Abstract
The replicative life span (RLS) of Saccharomyces cerevisiae has been established as a model for the genetic regulation of longevity despite the inherent difficulty of the RLS assay, which requires separation of mother and daughter cells by micromanipulation after every division. Here we present the mother enrichment program (MEP), an inducible genetic system in which mother cells maintain a normal RLS—a median of 36 generations in the diploid MEP strain—while the proliferative potential of daughter cells is eliminated. Thus, the viability of a population over time becomes a function of RLS, and it displays features of a survival curve such as changes in hazard rate with age. We show that viability of mother cells in liquid culture is regulated by SIR2 and FOB1, two opposing regulators of RLS in yeast. We demonstrate that viability curves of these short- and long-lived strains can be easily distinguished from wild type, using a colony formation assay. This provides a simplified screening method for identifying genetic or environmental factors that regulate RLS. Additionally, the MEP can provide a cohort of cells at any stage of their life span for the analysis of age-associated phenotypes. These capabilities effectively remove the hurdles presented by RLS analysis that have hindered S. cerevisiae aging studies since their inception 50 years ago.
THE budding yeast Saccharomyces cerevisiae is a popular model system for studying fundamental processes of cellular aging (reviewed in Steinkraus et al. 2008). Analyses over the past 50 years have led to the idea that budding yeast can be used to study three types of cellular aging. Replicative aging describes the division potential of individual cells and relies on the asymmetric cell divisions of budding yeast that yield distinct mother and daughter cells. Replicative life span (RLS) is defined as the number of times an individual cell divides before it undergoes senescence (Mortimer and Johnston 1959). Chronological aging describes the capacity of cells in stationary phase (analogous to G0 in higher eukaryotes) to maintain viability over time, which is assayed by their ability to reenter the cell cycle when nutrients are reintroduced (Longo et al. 1996). Finally, budding yeast have been used to study clonal senescence, which is analogous to the Hayflick limit imposed on mammalian tissue culture cells and characterized by a finite number of times a population of cells can divide. Although wild-type yeast populations do not senesce, this phenomenon has been observed in mutant strains such as those lacking telomerase components (Lundblad and Szostak 1989; Singer and Gottschling 1994).
While genetic screens have been applied to examine clonal and chronological aging (Lundblad and Szostak 1989; Powers et al. 2006; Murakami et al. 2008), they have been limited in their application to studying replicative aging (Kaeberlein and Kennedy 2005; Kaeberlein et al. 2005b). This limitation arises from the arduous nature of isolating replicatively aged yeast cells. The current “gold standard” for isolating aged mother cells is by micromanipulation, where daughter cells are counted and removed after every division (Park et al. 2002). Although micromanipulation is currently the only method capable of accurately measuring RLS in yeast, it is severely constrained by the small number of cells that can be analyzed. Thus, genetic analysis of the regulation of RLS has been limited to a candidate gene approach (reviewed in Steinkraus et al. 2008).
True genetic analysis of RLS will require large populations of aged cells. However, there are two confounding issues that make isolation of aged individuals difficult. First, single-cell pedigree analysis has shown that age-associated phenotypes, such as replicative life span potential, segregate asymmetrically between mother and daughter cells, rendering age-associated phenotypes nonheritable (Egilmez and Jazwinski 1989; Kennedy et al. 1994). Thus, daughter cells are generally “reset” to a young state with every generation. Second, when age is measured in terms of cell divisions, an unfractionated population is predominately young. The fraction of the population at an age of n cell divisions is ∼1/2n. Individual cells that reach the median RLS, which is ∼26 generations for haploid cells of the S288C strain background (Kaeberlein et al. 2005a), represent an insignificant fraction of the total population. In fact, it is unlikely that any cell reaches such an advanced age because nutrient depletion will limit the division potential of the population (Dickinson and Schweizer 1999).
As an alternative to micromanipulation, methods were developed to isolate aged cells from liquid cultures (Smeal et al. 1996; Sinclair and Guarente 1997; Chen and Contreras 2007). However, due to the exponential growth of progeny cells, these populations are technically limited to 7–12 generations before nutrient depletion interferes with replicative aging. While sequential rounds of growth and purification are possible, the inability to continuously follow an undisturbed cohort of cells prevents the measurement of RLS by these methods. Instead, purification methods are primarily used for the examination of molecular changes associated with aging cells. Unfortunately, low yields and loss of viability due to purification methods diminish their utility for analyzing phenotypes that affect cells of advanced age. As an alternative to purification from natural populations, a strategy to genetically regulate the replicative capacity of daughter cells and avoid the limits imposed by exponential growth has been described (Jarolim et al. 2004). While this system effectively prevents division of daughter cells, it unintentionally decreases the median RLS of mother cells to four cell divisions, thus restricting its usefulness.
Here we describe the development of a novel genetic selection against newborn daughter cells, the “mother enrichment program” (MEP), which restricts the replicative capacity of daughter cells while allowing mother cells to achieve a normal RLS. We demonstrate that upon induction of the selection, the viability of MEP strains growing in liquid culture is determined by the RLS of the initial population of mother cells. MEP cultures therefore allow the comparison of RLS between strains without the need for micromanipulation. Additionally, because MEP cultures are not subject to nutrient limitation, single-step affinity purification of aged cells can be achieved at any point during their life span. Together, these capabilities substantially resolve the technical hurdles that have made replicative aging studies in S. cerevisiae exceptionally challenging.
MATERIALS AND METHODS
All strains used in this work are in the S288C (BY) background. Strain and plasmid construction is described in supporting information, File S1. Oligonucleotides, strains, and plasmids described in this work are listed in Table S1, Table S2, and Table S3, respectively.
