRNA expression. (A) Jurkat cells were panned with the noted antibody combinations, and total RNA was harvested for Northern analysis at the indicated time points. (B) Curves display relative GAPDH-normalized, uPAR mRNA densitometric units and are representative of four separate experiments, with mean values ± SE.
LFA-1 Engagement Stabilizes Activation mRNAs.
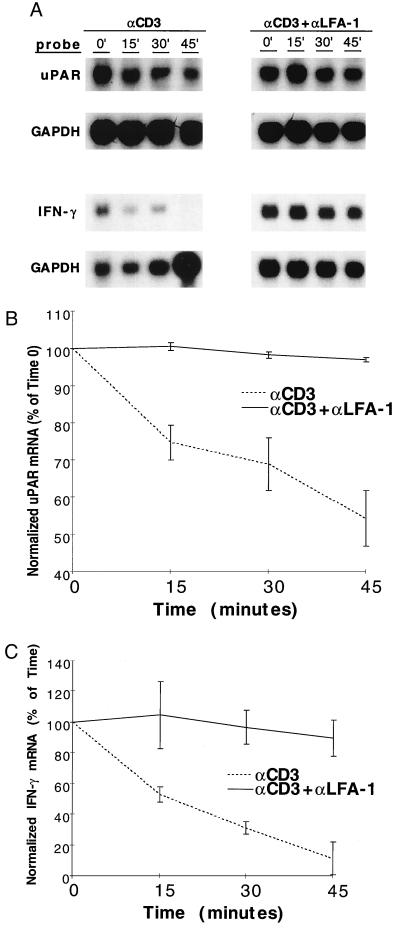
To address the effect of LFA-1 engagement on uPAR mRNA stability, Jurkat cells were arrested transcriptionally with DRB after induction of the initial transcripts. DRB was added 3 h after antibody treatment, after which RNA was harvested at various time points to determine the decay curves. Fig. 2 A and B demonstrate that induced uPAR mRNA decays rapidly in anti-CD3-treated cells, falling to 60% of maximal levels within 45 min of DRB treatment. However, the addition of anti-LFA-1 mAbs stabilized the mRNA, with no significant reduction in message level at the same 45-min time point. The data are represented out to 45 min because it is within these early time points that the most dramatic differences in uPAR mRNA stability are observed. This result is consistent with prior reports in which other costimulatory signaling events, namely CD28-mediated, impart dramatic stabilization of cytokine and proto-oncogene mRNAs at early time points (11).
Figure 2.
Effect of LFA-1 engagement on T cell activation mRNA degradation. (A) Jurkat cells (for uPAR) or peripheral T cells (for IFN-γ) were treated with anti-CD3 or anti-CD3 plus anti-LFA-1 antibodies, panned, and incubated × 3 h at 37°C, after which transcription was arrested by the addition of DRB (0.2 mM). Antibody concentrations were as per Materials and Methods section, except that anti-CD3 0.5 μg/107 cells (five times standard protocol) were used to generate higher uPAR mRNA levels with anti-CD3 alone, allowing more easily interpretable decay curves. Total RNA was harvested at the indicated time points and used for uPAR, IFN-γ, and GAPDH Northern blots. Data displayed are representative of results from five separate experiments. Northern signals were densitometrically analyzed and displayed as % of maximal (time 0) GAPDH-normalized, densitometric units with mean values ± SE, for uPAR (B) and IFN-γ (C).
To evaluate whether LFA-1 engagement might affect other T cell activation transcripts, a variety of induced cytokine mRNAs were assessed in similar degradation assays. As shown in Fig. 2 A and C, the half-life of induced IFN-γ transcripts was dramatically prolonged by LFA-1 mediated activation. Similar results were obtained for induced granulocyte–macrophage colony-stimulating factor and tumor necrosis factor mRNAs but not c-myc (data not shown). These results demonstrate that β2 integrin adhesion receptor engagement enhances T cell activation molecule expression through mRNA stabilization. An additional effect of LFA-1 on transcription has not been excluded.
uPAR mRNA Degradation Is Regulated by an AU-Rich Element (ARE) in its 3′ UTR.
