Abstract
Most elite wheat varieties cannot be crossed with related species thereby restricting greatly the germplasm that can be used for alien introgression in breeding programs. Inhibition to crossability is controlled genetically and a number of QTL have been identified to date, including the major gene Kr1 on 5BL and SKr, a strong QTL affecting crossability between wheat and rye on chromosome 5BS. In this study, we used a recombinant SSD population originating from a cross between the poorly crossable cultivar Courtot (Ct) and the crossable line MP98 to characterize the major dominant effect of SKr and map the gene at the distal end of the chromosome near the 5B homeologous GSP locus. Colinearity with barley and rice was used to saturate the SKr region with new markers and establish orthologous relationships with a 54-kb region on rice chromosome 12. In total, five markers were mapped within a genetic interval of 0.3 cM and 400 kb of BAC contigs were established on both sides of the gene to lay the foundation for map-based cloning of SKr. Two SSR markers completely linked to SKr were used to evaluate a collection of crossable wheat progenies originating from primary triticale breeding programs. The results confirm the major effect of SKr on crossability and the usefulness of the two markers for the efficient introgression of crossability in elite wheat varieties.
DURING domestication and selection of a number of important crop species, diversity has eroded resulting in increased vulnerability to biotic and abiotic stresses while also jeopardizing the potential for sustained genetic improvement of elite cultivars over the long term (Tanksley and McCouch 1997; Fu and Somers 2009). The reintroduction of the remarkable diversity present in the different gene pools into elite varieties through intra- and interspecific crosses (primary and secondary gene pools) and intergeneric crosses (tertiary gene pools) has been practiced for decades in cereals (Feuillet et al. 2008). Despite some highly significant successes, including the incorporation of dwarfing and disease-resistance genes that fueled the Green Revolution, introgression remains laborious and, for complex characters, largely unfulfilled. Wheat (Triticum aestivum L.) has been crossed with a wide range of related species from the Triticeae tribe (Jiang et al. 1994), such as Aegilops, Agropyron, Haynaldia, Secale, and Hordeum, which represent a reservoir of interesting alleles for improving wheat resistance to biotic (diseases, insects) and abiotic stresses (cold, salinity, and drought) as well as for quality traits such as grain protein content (Fedak 1985). Intergeneric crosses have resulted in the transfer of desirable rye (Secale cereale L.) characteristics into wheat (Florell 1931) with one of the best examples being the 1BL/1RS chromosomal translocation that provided novel race-specific resistance to rust diseases, improved adaptation and stress tolerance, superior aerial biomass, and higher kernel weight to wheat varieties (Zarco-Hernandez et al. 2005). However, most of the adapted wheat germplasm is not crossable with alien species thereby restricting the panel of lines that can be used for alien introgression in wheat breeding (Krolow 1970) or for the production of primary triticale, a man-made wheat–rye hybrid.
Beginning in the early 1900s, researchers were producing experimental crosses between bread wheat, T. aestivum L. (2n = 6x = 42) as a recipient, and rye, S. cereale L. (2n =14) as the pollen donor (Backhouse 1916). Genetic studies conducted by Lein (1943) showed that dominant alleles of two genes, named Kr1 and Kr2, are responsible for the poor crossability between bread wheat and rye. Kr1 and Kr2 genes were localized roughly on chromosome 5B and 5A, respectively (Riley and Chapman 1967) and subsequently located more precisely on the long arms of these two chromosomes (Lange and Riley 1973; Sitch et al. 1985). Further studies indicated that the dominant alleles driving incompatibility of crossing wheat with rye act by actively inhibiting the production of intergeneric hybrids (Riley and Chapman 1967; Lange and Wojciechowska 1976; Jalani and Moss 1980, 1981; Cameron and Reger 1991 ). Other crossability genes, such as Kr3 on chromosome 5D (Krolow 1970) and Kr4 on chromosome 1A (Zheng et al. 1992), were identified later. Finally, a study elucidated that chromosome 1A, derived from the Chinese (Sichuan) tetraploid wheat T. turgidum L. cv. Ailanmai, carries a recessive allele for high crossability with rye (Liu et al. 1999). Genetic studies also indicated that Kr genes have different effects on wheat–rye crossability. For example, by testing the cultivars Chinese Spring (CS), Hope, and the substitution lines CS/Hope 5B and CS/Hope 5A, Riley and Chapman (1967) demonstrated that Kr1 has a stronger effect than Kr2, whereas Kr3 seemed weaker than the two other genes (Krolow 1970).
To further explore the mechanisms controlling crossability in wheat, Snape et al. (1979) performed crosses between the wild barley Hordeum bulbosum and the wheat cultivars Chinese Spring and Hope as well as 21 substitution lines carrying individual chromosomes of Hope in the background of Chinese Spring. The results revealed that the Kr1 and Kr2 genes on chromosomes 5B and 5A that govern crossability between wheat and rye also are involved in controlling crossability between wheat and barley, although the percentage of crossability observed was significantly lower than with rye. Using crosses between Chinese Spring, Hope, and the entire series of substitution lines with the cultivated barley (H. vulgare L.) cv. Betzes, Fedak and Jui (1982) also suggested that the homeologous alleles of the Kr genes on chromosome group 5 of Chinese Spring (5A, 5B, and 5D) favor crossability with additive effects.
In 1998, a new locus, named SKr, controlling crossability between wheat and rye was detected using a mapping population of 187 double haploid (DH) lines produced by anther culture from F1 hybrids of a cross between the noncrossable (NC) French wheat cv. Courtot (Ct) and the Chinese crossable (C) cv. Chinese Spring (Tixier et al. 1998). SKr was identified as a major QTL located on the distal end of the short arm of chromosome 5B within a confidence interval ranging from 8.7 to 20.9 cM (Lamoureux et al. 2002). In this population, the effect of SKr was stronger (22.1% of heritability) than the one of a QTL identified on 5BL (supposedly Kr1, 5.5% of heritability), whereas no significant effect was detected on 5AL (for Kr2). Moreover, the results indicated a 95% crossability rate for cv. Chinese Spring and ∼10% for Courtot, suggesting that the Courtot genotype is Kr1Kr1/kr2kr2, whereas Chinese Spring would be kr1kr1/kr2kr2.