Fluctuation analysis:
Twenty parallel 1-ml YEPD cultures of UCC5181 (haploid) and UCC5185 (diploid) MEP were grown to saturation overnight. Samples of each culture (∼2 × 106 cells) were plated to 150-mm YEPD + 1 μm estradiol plates and incubated at 30°. Visible colonies after 48 hr were scored as estradiol-resistant mutants. Mutation rates were calculated and corrected for sampling error using the MSS-maximum-likelihood estimator method from the FALCOR fluctuation analysis calculator (http://www.mitochondria.org/protocols/FALCOR.html) (Hall et al. 2009).
Life span measurement by micromanipulation:
Cells from logarithmically growing YEPD liquid cultures were applied to YEPD plates and incubated at 30°. Founder cells were selected for life span analysis by selecting the first daughter of a newborn daughter cell. Cells were monitored for cell divisions every 2 hr until lysis. To measure RLS on estradiol-containing YEPD plates, a portion of agar was cut away and replaced by an island of fresh YEPD agar without estradiol (Figure S1). Founding cells were isolated as described above on the YEPD island. For mother cell analysis, founding cells were allowed to divide twice before being transferred to the estradiol-containing portion of the plate for life span analysis. For daughter cell analysis, founding cells were transferred immediately to the estradiol-containing portion of the plate for life span analysis.
Liquid aging assay:
Cells grown overnight in YEPD medium were diluted 1:50 to inoculate fresh YEPD cultures and incubated at 30° for 3 hr. Cultures were counted and used to inoculate 25 ml of prewarmed YEPD to a cell density of 2 × 103 cells/ml. 17β-Estradiol (Sigma, St. Louis) was added to a final concentration of 1 μm and cultures were incubated at 30° for 120 hr. At each time point, samples were taken and cells were pelleted by centrifugation at 800 × g. All but 100 μl of the supernatant was removed; cells were washed once with 1 ml of fresh YEPD and pelleted again. Cells were resuspended in 500 μl YEPD and plated to YEPD. Colonies were counted after 3 days incubation at 30°. Viability was calculated as CFUs per milliliter and expressed as percentage of viability compared to CFUs per milliliter at the 0- or 4-hr time points. When comparing viability between MET15 and met15Δ strains, each strain was inoculated to 1 × 103 cells/ml and treated as described above, except plating was done to Pb(NO3)2 media (Cost and Boeke 1996) and colony color was scored after 5 days incubation at 30°.
Purification of aged cells:
Cells were harvested from logarithmically growing YEPD cultures, washed twice in PBS, and resuspended in PBS at 5 × 107 cells/ml. Sulfa-NHS-LC-biotin (Pierce Chemical, Rockford, IL) was added to a final concentration of 3 mg/ml and incubated at room temperature protected from light on a Labquake (Thermolyne) for 30 min. Cells were pelleted, washed three times with YEPD, and used to inoculate a 500-ml YEPD culture at 2 × 104 cells/ml. Cultures were incubated 2 hr at 30° prior to the addition of estradiol to a final concentration of 1 μm. At each time point, 100 ml of the culture was harvested; cells were pelleted by centrifugation and washed twice in PBS. Cells were resuspended in 500 μl PBS and fixed with 4% paraformaldehyde for 10 min. After fixation, cells were washed twice with PBS, resuspended in 500 μl PBS, and incubated with 50 μl streptavidin-coated magnetic beads (MicroMACS, Miltenyi Biotec) for 30 min at room temperature on a Labquake. Cells were pelleted and washed twice with PBS and then resuspended in 8 ml PBS and loaded onto a LS MACS column (Miltenyi Biotec). Columns were washed with 8 ml PBS, removed from the magnetic field, and eluted with 8 ml PBS. Purified cells were pelleted and resuspended in 500 μl PBS + 1 μl fluorescein–avidin (Pierce) and incubated at room temperature for 90 min on a Labquake. Calcofluor (Sigma) was added to a concentration of 0.1 mg/ml and incubated for 5 min before cells were pelleted and washed three times in PBS. Final stained cell pellets were resuspended in 100 μl PBS and spotted to polylysine-coated microscope slides. Individual fluorescein-avidin stained cells were imaged with a Deltavision microscope for bud scar counting.