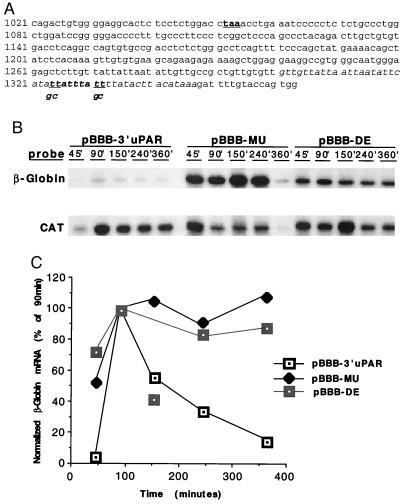
Different classes of highly conserved AU-rich motifs have been identified in the 3′ UTR of short-lived mRNAs. Among these motifs, the nonamer UUAUUUA(U/A)(U/A) has been identified as the shortest AU-rich sequence capable of independently mediating mRNA degradation (12, 13). The uPAR mRNA has an AU-rich region of ≈50 nt in length, located ≈250 nt downstream of the stop codon (see cDNA sequence, Fig. 4A). Within the ARE, exists the UUAUUUAUU nonamer overlapping with another sequence, UUAUUUUAUA, both located very close to the poly(A) tail. To determine whether the uPAR 3′ UTR could effect an alteration in mRNA stability, an mRNA reporter system was used in a series of Jurkat transfection experiments. The Jurkat cells were serum-starved after transfection, after which mRNA was harvested at various time points after transient serum enhancement of transcription. Fig. 3 displays the mRNA levels detected by Northern analysis out to 6 h after serum induction. It has been shown previously that there is some constitutive c-fos promoter activation in T lymphocytes (18). Thus, given the marked stability of the wild-type β-globin mRNA, there is an insignificant induction by serum in Jurkat. However, it is evident that the existing β-globin message is stable in the pBBB-transfected cells (Fig. 3 A and B). Given the presumed instability of the mRNA derived from the pBBB 3′ uPAR construct, leading to much lower constitutive message levels, the serum inducibility is apparent, with a peak accumulation of the reporter mRNA at 1.5 h after the addition of serum. More importantly, the uPAR 3′ UTR conferred marked instability to the β-globin mRNA, which was undetectable at 6 h.
Figure 4.
Degradation of β-globin mRNA containing wild-type or mutant uPAR 3′ UTR. (A) The 3′ uPAR cDNA, with stop codon (taa) bold and underlined. Sequence 1084–1347 was subcloned into pBBB to generate pBBB-3′ uPAR. AU-rich region is italicized and deleted in pBBB-DE. The nonameric degradation motif is italicized and bolded. The underlined bases (t) in this sequence were mutated to g and c, as indicated, to generate pBBB-MU. (B) Jurkat cells were cotransfected with pEF-BOS-CAT and either pBBB 3′ uPAR, pBBB-MU, or pBBB-DE. Cells were fetal bovine serum-stimulated after a 24-h serum starvation, after which total RNA was harvested at the indicated time points for ribonuclease protection assay. Findings were similar in three separate experiments. (C) β-globin RPA signals from B were densitometrically analyzed and normalized to CAT signals. Isolated data point in the pBBB-DE curve (150 min) represents an aberrant experimental sample, not reproduced in other similar experiments.
Figure 3.
Effect of uPAR 3′ UTR on degradation of a stable mRNA. (A) Jurkat cells were cotransfected with the pEF-BOS-CAT normalization construct and either pBBB or pBBB 3′ uPAR. Cells were fetal bovine serum-stimulated after a 24 h serum starvation, after which poly(A)+ RNA was isolated at the indicated time points for sequential β-globin and CAT Northern analyses. The larger β-globin mRNA size in the pBBB 3′ uPAR samples reflects the additional 263 nt of 3′ uPAR cloned into the pBBB construct. Results are representative of six separate experiments. (B) β-globin Northern signals from A were densitometrically analyzed and normalized to CAT signals. Data represent percentage of maximal (90 min) counts.
To pinpoint the critical region of the uPAR 3′ UTR effecting reporter mRNA degradation, two mutations of the pBBB 3′ uPAR were made. pBBB-DE (see Fig. 4A) has the entire ARE (1301–1347) deleted whereas pBBB-MU (see Fig. 4A) includes substitution mutations of the two uridine residues for guanosine and cytidine at both ends of the critical nonamer. Fig. 4 B and C demonstrate that both mutations abolish the rapid degradation imposed by the uPAR 3′ UTR and support that the nonameric sequence within the AU-rich region is a key element in mRNA degradation. Comparing cloned uPAR cDNAs from human, cow, rat, and mouse, we identified both a conserved 3′ UTR AU-rich region and a common UUAUUUA(U/A)(U/A) motif very close to the poly(A) tail. The highly conserved ARE and nonameric sequence indicate that regulation of mRNA degradation may be a commonly shared mechanism in the induction of uPAR protein across many species.