Here, we report the construction of a high resolution genetic map at the SKr locus using a single seed descent (SSD) population derived from a cross between Courtot and a crossable DH line (MP98) that allowed us to assess the crossability phenotype as a single “Mendelian factor” and to demonstrate the major dominant effect of SKr on crossability. Synteny between wheat, barley, and rice was used to increase the density of markers and reduce the genetic interval around the SKr gene to 0.3 cM. BAC contigs from Chinese Spring were established with closely linked and cosegregating markers to lay the foundation for positional cloning of the gene. Finally, a SSR marker cosegregating with SKr was developed and its value for the exploitation of SKr in breeding was assessed in a collection of crossable lines used to produce primary triticale.
MATERIALS AND METHODS
Plant material:
A mapping population was developed by crossing the noncrossable wheat cv. Courtot (Ct) with the highly crossable cv. Chinese Spring (CS). F1 hybrids were used to produce a population of 187 doubled haploid (DH) by anther culture and colchicine treatment (Felix et al. 1996; Cadalen et al. 1997). A highly crossable DH line (MP98) that carried Chinese Spring alleles at the SKr locus (D. Lamoureux, unpublished data) then was backcrossed with Ct followed by six generations of selfing to generate a SSD population of 618 individuals referred to hereafter as MP98 × Ct.
Crossable wheat progenies employed to increase the genetic basis of wheat in triticale breeding programs were evaluated to determine the usefulness of the SSR markers cfb306 and cfb341. The markers were tested on 11 lines originating from backcrosses between six noncrossable breeding varieties (Genesis, Balthazar, Oratorio, Altria, FD92017, and Ornicar) and the substitution line Ct(FukuhoKomugi 5B) referred to hereafter as Ct(FK 5B). In this latter line, chromosome 5B of the noncrossable Ct has been replaced by the 5B chromosome of the crossable Japanese cultivar FukuhoKomugi (FK) (Gay and Bernard 1994). Two backcrosses were carried out with a self-fertilization generation between each backcross. A large number of F2 plants were selected and crossed with rye for identifying the crossable vs. noncrossable progenies. For each F2 selection step and for each variety, 12 plants were tested for crossability using rye cv. Dankowskie Nowe. One or more compatible progeny was tested with the two SSR markers cfb306 and cfb341. In addition, 66 varieties and landraces originating from different countries [i.e., France (14 varieties), China (19 varieties and 6 landraces), India (1 variety), Japan (12 varieties), Mongolia (1 variety), Pakistan (2 varieties), and Algeria (11 varieties)] were tested to estimate the polymorphism rate of the two SSR markers. Finally, the Chinese Spring N5B/T5D (nullisomic 5B/tetrasomic 5D) line was used to identify markers specific for chromosome 5B, while the Chinese Spring deletion lines 5BS6-0.81–1.00, 5BS5-0.71–0.81, 5BS8-0.56–0.71, and 5BS4-0.43–0.56 were used to physically locate markers on the short arm of chromosome 5B.
Crossability evaluation:
The percentage of crossability of a wheat plant with rye was determined as follows: two to four wheat ears with 20 florets per ear (and sometimes more for the 11 wheat crossable progenies) were emasculated 2−3 days prior to anthesis. Apical and basal spikelets as well as all except the outermost florets of the remaining spikelet were removed to achieve uniform maturity throughout the ear. When receptive, stigmas were pollinated with fresh pollen collected from the rye cv. Dankowskie Nowe. The experiments were conducted in a greenhouse under natural photoperiod between March and July for the MP98 × Ct SSD plants. For the 11 crossable wheat progenies, means of data from greenhouse and from field conditions were used. Before and after pollination, emasculated ears were covered with bags to avoid uncontrolled pollination. Crossability was expressed as the ratio (in %) of seeds obtained 50 days after pollination divided by the number of pollinated florets for each ear examined. An average crossability percentage was then calculated for each plant.
BAC library screening:
The T. aestivum cv. Chinese Spring BAC library (Allouis et al. 2003; http://cnrgv.toulouse.inra.fr/en/library/genomic_resource/Tae-B-Chinese spring) was screened by PCR using genome-specific primers corresponding to the ATPase1-5B gene/GBR0233 marker (F-5′ CAGTGCCGTGCTTACCAGC 3′ and R-5′ AGCGTGTGCCCACTTGAGCT 3′). The primers were designed from the sequence of the Genoplante EST contig CTG_WHP_856.1-G356.103K22R011024 (http://urgi.versailles.inra.fr/GnpSeq/). In addition, genome-specific primers were designed to amplify the cfb306 (F-5′ TAAAGCGGATGGGTCTTGTT 3′ and R-5′ ATAAGATTACCTCGGGTGAA 3′) and cfb309 (F-5′ TAGGGCATATTTCCAACACT 3′ and R-5′ TAAGTCCGCGTATTAGCATT 3′) markers. The primers were designed from the sequence of the BAC clone 1793L02 [GenBank accession (acc.) no. CT009585; Chantret et al. 2005]. For the ATPase1-5B/GBR0233 marker, PCR reactions were performed in a final volume of 10 μl containing 4 μl of diluted BAC DNA pools added to a mix containing 2 μl of buffer 10×, 0.5 μl of MgCl2 (25 mm), 0.05 μl (5 units/μl) of GoTaq Flexi DNA polymerase (Promega), 0.2 μl of 10 mm dNTP mix, 0.125 μl of each primer (10 μm), and 3 μl of H2O. Forty cycles of 30 sec at 95°, 45 sec at 65°, and 1 min 30 sec at 72°, followed by a final elongation of 5 min at 72° were performed. For the cfb306 and cfb309 SSR markers, real-time PCR reactions were performed on the LightCycler 480 DNA SYBR Green I Master kit (Roche Applied Science) in a final reaction volume of 10 μl containing 4 μl of diluted BAC DNA pools, 5 μl of premix SYBR2X, 0.125 μl of each primer, and 0.75 μl of H2O. Forty cycles of 20 sec at 95°, 20 sec at 55° for cfb306, and 65° for cfb309, and 20 sec at 72°, were followed by a final elongation of 5 min at 72°.