RESULTS
A Cre-lox mediated selection against daughter cells:
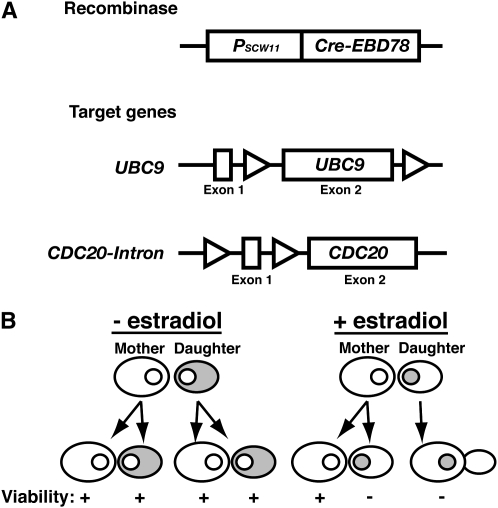
To generate a population of replicatively aged S. cerevisiae cells, we focused on creating a system that would allow a mother cell to divide, but eliminate that potential in her daughters. We imposed several requirements on this system. First, it must accurately distinguish between mother and daughter after each cell division to avoid prematurely limiting the mother's life span. Second, it must effectively prevent daughter cells from producing progeny. This must be accomplished within a single cell division, before the daughter cell develops into a mother cell upon exiting its first mitosis. Finally, the selection must be conditional, so that a strain can be propagated. The MEP, outlined in Figure 1, achieves the stated goals by using Cre-lox recombination to disrupt two essential genes, UBC9 and CDC20, in daughter cells (Figure 1A). We chose disrupton of essential genes as our selection mechanism to allow a combinatorial approach: By combining multiple targets we could improve the stringency of the selection. UBC9 encodes the SUMO-conjugating enzyme, while CDC20 encodes an activator of the anaphase-promoting complex (APC). Both are required for the degradation of mitotic cyclins and other targets vital to cell cycle progression (Sethi et al. 1991; Seufert et al. 1995; Dieckhoff et al. 2004), so eliminating these genes by recombination results in daughter cells that permanently arrest in M-phase. Expression of the Cre recombinase is restricted by a daughter-specific promoter derived from SCW11 (PSCW11) (Figure 1A). Daughter-specific expression from PSCW11 is regulated by the transcription factor Ace2, which is asymmetrically distributed to daughter cell nuclei prior to cytokinesis (Colman-Lerner et al. 2001; Doolin et al. 2001). Cre activity is also post-transcriptionally regulated by fusion to the estradiol-binding domain (EBD) of the murine estrogen receptor (Cre-EBD78). The EBD sequesters the fusion protein in the cytoplasm until estradiol is introduced, at which point the fusion protein is transported into the nucleus where Cre can act upon its loxP DNA substrates (Figure 1B) (Cheng et al. 2000).
Figure 1.—
Components of the MEP. (A) Diagram of the MEP components. Cre-EBD78 is a novel version of a fusion protein between the Cre recombinase from bacteriophage P1 and the estrogen-binding domain of the murine estrogen receptor that is strictly dependent on estradiol for activity (see materials and methods). PSCW11-cre-EBD78 is integrated at the ho locus and specifically expressed in newborn cells, using a daughter-specific promoter derived from SCW11. Target genes were constructed at their endogenous loci by introduction of loxP sites (triangles). For UBC9, recombination between the loxP sites removes exon 2, representing 92% of the coding region. For CDC20, an intron derived from ACT1 and containing a loxP site was introduced to generate a 42-bp exon 1. Recombination between loxP sites removes exon 1, leaving the first in-frame start codon at methionine 197, which would generate a nonfunctional protein. (B) Illustration of the expected localization of Cre-EBD78 (shading) in response to estradiol. PSCW11-cre-EBD78 expression is restricted to the G1 phase of daughter cells. In the absence of estradiol the fusion protein is sequestered in the cytoplasm. Upon ligand binding, the fusion protein is translocated into the nucleus (small circle), where it can act on loxP target sites.
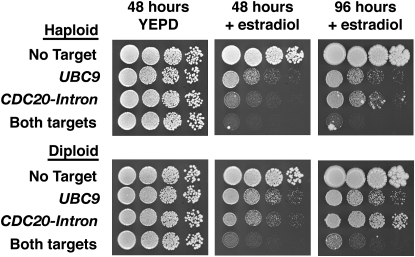
To test the inducible nature of the MEP, strains were placed on media with or without estradiol (Figure 2). Cells formed robust colonies within 2 days in the absence of estradiol. By contrast, growth was severely limited in the presence of estradiol. The estradiol effect was dependent upon the presence of the MEP: Strains lacking PSCW11-cre-EBD78 or loxP targets grew normally in the presence of estradiol (Figure 2 and data not shown). Examination of individual cells on estradiol plates revealed that they formed microcolonies of large dumbbell-shaped cells (data not shown). This is consistent with an M-phase arrest phenotype caused by the loss of either UBC9 or CDC20 (Sethi et al. 1991; Dieckhoff et al. 2004). Because Cre-lox recombination is a reversible reaction (Sauer and Henderson 1990), we did not expect selection based on a single target to be absolute. Consistent with this idea, we observed a synergistic growth defect on estradiol in cells that contained both target genes (Figure 2). Taken together, these results indicate that the MEP is effectively activated by estradiol and that UBC9 and CDC20 are efficiently disrupted by the Cre recombinase.
Figure 2.—
MEP induction by estradiol. Tenfold serial dilutions of haploid and diploid MEP strains were applied to YEPD plates with or without 1 μm estradiol. Plates were incubated at 30° and photographed at the times indicated. Each strain carries PSCW11-cre-EBD78 and the indicated loxP target genes.
In the course of our analyses, we occasionally encountered fast-growing cells that were resistant to estradiol-induced arrest (Figure 2). When these estradiol-resistant mutants were generated in liquid culture, their progeny quickly saturated the culture and foiled the intended purpose of the MEP. We used fluctuation analysis (Rosche and Foster 2000) to determine the rate at which estradiol-resistant mutations appeared in a population of cells to estimate an upper limit of the number of cells that can be analyzed with the MEP. Logarithmically growing haploid cells produced resistant mutations at a rate of 1.4 × 10−6 per cell division. This rate is ∼10-fold higher than expected for forward mutation of a single locus (i.e., cre-EBD78) (Kunz et al. 1998) and is consistent with previous reports that identified multiple loci in yeast required for EBD function (Gilbert et al. 1993; Kralli et al. 1995; McEwan 2001). In diploids, resistant mutations arose at a rate of 1.4 × 10−8 per division, indicating that many of the mutations were recessive. The upper boundary represented by the 95% confidence interval of these measurements (1.9 × 10−6 for haploids and 3.4 × 10−8 for diploids) provides an estimate that can be used to calculate the maximum number of cells that should be used to inoculate a liquid “aging” culture without risk of contamination by an estradiol-resistant mutant.