LFA-1 Engagement Stabilizes the 3′ UTR Degradation Signal-Containing Reporter mRNA.
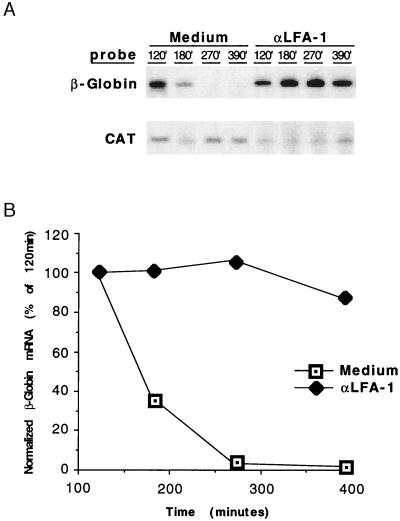
To directly determine whether β2 integrin engagement can overcome the mRNA-destabilizing effect of the uPAR ARE, Jurkat cells transfected with the pBBB-3′ uPAR were activated with anti-LFA-1 antibodies. Transfected, serum-starved cells were incubated with anti-LFA-1 antibodies and panned onto goat anti-mouse-coated dishes before serum enhancement of transcription. Fig. 5 demonstrates the marked and complete degradation of the uPAR 3′ UTR-bearing reporter mRNA in control cells, as shown above. However, the β-globin message remained stable for >6 h after serum addition in cells treated with anti-LFA-1 antibodies. Of note is that CD3-mediated activation (anti-CD3 antibody) was not required in these experiments because the reporter gene driven by the c-fos promoter is constitutively transcribed and enhanced by serum stimulation. The serum-driven transcription is in contrast with endogenous uPAR induction in T cells, which requires CD3 activation to apparently initiate transcription followed by the LFA-1 costimulus to stabilize the mRNA. To exclude the possibility that higher levels of β-globin transcripts in these LFA-1-activated cells were a consequence of enhanced transcriptional activation of the c-fos promoter, nuclear run-on experiments were performed on nuclei harvested from control and anti-LFA-1-stimulated cells. There was no detectable difference in transcription rate (normalized nuclear transcripts) between control and LFA-1-activated cells (data not shown). Thus, the major effect of LFA-1 engagement on the accumulated steady–state levels of the reporter mRNA is posttranscriptional, influencing decay mediated through 3′ UTR AREs.
Figure 5.
LFA-1 engagement stabilizes β-globin mRNA containing the uPAR 3′ ARE. (A) Jurkat T cells were cotransfected with pBBB-3′ uPAR and pEF-BOS-CAT. Cells were serum starved for 24 h, the last 1 h of which included panning with anti-LFA-1 mAbs. Cells were then fetal bovine serum-stimulated at 37°C and total RNA harvested at the indicated time points for RPA. Three separate experiments yielded identical results. (B) β-Globin RPA signals from A were densitometrically analyzed and normalized to CAT signals.
DISCUSSION
Although the exact mechanism(s) whereby costimulatory molecules exert their effects has not been established, it has been shown recently that engagement of the T cell costimulator CD28 can induce stabilization of a variety of cytokine mRNAs, including interleukin 2, interferon-γ, tumor necrosis factor, and granulocyte–macrophage colony-stimulating factor (11). Cell adhesion also has been shown to impart RNA stabilization signals. In a recent study, monocyte adhesion was demonstrated to increase GROα mRNA half-life, apparently through modulation of protein binding to its 3′ UTR (19). In this report, we provide direct evidence that transmembrane signaling via LFA-1 can alter T cell activation gene expression levels by influencing mRNA decay. Specifically, LFA-1 engagement stabilizes the mRNA of an induced, activation-dependent, and functionally relevant T cell membrane molecule, uPAR. uPAR mRNA appears intrinsically unstable in T lymphocytes, based on its AU-rich region in the 3′ UTR. LFA-1-mediated activation can overcome this intrinsic instability, greatly augmenting the half-life of a reporter mRNA containing the uPAR 3′ ARE.