BAC DNA extraction and Shotgun BAC sequencing:
Shotgun BAC sequencing was carried out on the BAC clones 317L24 and 2163O14. BAC DNA was extracted by alkaline bacterial lysis (QIAGEN Plasmid Midi kit, Hilden, Germany) and 200 μl of BAC DNA was then subjected to shearing using the Hydroshear (Genemachines; Thorstenson et al. 1988). The resulting fragments were separated on 1% agarose gel. DNA fragments between 500 bp and 3 kb were purified with the GFX PCR and gel purification kit (Amersham Biosciences, Little Chalfont, UK) and 2 μl of DNA was ligated into the pCR-Blunt-II-TOPO vector following the manufacturer's instructions (Invitrogen, Carlsbad, CA). The ligation product was then transformed into Electro-competent Top 10 Escherichia coli cells. Transformants were selected on agar medium containing 50 μg/ml kanamycin and 192 subclones (for 317L24 BAC clone) and 288 subclones (for BAC clone 2163O14) were picked for further DNA analysis. Subclones were sequenced from both ends with the SP6 (5′ ATTTAGGTGACACTATAGAA 3′) and T7 (5′ TAATACGACTCACTATAGGG 3′) primers, and the sequence was assembled using the Pred/Phrap/Consed programs (Ewing and Green 1998; Ewing et al. 1998). Vector sequences were clipped using CROSS_MATCH and the E. coli sequences were masked by comparing the obtained sequences with E. coli K12 sequence (GenBank acc. no. U00096).
Marker analysis and genetic mapping:
PCR reactions for SSR markers were performed under the following conditions: 3 μl of DNA (10 ng/μl) was added to 2 μl of buffer 10× containing 31 mm MgCl2, 4 μl of solution Q, 0.8 μl of Taq polymerase (QIAGEN), 0.4 μl of 10 mm dNTP mix, 1 μl of each primer (10 μm) in a total volume of 20 μl. Thirty cycles of 30 sec at 95°, 30 sec at 55°–65° depending on the primers Tm, and 30 sec at 72°, followed by a final elongation of 5 min at 72° were performed in a thermal cycler (MJ Research PTC-225). The SSR markers were detected by nonradioactive silver nitrate staining after 2-hr migration on an acrylamide gel 6% (Sigma–Aldrich) at 2000 V. The insertion site-based polymorphism (ISBP) markers were amplified as described in Paux et al. (2006).
For RFLP analysis, 20 μl of genomic DNA (1 μg/μl) from the parental lines was digested with five restriction enzymes: EcoRI, EcoRV, DraI, HindIII, and BamHI (New England Biolabs, Beverly, MA) (4 units enzyme/μg DNA). The mix was completed with 3.5 μl specific enzyme buffer and H2O to reach a final volume of 35 μl and incubated at 37° for 4 hr. Digested genomic DNA then was migrated on agarose gel 1% at 35 V for 16 hr. Alkaline transfer was performed during 24 hr on a nylon membrane (Bio-Rad, Hercules, CA) before Southern hybridization with 32P radioactively labeled probes (Megaprime DNA Labeling System kit, Amersham Biosciences, Little Chalfont, UK). A set of 18 barley ESTs and 3 RFLPs mapped in distal position on 5HS (Stein et al. 2007) was kindly provided by N. Stein (Institute of Plant Genetics and Crop Plant Research-IPK, Gatersleben, Germany). The primer pair F-5′ TCCTACCTCCATTCCCCTTT 3′ and R-5′ TCAAAATGAATCGGAAGGGT 3′ was used to amplify the STS-E41M48 marker utilizing the PCR conditions described in Williams et al. (2003).
Linkage analysis was performed with Mapmaker/exp v3.0b (Lander et al. 1987) with a LOD 3.0 and the Kosambi centimorgan (cM) function (Kosambi 1944). To check for the effect of the Kr1 and Kr2 genes on crossability in the F6 MP98 × Ct population, SSR markers located near the Kr1 and Kr2 loci on chromosomes 5BL (gwm213, gwm371, and gwm499) and 5AL (gpw2136, gwm617, gpw7007, and gwm666) 5AL, respectively, were evaluated. A coefficient of correlation was then calculated by replacing the Courtot allele (A) by 0 and the MP98 allele (B) by 1. The heterozygous alleles were considered as missing data.
RESULTS
SKr behaves as a major dominant gene in the MP98 × Ct SSD population:
The SKr gene was located previously by QTL mapping in a DH population of 187 individuals in a confidence interval of 8.7–20.9 cM at 12.7 cM from the distal end of chromosome 5BS containing the dl103, fba367, and gpw1072 markers (Tixier et al. 1998; Lamoureux et al. 2002). A DH line, MP98, showing high crossability, was then backcrossed with cv. Courtot to develop a population of 618 individuals by SSD. All individuals were phenotyped for their crossability to rye and genotyped with markers for SKr on 5BS and for the Kr1 and Kr2 QTL on 5BL and 5AL, respectively (D. Lamoureux, unpublished data). Fifty individuals with recombination between the markers dl103 and gpw1072 on 5BS were selected for further fine mapping analysis and to determine more precisely the effect and location of SKr.