Validation of the MEP by single-cell analysis:
To examine the specificity of the MEP, micromanipulation was used to track the fate of mothers and daughters in the absence or presence of estradiol. The median RLS of the diploid MEP strain UCC5185 in the absence of estradiol was not significantly different from that of a diploid parent strain (UCC8600) lacking the MEP components (36 vs. 37 generations, Figure 3A). This indicates that components of the MEP do not affect the RLS of mother cells in the absence of estradiol.
Figure 3.—
Validation of the MEP by microdissection. (A) Survival curves of diploid parent (UCC8600, n = 147, median = 37) and MEP (UCC5185, n = 90, median = 36) strains generated by micromanipulation on YEPD without estradiol. Median life spans are not significantly different (Mann–Whitney nonparametric t-test, P = 0.3904). (B) Survival curves of MEP strain UCC5185 generated by micromanipulation on YEPD with 1 μm estradiol (+ED, n = 39, median = 36) or without it (−ED, n = 90, median = 36). The survival curve for +ED includes only naive mother cells that did not exhibit rapid M-phase arrest upon transfer to estradiol. (C) The distribution of the number of cells per microcolony generated by daughter cells born during +ED RLS analysis presented in B. (D) Survival curves of UCC5185 naive daughters (n = 118, median = 2) transferred to YEPD + 1 μm estradiol and their resulting progeny (granddaughters, n = 269, median <1). The two leaky daughters have been censored from the analysis. In each case, the RLSs of cells that failed to complete a single division were scored as 1 generation.
The RLSs of cells on media containing estradiol were also measured. Typically an RLS measurement begins with newborn daughter cells that, after a single division, become mother cells. A modification of the RLS protocol was made to measure the RLS of naive (i.e., no prior exposure to estradiol) mother and daughter cells by constructing plates that contained an island lacking estradiol and a larger zone containing 1 μm estradiol to perform the RLS analysis (Figure S1).To generate naive mother cells, newborn daughters were allowed to complete two divisions on the island lacking estradiol before transfer to the RLS zone. Naive mother cells displayed either of two responses upon exposure to estradiol: Approximately 15% of the mothers showed a terminal M-phase arrest phenotype and died quickly (data not shown), which is consistent with these young mothers having experienced elimination of CDC20 or UBC9 by Cre-lox recombination. This response to estradiol was not observed in the parent strain UCC8600 that lacks the MEP (median RLS = 35, n = 59, data not shown), indicating that these early deaths were not a general effect of estradiol on yeast. We postulate that M-phase arrest in naive mothers—individuals that were recently daughter cells—is the result of perdurance of Cre-EBD mRNA or protein. For the other 85% of mother cells, no terminal M-phase arrest was observed and these mothers had life spans that were indistinguishable from those of cells grown in the absence of estradiol (median RLS = 36 generations, Figure 3B), indicating that the majority of mother cells were unaffected by estradiol throughout their life spans. Furthermore, this result suggests that there is no loss of mother–daughter asymmetry in expression from PSCW11 in aging cells.
By contrast, the effect of estradiol on the daughters born to this cohort of mother cells was profound. All 472 daughters born on estradiol had a severely limited replicative capacity and formed microcolonies of M-phase arrested cells. The majority failed to complete more than a single division (Figure 3C and Figure S2). We saw a negative correlation between age of the mother cell and division potential of the daughter (Nonparametric correlation, P < 0.0001)(Figure S3), supporting the conclusion that the effectiveness of the MEP does not deteriorate with age. Thus, upon induction of the MEP, most mother cells live a normal life span in the presence of estradiol, while the majority of their daughters succumb to Cre-mediated recombination and arrest within a single generation.
Approximately 8% of daughter cells on estradiol formed microcolonies of >10 M-phase arrested cells. These microcolonies could be the result of phenotypic lag in the MEP selection; it may take several cell divisions to effectively deplete Cdc20 or Ubc9 after deletion of the genes in some daughter cells. Alternatively, microcolonies could be the result of individuals that escaped the MEP selection and became productive mother cells, a phenomenon we term “leakiness.” To distinguish between these possibilities, we determined the fate of naive daughter cells upon exposure to estradiol. Naive daughter cells were isolated on an island lacking estradiol and immediately transferred to the RLS zone. Of 120 naive daughter cells transferred, 29 arrested in M-phase and never divided. The remaining 91 cells completed their first division and developed into mother cells. With the exception of 2 individuals, these newly formed mothers displayed an aberrant cell cycle, with an average division time of 3.1 hr and a prolonged G2–M phase. Their median RLS was two generations and ended with a terminal M-phase arrest (Figure 3D), indicating that these cells were succumbing to Cre-lox recombination. We monitored the fate of progeny cells (“granddaughters”) born to these mothers and found 61% of granddaughters arrested without completing a single division (median RLS <1 generation, maximum = 4, n = 269, Figure 3D). We conclude that the vast majority of daughter cell divisions are the result of phenotypic lag rather than leakiness.
Two naive daughter cells of 120 (1.7%) failed to display an M-phase arrest upon exposure to estradiol and became mother cells that divided at a normal rate and achieved normal life spans. These cells produced daughters that responded to estradiol and quickly arrested, indicating that they were “leaky” mothers that had escaped the MEP selection rather than estradiol-resistant mutants. At this rate of leakiness, we expect an average mother cell to generate 0.6 viable progeny throughout her life span. However, the leakiness rate is likely to be lower in daughters that are born in the presence of estradiol. For example, in the experiment where naive daughters were transferred to estradiol, 41% arrested without completing >1 division (Figure 3D). In contrast, 63% of daughter cells born on estradiol media during the measurement of naive mother cell RLS arrested without completing >1 division (Figure 3C). This indicates that the replicative capacity of daughter cells born in the presence of estradiol is very low.