mRNA degradation is a regulated process that is initiated by shortening of the 3′ poly(A) tails in the decay of cytokine and labile early response gene mRNAs. Functional ARE motifs mediate deadenylation as the first decay step but exhibit distinct reaction kinetics (20). Two types of motifs, coupled to or located in U-rich regions, have been characterized and may be differentially regulated. Type I motifs have multiple copies of AUUUA whereas the sequence UUAUUUA(U/A)(U/A) defines type II motifs and facilitates binding of specific protein factors to AREs (20). The decameric sequence UUAUUUUAUU is also thought to be a functional degradation motif (21). As noted above, the ARE and, more specifically, the type II UUAUUUAUU nonamer is conserved in uPAR genes from multiple species. This element is highly destabilizing but overcome by LFA-1 engagement, suggesting that signals imparted via LFA-1 activation modulate the effect imposed by this element. LFA-1 clustering induces a cascade of intracellular signaling events, including PLCγ1 activation with consequent IP3 and DAG generation, leading to Ca2+ mobilization and protein kinase C activation, which have been shown to regulate type II AREs (22, 23). It is possible that protein kinase C activation induces stabilizing or inhibits destabilizing factors, which directly bind to the critical motif, thereby prolonging uPAR mRNA half-life. This model is consistent with our prior result that the protein kinase C inhibitor bisindolylmaleimide abrogates T cell membrane uPAR expression and increases uPAR steady-state mRNA levels induced by anti-CD3 plus anti-LFA-1 (10).
The mRNA stabilization induced by LFA-1 engagement appears to be a general mechanism whereby adhesion receptor-mediated costimulation enhances T cell activation responses. In addition to the prolongation of uPAR mRNA half-life, labile transcripts encoding several cytokines also were stabilized by LFA-1 engagement. These include transcripts for IFN-γ, granulocyte–macrophage colony-stimulating factor, and tumor necrosis factor. Of note is that the induced stabilization of these cytokine mRNAs appears more dramatic than that observed for uPAR. uPAR mRNA is intrinsically less unstable than the noted cytokine transcripts, likely because of the greater number of type II degradation motifs expressed in the cytokine 3′ UTRs. Thus, the stabilization imparted through these motifs is quantitatively greater for the less stable transcripts. Other labile, CD3-induced transcripts that do not contain type II motifs, such as c-myc, were not significantly stabilized by LFA-1 engagement, further suggesting that these sequences are specific targets of induced stabilization.
Although our work emphasizes the role of this type II ARE motif in uPAR expression, there may be other important posttranscriptional, dynamically regulatable mechanisms. A recent report revealed a determinant of mRNA turnover within the coding region (nucleotides 195–246) of uPAR (24). This sequence was found to constitutively bind a 50-kDa protein isolated from a mesothelioma cell line, termed uPAR mRNABp. Inflammatory mediators such as tumor necrosis factor, transforming growth factor-β, lipopolysaccharide, and phorbol ester all augment uPAR mRNA levels and abrogate the uPAR mRNA–uPAR mRNABp interaction. Thus, it is likely that the control of uPAR mRNA destabilization occurs via multiple elements and, perhaps, multiple distinct mechanisms.
Integrins, through interactions with matrix ligands, have been shown to influence gene expression, largely through kinase cascades modulating transcription factor complexes (25, 26). β1 integrin engagement can synergize with growth factor responses in fibroblasts, leading to stabilization of various α integrin mRNAs (27). We have previously shown that, as with LFA-1 activation, β1 integrin engagement also results in T cell uPAR induction (10). We now describe a mechanism whereby leukocyte integrins, in addition to their role in cell–cell adhesion, can costimulate T cells to express a functionally relevant activation molecule, through stabilization of its otherwise rapidly degraded mRNA. This process may define an important role for adhesion receptors on immune cells, leading to prolonged expression of early response and cytokine genes during immune responses.
Acknowledgments
We gratefully acknowledge David Sher, Lynn O’Donnell, and Cornelius Watson for technical assistance. We also thank all those individuals who kindly provided cell lines and reagents. We thank Xinhao Fan and Joan Steitz for helpful discussions, and we acknowledge Dana Brenckle for assistance with manuscript preparation. We are also grateful to Rita Girdzis and the Yale Pheresis Unit for assistance with leukopheresis. This work was supported by National Institutes of Health Grant HL43331 (J.R.B.), the Associazione Italiana Ricerca sul Cancro (R.P. and F.B.), the European Community Concerted Health Action BMH4-CT95-0875 and Telethon grant (R.P.), and the AIDS Fund of the Italian Ministry (F.B.). J.R.B. is a Raymond and Beverly Sackler Foundation Scholar. M.C. is a Raymond and Beverly Sackler Foundation Research Scientist.