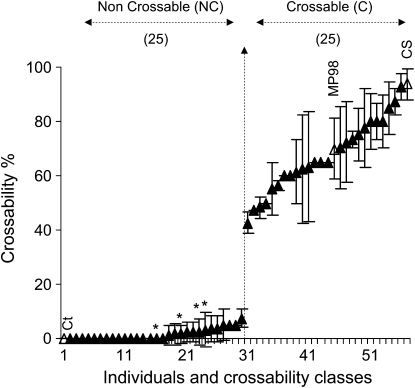
The 50 SSD recombinants as well as the parents of the mapping population MP98 and Courtot were crossed with rye to reevaluate their crossability. MP98 revealed an average value of 70 ± 11.32% while Courtot showed a complete lack of crossability with no seed (0%) obtained after pollination with rye. Two clear classes were observed in the recombinant SSD (Figure 1): 25 individuals were NC with 15 of them at 0%, 1 at 1.25%, 1 at 1.67%, 2 at 2.5%, 2 at 3.75%, 3 at 5%, and 1 individual with 7.5% of crossability. Among the 25 crossable (C) SSD, 16 individuals had crossability values from 58.68 to 81.32%, which corresponds to the average and standard deviation crossability values of the crossable MP98 parent (Figure 1), whereas the nine other recombinants showed either smaller (42.5, 47.5, 48.33, 50, 55, and 56.43%) or larger (85, 87.5, and 92.5%) average crossability values (Figure 1). Previous analysis (Tixier et al. 1998) suggested that Courtot carries the Kr1Kr1/kr2kr2 alleles and Chinese Spring the kr1/kr1/kr2kr2 alleles. Here, two main classes can be distinguished in the SSD population thereby suggesting that either SKr has a stronger effect than Kr1 and the variations observed around the parental phenotypes originates from residual Kr1 alleles or that Courtot carries a weak or recessive kr1 allele and that, in this population, only the effect of SKr can be studied. To address this question, the parental lines and the 50 SSD individuals were genotyped with seven new SSR markers located near the Kr1 (gwm213, gwm371, and gwm499) and Kr2 loci (gpw2136, gwm617, gpw7007, and gwm666) on 5BL and 5AL, respectively. An analysis of correlation between the phenotype and the genotypes at the SKr, Kr1, and Kr2 loci (supporting information, Table S2) did not indicate any significant correlation between crossability and the allelic composition at the Kr1 or Kr2 loci (Table S2). This suggests that Courtot may not carry a strong allele of Kr1 and that other minor Kr genes at other locations in Courtot are responsible for the slight phenotypic variations, thereby confirming the previous observations of allelic variations for the crossability genes (Falk and Kasha 1983).
Figure 1.—
Distribution of the crossability (in %) in the 50 recombinant SSD individuals selected at the SKr locus. Standard deviation and confidence intervals were calculated for each individual. The number of plants found in each of the two crossability classes, noncrossable (NC) and crossable (C) is indicated above the graph. The crossability values are illustrated for the reference parental lines Ct (0%), MP98 (70 ± 11.32%), and Chinese Spring (CS, 93.3 ± 5.66%) (open triangles). The four asterisks denote the individuals showing residual heterozygosity at the SKr locus.
As expected for a single major gene in a SSD population after six generations of selfing, the two phenotypic classes showed a 1:1 (25:25) (X2 = 0; P < 0.01, 1 d.f., NS) segregation. Interestingly, during fine mapping analysis, we detected four SSD lines with residual heterozygosity for the markers cosegregating and flanking the SKr gene on 5BS. All of them were noncrossable (Figure 1) and carried the Chinese Spring alleles for the Kr1 markers demonstrating that SKr has a dominant effect on the inhibition of crossability. Thus, our results indicate that SKr is a single major dominant gene inhibiting crossability. The capability of assessing SKr as a single genetic locus in the SSD population enabled us to localize further its position on the genetic map of chromosome 5BS.
High-density genetic mapping at the SKr locus using colinearity with rice and barley:
To increase the number of markers at the SKr locus and identify the location of the gene in a defined genetic interval, markers mapped on wheat chromosome 5BS in the public databases were tested for polymorphism between MP98 and Courtot, the parents of the SSD mapping population. Eight markers, including four microsatellites, gwm234, gpw4098, wmc149, and gwm443; three RFLP markers, fba367, fbb276 (with DraI restriction enzyme), and bcd873 (with BamHI restriction enzyme); and a STS marker (E41M48), revealed polymorphism between the two cultivars. Linkage analysis using the 50 SSD recombinant individuals showed that these new markers are all proximal to the SKr gene (Figure 2A). To further increase the density of markers, find a distal marker to SKr, and identify a colinear region in rice, we first investigated the barley EST genetic map (Stein et al. 2007) that consists of >1000 ESTs, including 117 on chromosome 5HS, instead of the 50 wheat ESTs that are only physically assigned to the most distal deletion bin of chromosome 5BS (5BS6-0.81–1.00; http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi; Linkiewicz et al. 2004) and are therefore less informative for identifying distal markers. Twenty-one barley markers (18 ESTs and three RFLPs) located in the distal region of 5HS (Figure 2B) were used as RFLP probes to test for polymorphism between MP98 and Courtot as well as for 5BS specificity using the nullisomic–tetrasomic line N5B/T5D and the 5BS6-0.81–1.00 deletion line. Twelve of these were specific to 5BS (Table S1); however, only 2 EST markers, GBR0233 and GBR1541 (GenBank acc. no. BU970461 and AL501780, respectively) exhibited polymorphism between the two parental cultivars and were mapped on the 50 SSD recombinant population. GBR1541 was mapped on the proximal side of SKr close to the RFLP fbb276, whereas GBR0233 was mapped at the end of chromosome 5BS on the distal side of the SKr locus (Figure 2A). Alignment of the GBR0233 sequence against the wheat EST database identified an EST (GenBank acc. no. BE606637; E-value 3e-69, score of 258 bp) assigned to the distal deletion bin (5BS6-0.81–1.00) of chromosome arm 5BS. Interestingly, BE606637 also belonged to a larger EST contig (NSFT03P2_Contig17621) of 2308 bp that contained another EST (GenBank acc. no. BE636954) assigned to the same distal 5BS deletion bin. Thus, these results confirmed the usefulness of using barley as a vehicle for wheat mapping and provided a distal marker to the SKr gene on 5BS.
Figure 2.—
Genetic maps and syntenic relationships at the SKr locus in wheat and barley. (A) Wheat 5BS genetic map (MP98 × Ct). (B) 5HS barley genetic map (Stein et al. 2007).