Aging cells in liquid cultures via the MEP:
Given its successful performance on solid medium, we next examined the effectiveness of the MEP in liquid culture, where it has its greatest potential as a resource for studying yeast replicative aging. To avoid generation of estradiol-resistant mutants during the experiment, 25 ml YEPD cultures containing 1 μm estradiol were innoculated at a cell density of 2 × 103 cells/ml. We found that cultures at this starting cell density do not exceed ∼106 cells/ml after 120 hr of incubation and thus should not impinge on the growth of mother cells through nutrient depletion (data not shown). Samples of cells were harvested at regular intervals, washed, and plated to media lacking estradiol. Formation of colonies on these plates demonstrated that the MEP was reversible; once estradiol was removed, if a mother cell still had replicative capacity, her daughters divided normally to form a colony.
MEP cells demonstrated a rapid loss of viability within the first 4 hr of exposure to estradiol (Figure 4A). This was expected since daughters constituted approximately half of the initial inoculum. These daughter cells are immediately arrested by estradiol, leaving only a cohort of mother cells in the viable population. After arrest of the daughter population, there was a 16-hr period with little change in viability, demonstrating a low hazard rate of death in young mothers (Machin 2006). This phase was followed by a rapid decline in viability over ∼48 hr, indicating an increasing hazard rate in the aging population. This change in the hazard rate in liquid culture parallels the changes in hazard rate observed when measuring the RLS of MEP strains on solid media containing estradiol (Figure 3A and Figure S4) and is stereotypical for survival curves in yeast and other species (Pohley 1987; Kennedy et al. 1994). These similarities suggest that cells aging in liquid culture exhibit the same pattern of age-associated changes in hazard rate as individual cells aging on solid media.
Figure 4.—
Aging cells in liquid culture with the MEP. (A) Viability curve of diploid MEP strain UCC8848 in liquid YEPD with 1 μm estradiol, normalized to the 0-hr time point. Viability as represented by CFUs per milliliter was monitored by harvesting samples at the indicated time points and washing and plating cells to media lacking estradiol. Values represent the median of 12 independent cultures with 95% confidence intervals indicated by error bars. (B) A flow chart of steps for purifying and measuring age of founding mother cells from a liquid MEP culture. LHC-Biotin was covalently cross-linked to the cell walls of logarithmically growing UCC5185 cells. Cells were incubated in the absence of estradiol for 2 hr to allow all labeled cells to become mothers, and estradiol was then added to a final concentration of 1 μm. At intervals, samples were removed for purification with streptavidin-coated magnetic beads and stained with fluorescein-avidin and calcofluor white for bud scar counting. (C) The distribution of bud scars on purified cells is presented after binning into five-generation intervals: 20 hr, n = 30, mean = 12; 45 hr, n = 18, mean = 22; 70 hr, n = 21, mean = 31; 95 hr, n = 26, mean = 35.
We next determined whether individual mother cells in a liquid MEP culture continued to divide throughout the experimental time course. Many of the cell wall components that a cell is born with remain intact throughout its life span (Ballou 1982). We took advantage of this attribute and cross-linked biotin to the cell wall of the starting population of mother cells (Smeal et al. 1996). The biotin tag permitted sampling and purification of founder cells at intervals throughout the time course. Purified cells were stained with fluorescein-avidin, to confirm that an individual cell was part of the initial mother population, and with calcofluor white to count bud scars to determine replicative age (Figure 4B) (Park et al. 2002). Both the median and the maximum age of the founding mother population increased throughout the time course and were similar to values obtained by micromanipulation (Figure 4C and data not shown). Thus, individual mothers can achieve a normal RLS in the presence of estradiol in liquid cultures.
The MEP can distinguish between mutants with altered RLS in liquid cultures:
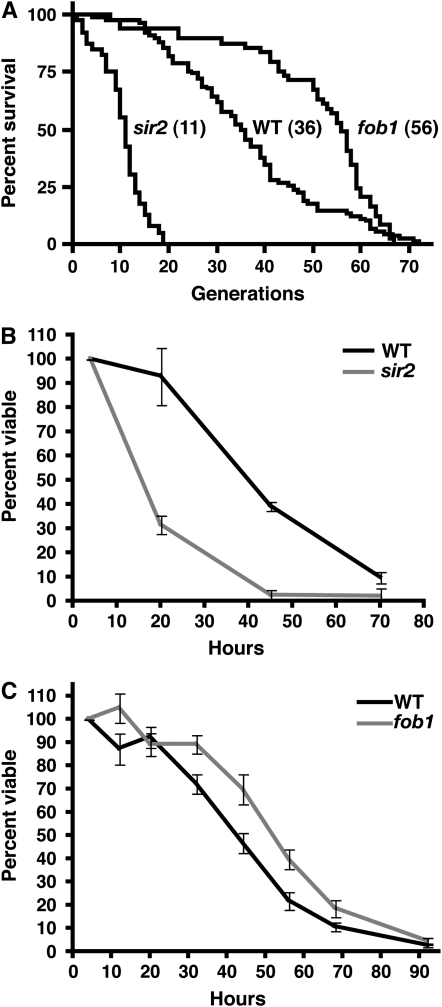
The results presented above indicate that aging of a MEP strain in liquid culture recapitulates the characteristics of replicative aging on solid medium. To further examine the relationship between RLS and viability in liquid culture, we constructed MEP strains deleted for either SIR2 (UCC8836) or FOB1 (UCC526), which are well characterized for their effects on yeast RLS (Defossez et al. 1999; Kaeberlein et al. 1999; reviewed in Steinkraus et al. 2008). Consistent with previous studies using haploid cells (Kaeberlein et al. 2005a), these mutations affect RLS on solid media in the diploid MEP background (Figure 5A, −69% (sir2Δ) and +56% (fob1Δ) change in median RLS compared to UCC5185). To compare viability of mutant strains to wild type within a single culture, we took advantage of a colony color phenotype in MET15 mutants (Cost and Boeke 1996). First, we constructed diploid MEP strains that were homozygous MET15 or met15Δ. Estradiol cultures were inoculated with an equal number of cells of each type, and both viability and MET15 status were monitored by plating to solid media containing Pb+2. We observed no effect of the MET15 allele on viability under these conditions (data not shown). Next, we compared a wild-type MET15 diploid with a sir2Δ met15Δ strain. We observed a rapid decline in viability in the sir2Δ strain, with no evidence of a low hazard of death in young cells (Figure 5B). This was consistent with the hazard rate observed by measuring RLS on solid media (Figure 5A). The reduction of ∼60% in median viability of the sir2Δ strain compared to wild type was also consistent with the reduction in RLS observed on solid media. These results indicate that viability in liquid culture can easily distinguish between strains with normal and short life spans.