ABBREVIATIONS
- LFA-1
lymphocyte function-associated antigen-1
- uPAR
urokinase plasminogen activator receptor
- DRB
5,6-dichloro-1-β-d-ribobenzimidazole
- 3′ UTR
3′-untranslated region
- ARE
AU-rich elements
- RPA
ribonuclease protection assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CAT
chloramphenicol acetyltransferase
References
- 1.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz R H. Cell. 1992;71:1065–1068. doi: 10.1016/s0092-8674(05)80055-8. [DOI] [PubMed] [Google Scholar]
- 3.Fan S-T, Brian A A, Lollo B A, Mackman N, Shen N L, Edington T S. Cell Immunol. 1993;148:48–59. doi: 10.1006/cimm.1993.1090. [DOI] [PubMed] [Google Scholar]
- 4.Damle N K, Klussman K, Leytze G, Myrdal S, Aruffo A, Ledbetter J A, Linsley P S. J Immunol. 1994;152:2686– 2697. [PubMed] [Google Scholar]
- 5.Van Seventer G A, Shimizu Y, Horgan K J, Shaw S. J Immunol. 1990;144:4579–4586. [PubMed] [Google Scholar]
- 6.Hernandez-Caselles T, Rubio G, Campanero M R, del Pozo M A, Muro M, Sanchez-Madrid F, Aparicio P. Eur J Immunol. 1993;23:2799–2806. doi: 10.1002/eji.1830231112. [DOI] [PubMed] [Google Scholar]
- 7.Meredith J E, Jr, Winitz S, Lewis J M, Hess S, Ren X D, Renshaw M W, Schwartz M A. Endocr Rev. 1996;17:207– 220. doi: 10.1210/edrv-17-3-207. [DOI] [PubMed] [Google Scholar]
- 8.Pardi R, Bender J R, Dettori C, Giannazza E, Engleman E G. J Immunol. 1989;143:3157–3166. [PubMed] [Google Scholar]
- 9.Kanner S B, Grosmaire L S, Ledbetter J A, Damle N K. Proc Natl Acad Sci USA. 1993;90:7099–7103. doi: 10.1073/pnas.90.15.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi E, Ferrero E, Fazioli F, Mangili F, Wang J, Bender J R, Blasi F, Pardi R. J Clin Invest. 1996;98:1133–1141. doi: 10.1172/JCI118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsten T, June C H, Ledbetter J A, Stella G, Thompson G B. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 12.Lagnado C A, Brown C Y, Goodall G J. Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubiaga A M, Belasco J G, Greenberg M E. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shyu A B, Greenberg M E, Belasco J G. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 15.Shaw G, Karmen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 16.Siu G, Wurster A L, Lipsick J S, Hendrick S M. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson C A, Petzelbauer P, Zhou J, Pardi R, Bender J R. J Immunol. 1995;154:3222–3233. [PubMed] [Google Scholar]
- 18.Schneider-Schaulies J, Schimpl A, Wecker E. Eur J Immunol. 1987;17:713–718. doi: 10.1002/eji.1830170521. [DOI] [PubMed] [Google Scholar]
- 19.Sirenko O I, Lofquist A K, Demaria C T, Morris J S, Brewer G, Haskill J S. Mol Cell Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Y A, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 21.Ma W-J, Cheng S, Campbell C, Wright A, Furneaux H. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 22.Wondnar-Filipowicz A, Moroni C. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai Y, Akahane K, Pluznik D H, Cohen R B. J Immunol. 1993;150:4386–4394. [PubMed] [Google Scholar]
- 24.Shetty S, Kumar A, Idell S. Mol Cell Biol. 1997;17:1075–1083. doi: 10.1128/mcb.17.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roskelly C D, Stebrow A, Bissell M J. Curr Opin Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thieblemont N, Haeffner-Cavaillon N, Haeffner A, Cholley B, Weiss L, Kazatchkine M D. J Immunol. 1995;155:4861–4867. [PubMed] [Google Scholar]
- 27.Xu J, Clark R A. J Cell Biol. 1996;132:239–249. doi: 10.1083/jcb.132.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]