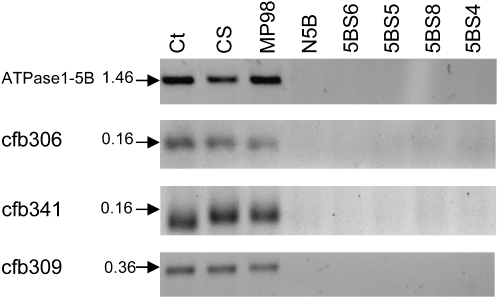
To convert the GBR0233 RFLP marker into a PCR-based marker for future high-density mapping, the longest possible wheat homologous cDNA sequence in the databases was identified as a unigene EST contig (CTG_WHP_856.1-G356.103K22R011024) of 2707 bp showing 97% of identity (E-value 0.0) to the NSFT03P2_Contig17621 sequence. CTG_WHP_856.1-G356.103K22R011024 is a consensus sequence resulting from the assembly of several ESTs obtained from the wheat cultivar Recital (http://urgi.versailles.inra.fr/GnpSeq/). In hexaploid wheat, such unigene EST contigs usually correspond to consensus sequences formed from the three homeologous A, B, and D copies of a gene. Single nucleotide polymorphisms (SNPs) were identified between three types of sequences and specific primer pairs were designed for each of them to distinguish the putative 5B homeologous copy. One of the primer pairs (F-5′ CAGTGCCGTGCTTACCAGC 3′ and R-5′ AGCGTGTGCCCACTTGAGCT 3′) amplified a product of 1464 bp (named ATPase1-5B) specific for chromosome 5B as shown, by nulli–tetrasomic and deletion mapping (Figure 3). PCR products of the same size were amplified from CS, MP98, and Courtot (Figure 3) and further sequenced [GenBank acc. no. FJ666342 (CS), FJ805255 (Ct), and FJ805254 (MP98)]. Sequence alignment showed 100% identity between the parental sequences even in the intronic regions, thereby hampering the development of a SNP-based marker for the GBR0233 locus.
Figure 3.—
Deletion mapping of four markers flanking SKr. Markers were amplified by PCR on Chinese Spring (CS) as a reference, on the nullisomic–tetrasomic line N5B-T5D (N5B), and on the deletion lines 5BS6-0.81–1.00 (5BS6), 5BS5-0.71–0.81 (5BS5), 5BS8-0.56–0.71 (5BS8), and 5BS4-0.43–0.56 (5BS4). The two parental lines Courtot (Ct) and MP98 were analyzed for polymorphism. The sizes of the amplicons, run on agarose gel 2%, are shown (in kilobases) on the left side of the figure.
The six new markers mapped at the SKr locus were incorporated with the phenotypic data into a linkage analysis providing, for the first time, a more detailed position for the SKr gene at the end of chromosome 5BS (Figure 2A). The results demonstrated that the two markers, gpw1072 and dl103, originally used for selecting the 50 SSD recombinants, are actually proximal to the SKr gene. Therefore, the recombinants selected on this basis did not include all potential recombination events in the distal part of the locus and the genetic distances estimated between SKr and the markers distal to dl103 are, likely, underestimated (Figure 2A). Nevertheless, this first recombinant map was very useful in determining the relative order of the markers and in identifying markers flanking the SKr gene for future map-based cloning with new populations.
Comparison of the two mapped EST sequences GBR0233 and GBR1541 as well as the other barley and wheat ESTs revealed the relationship between the distal region of chromosome 5B and 5HS and the rice pseudomolecules. The analysis identified homologous genes on 6 different rice chromosomes (r1, r2, r3, r9, r10, and r12) with a majority on chromosome 12 (Table S1) as observed previously by Sarma et al. (2000) and Linkiewicz et al. (2004). Os12g44150, the rice homolog to GBR0233, which is the closest marker to SKr, is annotated as a plasma membrane ATPase 1 located at the distal end of chromosome 12L, whereas the GBR1541 homologous sequence is annotated as an Enolase2 gene on chromosome 10. Thus, these results suggest that the orthologous region to SKr in rice is on chromosome 12L.
The 5B homeologous GSP locus is linked closely to the crossability gene SKr:
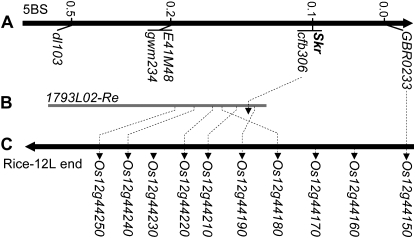
The rice homologous gene to GBR0233 near SKr, Os12g44150, is separated by only two genes in the rice genome annotation from a group of genes (Os12g44180–Os12g44250) that were identified previously as orthologous to the regions carrying the grain softness protein (GSP) gene on the short arm of wheat chromosomes group 5 (Sourdille et al. 1996; Turner et al. 1999; Igrejas et al. 2002; Chantret et al. 2004) (Figure 4) and the short arm of chromosome 5H in barley (Caldwell et al. 2004). To assess whether SKr is in the vicinity of the GSP locus on 5B and derive new markers, SSR and insertion site-based polymorphism (ISBP) markers (Paux et al. 2006) were designed from the sequence of the BAC clone 1793L02 (GenBank acc. no. CT009585) identified by Chantret et al. (2005) on chromosome 5B of cv. Renan. Out of 17 ISBP and 13 SSR markers, 5 SSR markers (cfb301, cfb302, cfb303, cfb306, and cfb309) and 9 ISBPs (cfp187, cfp188, cfp190, cfp191, cfp192, cfp193, cfp194, cfp195, and cfp1005) were specific for 5B (data not shown). Cfb306, the only marker showing polymorphism between Ct and MP98 on acrylamide gel, was found in complete segregation with SKr in the 50 recombinant SSD population (Figures 2A and 4), thereby indicating a very close linkage between the SKr and the GSP loci on chromosome 5BS. As observed for the GBR0233 marker, the polymorphism in the SKr region appeared extremely low, suggesting selective constraints in this region during domestication and/or selection. Finally, these results indicate very high microcolinearity between wheat 5BS and rice chromosome 12L in the SKr region (Figure 4) and suggest that the region around SKr which represents only 54 kb in rice may be of a reasonable size and thus amenable to efficient map-based cloning in wheat.