Figure 5.—
Differences in RLS measured with the MEP. (A) Survival curves of diploid MEP strains generated by micromanipulation on YEPD without estradiol (wild-type UCC5185, n = 90, median = 36; fob1Δ UCC526, n = 49, median = 56; sir2Δ UCC8836, n = 40, median = 11). (B) Viability of wild-type MET15 (UCC8848) vs. sir2Δ met15Δ (UCC8849) diploid MEP strains in liquid culture measured as described in Figure 4A. Values represent the median of three independent cultures normalized to the 4-hr time point, with error bars indicating 95% confidence intervals. (C) Viability of wild-type vs. fob1Δ diploid MEP strains in liquid culture. Values represent the median of six wild-type MET15 (UCC8848) vs. fob1Δ met15Δ (UCC8850) and six wild-type met15Δ (UCC8861) vs. fob1Δ MET15 (UCC8863) cultures, with error bars indicating 95% confidence intervals.
We next examined the viability in liquid culture of a long-lived fob1Δ strain. We found that the fob1Δ strain showed an increase in median viability in liquid culture of ∼21% compared to a cocultured wild-type strain (Figure 5C). While the magnitude of this increase was smaller than that observed on solid media, the fob1Δ strain yielded significantly higher viability than wild type at each time point between 32 and 68 hr (paired t-test, P < 0.01). These results indicate that viability of MEP strains in liquid culture can be used to distinguish between strains with normal and long life spans. Additionally, they indicate that life span in liquid culture is regulated by some of the same genetic pathways that regulate RLS on solid media.
DISCUSSION
Benefits of the MEP compared to other systems:
The MEP provides a new tool that greatly expands the potential for applying S. cerevisiae genetic, biochemical, and cell biological approaches to examine the fundamental processes of replicative aging. Previously, methods for isolating replicatively aged yeast cells for analysis have followed two complementary approaches. The gold standard is based upon following single cells by micromanipulation (Park et al. 2002). This method allows the accurate determination of an individual's life span, but only ∼100 cells can be followed in an experiment by a single researcher. Micromanipulation is the method by which all genetic analyses of replicative life span have been carried out (Steinkraus et al. 2008). However, because of the arduous nature of the assay, most genetic analyses have followed a candidate gene approach; only recently was an unbiased genetic screen attempted (Kaeberlein and Kennedy 2005; Kaeberlein et al. 2005b). Given the magnitude of effort required for this screen, it is unlikely to be duplicated with varying environmental conditions or genetic backgrounds—elements that are known to critically affect the aging process (Lin et al. 2002; Kaeberlein et al. 2005a). The MEP has overcome the limitations of measuring RLS by micromanipulation: It is easy to perform and does not require continuous monitoring. As shown here, it can readily distinguish between genetic alterations that either increase or decrease life span.
The complement to micromanipulation is purification of aged cells from liquid cultures (Smeal et al. 1996; Sinclair and Guarente 1997; Chen and Contreras 2007). These methods are not applicable to the measurement of RLS, but instead provide larger populations for the analysis of age-associated phenotypes. Unfortunately, the age of cells in liquid cultures is limited by the exponential proliferation of young progeny cells. Consequently, single-step purification strategies typically yield cells that are only 7–12 generations old. Purification strategies are also confounded by low yields, contamination by daughter cells, and extensive sample processing in PBS or other buffers. While older populations can be obtained through successive rounds of growth and purification, or the addition of a FACS sorting step (Chen and Contreras 2007), the problems facing purification are compounded with each round. The MEP has overcome the limitations imposed by progeny cells by efficiently eliminating daughter cell propagation. Aging of a cohort can be carried out for several days in a single culture that begins at a low cell density (e.g., ∼103 cells/ml, Figure 4A). Additionally, because the MEP is rapidly reversible upon removal of estradiol, purification of aged cells from their inviable progeny is not necessary for colony-based assays that monitor viability or other age-associated phenotypes.
A similar approach to eliminating daughter cells by restricting CDC6 expression to mother cells has been described (Jarolim et al. 2004). While daughter cell division is effectively prevented, the median RLS of mother cells is reduced by ∼75%. This has precluded its use in examining age-associated phenotypes. In contrast, the MEP is successful at preserving the normal RLS of mother cells (Figure 3B).