Figure 4.—
Genetic mapping at the SKr and the 5B homeologous GSP loci on 5BS and microcolinearity with chromosome 12L of rice. (A) Genetic map of the telomeric end of chromosome 5BS in the MP98 × Ct SSD recombinant population. (B) BAC clone 1793L02 of cv. Renan (Re) (Chantret et al. 2005) carries a microsatellite (cfb306) completely linked to SKr. (C) Schematic representation of the telomeric end of rice chromosome 12L carrying six genes conserved with BAC clone 1793L02 of T. aestivum cv. Renan.
Toward the establishment of a physical map spanning the SKr locus:
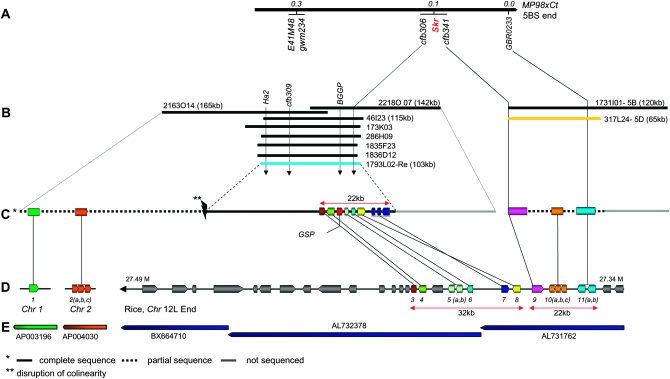
To initiate physical mapping at the SKr locus and establish a contig spanning the gene from cv. Chinese Spring, we used the cfb306 and cfb309 markers derived from BAC 1793L02 as well as the ATPase1-5B (derived from GBR0233) marker (Figure 5) to screen pools of the cv. Chinese Spring BAC library (Allouis et al. 2003) by PCR. Five positive BAC clones were identified. BACs 46I23 (115 kb), 173K03, and 1836D12 carried the two SSR markers cfb309 and cfb306, whereas BAC 2163O14 (165 kb) carried only cfb309, indicating that this BAC clone is located upstream of the three other BACs (Figure 5B). A single BAC clone 317L24 was identified with the ATPase1-5B specific primers. These results allowed us to construct an initial physical BAC contig in the SKr region (Figure 5).
Figure 5.—
Extended genetic and physical maps at the SKr locus and syntenic relationships with rice. (A) Genetic map of the SKr locus on wheat chromosome arm 5BS. (B) Physical map at the SKr locus on wheat chromosome 5BS. (C) Detailed representation of the BAC clones identified at the 5B homeologous GSP (1793L02 in blue, (Chantret et al. 2005, 2008) and SKr loci. The gene order on partially sequenced BAC 317L24 (in orange) corresponds to the order established on the genetic map for cfb341 and GBR0233. GSP-1 (grain softness protein). (D) Colinearity with genes located on rice chromosomes 1, 2, and 12L. (E) Rice BAC clones associated with the different wheat orthologous genes on chromosome 5BS. The 11 rice genes on chromosomes 1, 2, or 12 are annotated as follows: (1) Os01g14180: expressed protein; (2 a, b, c) Os02g13990: U2 small nuclear ribonucleoprotein A (U2A); (3) Os12g44250: vesicle-associated membrane protein; (4) OS12g44240: N-acetylglucosaminyltransferase; (5 a, b) Os12g44220: ATPase; (6) Os12g44210: ATPase, AAA family domain containing protein; (7) Os12g44190: ATPase 3; (8) Os12g44180: nodulin; (9) Os12g44170: pentatricopeptide; (10 a, b, c) Os12g44160: oxidoreductase; and (11 a, b) Os12g44150: plasma membrane ATPase. Gray: other genes present on rice chromosome 12.
Shotgun BAC sequencing of the BAC clone 317L24 was performed at 0.9× sequence coverage. After assembly, 21 contigs and 14 singletons representing 58467 bp of sequence were obtained and compared with the sequence of the rice pseudomolecules (http://rice.plantbiology.msu.edu/blast.shtml) using BLASTn. One contig (Ctg19) carries a gene corresponding to the plasma membrane ATPase marker GBR0233 orthologous to the rice gene Os12g44150 (Table 1). Interestingly, two other contigs (ctg20 and 09, Table 1) carry genes homologous to the rice genes Os12g44160 and Os12g44170 (Table 1), which are consecutive to Os12g44150 in the rice genome annotation. Thus, BAC 317L24 carries three wheat genes orthologous to three rice genes that are located immediately upstream of the six genes (Os12g44180 to -44250) identified previously as orthologous to the 5B homeologous GSP locus (Figure 5). To further investigate microcolinearity upstream the GSP-5B locus, we shotgun sequenced the BAC clone 2163O14 to 0.73× sequence coverage. Eighty-eight contigs and two singletons representing 122,365 bp of sequence were obtained and compared with the rice pseudomolecules using BLASTn. Two contigs (Ctg46 and Ctg101) carry genes corresponding to the rice genes Os01g14180 and Os02g13990 on rice chromosomes 1 and 2, respectively. These results indicate complete disruption of microcolinearity between the SKr/GSP-5B locus on wheat chromosome 5BS and rice chromosome 12L around the U2 small nuclear ribonucleoprotein A gene (Os02g13990) and upstream. Therefore, it is unlikely that beyond this limit the gene content of the equivalent region in rice will provide additional markers for high-density mapping at the SKr locus.
TABLE 1.
Comparative sequence analysis between the wheat genomic shotgun sequences from the BAC clones 317L24 (ctg: 19, 20, 09) and 2163O14 (ctg: 46, 101) and the rice genome sequence
| Sequence name | BAC clone | GenBank acc. no. | Size (bp) | TIGR gene | Rice chr. | Expect | Identity (%) | Annotation TIGR v.6 |
|---|---|---|---|---|---|---|---|---|
| Ctg19 | 317L24 | FJ666343 | 4484 | Os12g44150 | 12 | 1.5e-255 | 70 | Plasma membrane ATPase1 |
| Ctg20 | 317L24 | FJ666341 | 7208 | Os12g44160 | 12 | 8.0e-124 | 79 | Oxidoreductase |
| Ctg09 | 317L24 | FJ666346 | 1448 | Os12g44170 | 12 | 5.7e-163 | 82 | Pentatricopeptide |
| Ctg46 | 2163O14 | GQ219778 | 1293 | Os01g14180 | 1 | 2.1e-11 | 76 | Expressed protein |
| Ctg101 | 2163O14 | GQ219777 | 3091 | Os02g13990 | 2 | 1.5e-40 | 66 | U2 small nuclear ribonucleoprotein A |
In an attempt to develop additional markers in the distal part of the SKr gene, the shotgun sequences of BACs 317L24 and 2163O14 were used to design 10 and 20 ISBPs and 26 and 25 SSRs markers, respectively. Only 1 SSR marker named cfb331 (F-5′ TAAT TAGGGCCTGCTTCTGCT 3′ and R-5′ CAGATGCTTCCTTCATCCAAA 3′) from BAC 317l24 showed polymorphism between Courtot and MP98. The marker revealed two fragments (cfb331-5B and cfb331-5D) that were assigned to 5B and 5D by nulli–tetrasomic N5B-T5D/N5D-T5A analysis (data not shown). The 5B-specific locus was mapped in complete linkage with the SKr gene and the cfb306 SSR marker. Both fragments were cloned and sequenced (GenBank acc. nos. FJ666344 for cfb331-5B and FJ666345 for cfb331-5D). The results showed that cfb331-5D is completely identical to the BAC 317L24 shotgun sequence, whereas six SNPs were found with the cfb331-5B sequence. Thus, this indicates that BAC 317L24 originates from chromosome 5D and not from 5B and that it cannot be linked to the physical contig corresponding to the 46I23, 173K03, and 2163O14 BAC clones (Figure 5B). Nevertheless, this allowed us to design a 5B-specific STS marker renamed cfb341 (F-5′ TAATTAGGGCCTGCTTCTGCT 3′ and R-5′ TTCCTTCATCCAAAGAGACTGG 3′) (Figures 3 and 5A) that can be used now to rescreen the Chinese Spring BAC library. Interestingly, the cfb341 microsatellite is located within a gene (in contig 09, GenBank acc. no. FJ666346; Table 1) showing homology to the pentatricopeptide gene Os12g44170 in rice (Figure 5E and Table 1). Our results thus indicate that two markers (GBR0233 and cfb341) corresponding to two genes (plasma membrane ATPase1 and pentatricopeptide) separated only by 6 kb in rice are present on a single BAC clone (317L24) and are separated by at least one recombination event on 5BS. This suggests a high recombination rate in the distal SKr region and a physical-to-genetic distance ratio favorable for map-based cloning of the SKr gene (Figure 5).
To complete the physical map at the SKr locus on 5BS, a second screening of the Chinese Spring BAC library was initiated by hybridization. Four probes were used: Ha2 (low copy sequence located at 19.97 kb upstream of the cfb309 marker), BGGP (corresponding to N-acetylglucosaminyltransferase gene located at 16.71 kb upstream of the cfb306 marker), SG317L24P2-E01 (corresponding to the partial sequence of the pentatricopeptide gene carrying cfb341), and ATPase1-5B (the partial genomic sequence of the plasma membrane ATPase1 gene) (Figure 5). Four new BAC clones were identified in addition to the BAC clones found previously in the first PCR screening. BACs 286H09 and 1835F23 were obtained with both the Ha2 and BGGP probes. BAC 2218O07 (120 kb) was revealed with the BGGP probe only, whereas BAC 1731I01 (142 kb) was obtained with both the SG317L24P2-E01 and ATPase1-5B probes (Figure 5). The 5B origin of the 1731I01 BAC clone was confirmed by PCR using the 5B-specific cfb341 marker. With this result, we were able to establish a contig of ∼300 kb on the proximal side of SKr and identify a BAC clone (1731I01) of 120 kb on the distal side. BAC end sequencing of the 1731I01 clone showed that cfb341 is located at the end of the BAC clone and, therefore, that there is still a gap between the two physical contigs. Thus, with this work, we have identified new markers and established the foundation for designing additional ones by shotgun sequencing the 46I23 and 2218O07 BAC clones. These markers will be used to identify recombinants in novel high-density mapping populations to locate eventually the SKr gene within a genetic and physical interval for final map-based cloning.
The cfb306 and cfb341 markers are efficient tools for introducing high crossability alleles in elite wheat varieties:
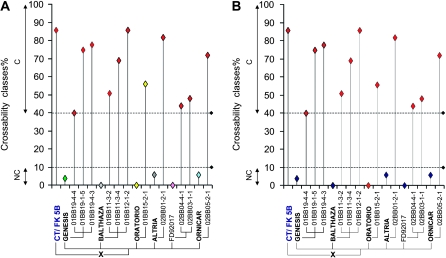
The cfb306 and cfb341 SSR markers are linked totally with the SKr gene in our population and, therefore, represent potentially good markers for introducing crossability alleles of SKr into breeding programs that aim at increasing the genetic basis of wheat for the production of primary triticale or for crossing with other related species. To evaluate their usefulness in such applications, BC2 lines produced from wheat elite varieties that serve as parents in primary triticale production were tested with the two markers. The BC2 panel consisted of 11 crossable progenies resulting from six noncrossable elite wheat varieties that had been crossed with Ct(FK 5B), a crossable substitution line carrying a chromosome 5B originating from the Japanese wheat accession FukuhoKomugi (FK) (Gay and Bernard 1994) (Figure 6). The results indicate complete association between the two SSR markers and crossability with rye except for one individual (01BB15-2-1 in Oratorio × Ct(FK 5B) that showed recombination between cfb306 and crossability (Figure 6) and no polymorphism for cfb341. However, phenotyping of this line indicated high variability (ranging from 6 to 58%) and these results need to be confirmed. Together, these data confirm that the two SSR markers cfb306 and cfb341 are located very close to the crossability gene SKr on chromosome 5BS and strongly support the genetic mapping data obtained with the 50 SSD recombinant population. They also revealed high allelic variability for the cfb306 marker with six different alleles found in seven different lines. In contrast, only two alleles were observed with cfb341 (Figure 6) and, to confirm this feature, a polymorphism test was carried out with the two markers on 66 wheat varieties and landraces originating from different regions of the world (data not shown). For cfb341, only two alleles were detected in 65 varieties and a third allele was found for a wheat variety originating from Pakistan. For cfb306, 18 alleles were found in the 66 lines confirming the extreme rate of polymorphism of this SSR marker and its interest for breeding.