Limitations of the MEP:
The MEP shares limitations common to other genetic selections. First, the individual components of the MEP must be introduced into a strain of interest. The MEP as presented here has been created in the S288C strain background, which is the basis for many comprehensive strain collections that are available [e.g., the nonessential gene deletion set (Winzeler et al. 1999)]. With the large molecular and genetic toolkits that have been built upon this strain background, studies of replicative aging in yeast should advance rapidly.
The second limitation is that mutations in MEP strains can arise that allow a mutant cell to avoid the selection. In addition to cre-EBD78 itself, genes involved in localization of the EBD (reviewed in McEwan 2001) or multidrug resistance (Gilbert et al. 1993; Kralli et al. 1995) are potential sources of estradiol resistance. We found that estradiol-resistant mutations arise at a rate of 1.4 × 10−6 per cell division in haploid cells and 1.4 × 10−8 per cell division in diploid cells. These rates, adjusted depending on the target age of the final population, can serve as a guide to determine how many cells can be evaluated in an experiment.
The third limitation is based on the observation that while the daughter cells and their progeny arrest irreversibly in M-phase, they do not immediately senesce. They can remain metabolically active in this arrested state, continuing to grow without dividing for ∼24 hr before lysis (data not shown). Upon exposure to estradiol, an average daughter cell produced 6.9 progeny cells (daughters and subsequent generations). If an average mother cell generates 36 daughter cells, this translates to ∼248 inviable progeny cells during her life span. We have been able to avoid nutrient limitation when measuring RLS by inoculating at low cell density (103–104 cells/ml). The actual starting density can be adjusted depending on the target age for the experiment; a higher initial inoculum can be used without risk of nutrient depletion when analysis is done after 1 or 2 days of aging.
Comparing RLS to viability in liquid culture:
For the past 50 years, RLS in S. cerevisiae has been measured using the metric of cell divisions rather than time, on the basis of the observation that perturbations such as temperature change drastically alter division rate without affecting the median number of generations (Muller et al. 1980). This metric also introduces an essential element for micromanipulation, since it allows one to interrupt a life span analysis nightly by moving the cells to 4°. Survival curves obtained by this method generally conform to a Gompertz distribution similar to that observed when monitoring survival in protected metazoan populations (Figure 3A and Kennedy et al. 1994; Kirkwood 2005).
With the MEP, we show that RLS can now be examined as a function of time. As long as the cell division rate remains constant within a cohort of mother cells, viability in liquid culture will be directly proportional to RLS. Because the MEP avoids fluctuations in temperature and nutrient availability, this assumption is generally true. We showed that with advancing time, mother cells in the original population advance in age (Figure 4C). When the life spans of three strains (wild type, sir2Δ, and fob1Δ) were examined by micromanipulation and MEP, we found that in both situations the median life span was changed in the same way: sir2Δ was much shorter, and fob1Δ was significantly longer, than wild type (Figure 5). However, the magnitude of the difference was somewhat smaller when measured with the MEP (sir2Δ, −69% vs. −60%; fob1Δ, +56% vs. +21%, for micromanipulation and MEP, respectively).
We offer a few explanations that could contribute to differences between life span measured using micromanipulation vs. the MEP. First, there may be intrinsic differences between growth on solid medium and that in liquid that affect the aging process. Second, cell division rate may change with increasing replicative age. Consistent with previous reports, we found that when cells approach the end of their life span on solid media, the time between cell divisions increases (Mortimer and Johnston 1959; Muller et al. 1980; and Figure S5, A and B). Differences in how division rates change with age between wild-type and mutant strains could lead to distortion of the MEP viability curves. While cell division rate (in terms of fitness) has been determined for the nonessential yeast gene deletion set for logarithmically growing (young) cells (Deutschbauer et al. 2005), age-dependent changes to these rates have not been systematically examined. Finally, the intrinsic cell division rate of mutants may differ, so that the relationship between time and age is not standard between wild-type and mutant strains. While there is no difference in division rate between young wild-type and fob1Δ cells (Chen and Contreras 2007 and data not shown), this may be an important consideration when examining other mutations that regulate RLS.
Further potential of the MEP:
Restriction of the replicative capacity of progeny cells both eliminates nutrient depletion in liquid cultures and effectively enriches for aging mother cells within a population. Thus, populations of aged cells can be generated at any point in their life span without the need for sequential rounds of purification (Figure 4C). By simply eliminating the losses inherent in sequential purifications, this advance improves S. cerevisiae as a model system for studying the molecular changes associated with aging. The uninterrupted aging of a large cohort of cells also unlocks the possibility of examining phenotypes, such as spontaneous genetic mutation, that occur at very low frequencies. By solving the limitations of both RLS measurement and aged cell purification, the MEP unlocks the burgeoning yeast molecular genetic toolbox for the examination of cellular aging. Given that a number of the processes of cellular aging may be conserved among eukaryotes (Smith et al. 2008; Steinkraus et al. 2008), the MEP offers a new opportunity for developing a greater depth of understanding of these processes.
Acknowledgments
We thank Zara Nelson and Elizabeth Hetrick for support with plasmid and strain construction and Fred van Leeuwen, Lazar Dimitrov, Daniel Lockshon, and Mark Gartenberg for providing plasmids for this work. This work was supported by postdoctoral fellowship PF-04-041-01-GMC from the American Cancer Society to D.L.L., by National Institutes of Health grants AG023779 and GM43893 to D.E.G., and by National Cancer Institute grant T32 CA09657.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.106229/DC1.
References
- Ballou, C. E., 1982. Yeast cell wall and cell surface, pp. 335–360 in The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chen, C., and R. Contreras, 2007. Identifying genes that extend life span using a high-throughput screening system. Methods Mol. Biol. 371: 237–248. [DOI] [PubMed] [Google Scholar]
- Cheng, T. H., C. R. Chang, P. Joy, S. Yablok and M. R. Gartenberg, 2000. Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res. 28: E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner, A., T. E. Chin and R. Brent, 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107: 739–750. [DOI] [PubMed] [Google Scholar]
- Cost, G. J., and J. D. Boeke, 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12: 939–941. [DOI] [PubMed] [Google Scholar]
- Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno et al., 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer et al., 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169: 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, J. R., and M. Schweizer (Editors), 1999. The Metabolism and Molecular Physiology of Saccharomyces cerevisiae. Taylor & Francis, Philadelphia.
- Dieckhoff, P., M. Bolte, Y. Sancak, G. H. Braus and S. Irniger, 2004. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51: 1375–1387. [DOI] [PubMed] [Google Scholar]
- Doolin, M. T., A. L. Johnson, L. H. Johnston and G. Butler, 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40: 422–432. [DOI] [PubMed] [Google Scholar]
- Egilmez, N. K., and S. M. Jazwinski, 1989. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J. Bacteriol. 171: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, D. M., D. M. Heery, R. Losson, P. Chambon and Y. Lemoine, 1993. Estradiol-inducible squelching and cell growth arrest by a chimeric VP16-estrogen receptor expressed in Saccharomyces cerevisiae: suppression by an allele of PDR1. Mol. Cell. Biol. 13: 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. M., C. X. Ma, P. Liang and K. K. Singh, 2009. Fluctuation AnaLysis CalculatOR (FALCOR): a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarolim, S., J. Millen, G. Heeren, P. Laun, D. S. Goldfarb et al., 2004. A novel assay for replicative lifespan in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 169–177. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., and B. K. Kennedy, 2005. Large-scale identification in yeast of conserved ageing genes. Mech. Ageing Dev. 126: 17–21. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., M. McVey and L. Guarente, 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., K. T. Kirkland, S. Fields and B. K. Kennedy, 2005. a Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 126: 491–504. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., R. W. Powers, K. K. Steffen, E. A. Westman, D. Hu et al., 2005. b Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kennedy, B. K., N. R. Austriaco and L. Guarente, 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127: 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood, T. B., 2005. Understanding the odd science of aging. Cell 120: 437–447. [DOI] [PubMed] [Google Scholar]
- Kralli, A., S. P. Bohen and K. R. Yamamoto, 1995. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc. Natl. Acad. Sci. USA 92: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, B. A., K. Ramachandran and E. J. Vonarx, 1998. DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics 148: 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez et al., 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418: 344–348. [DOI] [PubMed] [Google Scholar]
- Longo, V. D., E. B. Gralla and J. S. Valentine, 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271: 12275–12280. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Machin, D., Y. B. Cheung and M. K. B. Parmar, 2006 Survival Analysis, A Practical Approach. John Wiley & Sons, Chichester, UK.
- McEwan, I. J., 2001. Bakers yeast rises to the challenge: reconstitution of mammalian steroid receptor signalling in S. cerevisiae. Trends Genet. 17: 239–243. [DOI] [PubMed] [Google Scholar]
- Mortimer, R. K., and J. R. Johnston, 1959. Life span of individual yeast cells. Nature 183: 1751–1752. [DOI] [PubMed] [Google Scholar]
- Muller, I., M. Zimmermann, D. Becker and M. Flomer, 1980. Calendar life span versus budding life span of Saccharomyces cerevisiae. Mech. Ageing Dev. 12: 47–52. [DOI] [PubMed] [Google Scholar]
- Murakami, C. J., C. R. Burtner, B. K. Kennedy and M. Kaeberlein, 2008. A method for high-throughput quantitative analysis of yeast chronological life span. J. Gerontol. A Biol. Sci. Med. Sci. 63: 113–121. [DOI] [PubMed] [Google Scholar]
- Park, P. U., M. McVey and L. Guarente, 2002. Separation of mother and daughter cells. Methods Enzymol. 351: 468–477. [DOI] [PubMed] [Google Scholar]
- Pohley, H. J., 1987. A formal mortality analysis for populations of unicellular organisms (Saccharomyces cerevisiae). Mech. Ageing Dev. 38: 231–243. [DOI] [PubMed] [Google Scholar]
- Powers, R. W., M. Kaeberlein, S. D. Caldwell, B. K. Kennedy and S. Fields, 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20: 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche, W. A., and P. L. Foster, 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, B., and N. Henderson, 1990. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 2: 441–449. [PubMed] [Google Scholar]
- Sethi, N., M. C. Monteagudo, D. Koshland, E. Hogan and D. J. Burke, 1991. The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol. Cell. Biol. 11: 5592–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert, W., B. Futcher and S. Jentsch, 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., and L. Guarente, 1997. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Singer, M. S., and D. E. Gottschling, 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409. [DOI] [PubMed] [Google Scholar]
- Smeal, T., J. Claus, B. Kennedy, F. Cole and L. Guarente, 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84: 633–642. [DOI] [PubMed] [Google Scholar]
- Smith, E. D., M. Tsuchiya, L. A. Fox, N. Dang, D. Hu et al., 2008. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 18: 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus, K. A., M. Kaeberlein and B. K. Kennedy, 2008. Replicative aging in yeast: the means to the end. Annu. Rev. Cell. Dev. Biol. 24: 29–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]