Figure 6.—
Graphical representation of the phenotypes and genotypes for all parents and progenies of the different crosses with Ct(FK 5B). (A) Genotyping with the cfb306 marker. (B) Genotyping with the cfb341 marker. The crossability ratio (in %) is represented on the Y-axis, whereas the parental lines and the crossable BC2 progenies identified from each cross are indicated on the X-axis. The colored dots represent the different alleles for a given marker.
DISCUSSION
So far, wheat crossability genes have been identified either by deletion or substitution lines analyses (Lein 1943; Riley and Chapman 1967; Lange and Riley 1973; Sitch et al. 1985) with very few linkage analyses studies thereby making it difficult to discriminate precisely between the relative effects of the different Kr genes, especially for those located on the same chromosomes. The first evidence for the presence of a crossability gene on 5BS came from the telocentric mapping studies of Sitch et al. (1985) in which the crossability of wheat monotelosomic CS lines with Hordeum bulbosum revealed that a gene inhibiting crossability is present on the short arm of chromosome 5B in addition to the Kr1 gene already identified on 5BL. Subsequently, the SKr locus was identified as a major QTL (Tixier et al. 1998; Lamoureux et al. 2002) inhibiting crossability of wheat to rye on 5BS; however, at that time, it could not be mapped with precision and its relative effect to Kr1 that was revealed as a small QTL in the same study remained unclear. Here, using a SSD population, we were able to score SKr as a single Mendelian factor and demonstrate that it acts as a dominant gene inhibiting crossability. This confirms the first results of Riley and Chapman (1967) on the Kr1 mode of action wherein it was concluded that crossability is not promoted by recessive alleles but inhibited by dominant alleles. Later studies performed to determine the site of action of the Kr1 and Kr2 genes (Lange and Wojciechowska 1976; Jalani and Moss 1980; Jalani and Moss 1981) also demonstrated that dominant alleles from Hope, CS/Hope 5B, and CS/Hope 5A result in inhibition of the pollen tube growth between the style base and the embryo sac. Finally, Cameron and Reger (1991) showed that a soluble, dialyzed lysate extracted from the ovaries of Hope and CS/Hope 5B inhibits rye pollen tube elongation significantly more than a similar lysate from Chinese Spring ovaries. It remains to be proven if SKr also inhibits the rye pollen tube growth and acts through the same inhibitory pathway as the other Kr genes. Cloning and characterization of the SKr and Kr1 genes (G. Moore, personal communication) on chromosome 5B will help in determining whether the SKr and Kr1 genes on 5B act in additive or complementary ways.
Our results suggest also that in Courtot, SKr has a stronger effect than Kr1 on crossability of wheat to rye. Riley and Chapman (1967) showed that with 74.3% of crossability in Chinese Spring, 6.4% in CS/Hope 5B, 26.2% in CS/Hope 5A, and 0% in Hope, chromosome 5B of Hope that carries Kr1 has a stronger inhibition effect than chromosome 5A with Kr2. However, it is important to note that at that time, the effect of SKr on 5BS had not been identified. To our knowledge, it is not known whether Hope carries SKr; therefore, it is possible that the low crossability observed in the CS/Hope 5B substitution line also originates from SKr and that this was undermined so far in the absence of markers for SKr on 5BS. Alternatively, it is possible that Courtot carries a weak Kr1 allele and that the MP98 × Ct population is not suitable for comparing the relative strength of the two genes. This also could explain why SKr was identified as the main QTL for crossability in the original QTL analyses whereas Kr1 was detected only as a minor QTL (Tixier et al. 1998; Lamoureux et al. 2002). One of the difficulties in comparing the different crossability studies is that even if they used the same crossable line Chinese Spring, they were performed with different noncrossable varieties for which the allelic variation at the SKr and Kr loci was not assessed systematically. To address this question, it would be interesting in the first place to produce and compare ditelosomic 5BL and 5BS lines from Courtot and Hope. The markers developed in this study and future fine mapping at other Kr loci would be useful to better distinguish the relative effects of the different genes in the Hope × CS populations.
Together with previous studies (Krolow 1970; Falk and Kasha 1983), our results indicate allelic variability among the different crossability genes. At the Kr1 locus, for example, Falk and Kasha (1983) demonstrated that Atlas66 and Hope had stronger inhibitor alleles than Cheyenne and Chinese Spring. For SKr, we propose that Chinese Spring as well as Fukuhogomugi carry weak alleles the effect of which can be detected only by using ditelosomic mapping (Sitch et al. 1985) and that most of the noncrossable European wheat varieties, such as Courtot and the six other French varieties tested here, carry strong inhibiting SKr alleles. In a study of 1400 varieties, Zeven (1987) showed that most bread wheat varieties or landraces displaying high crossability with rye originate from China, Japan, East Siberia, and Iran. This raises interesting questions on the evolution of crossability in wheat.
Both tetraploid and hexaploid wheat possess genetic systems that regulate crossability with rye; however, it is not clear yet whether the same genes are involved in both species. Liu et al. (1999) mapped the major crossability genes in tetraploid wheat to chromosomes 1A, 6A, and 7A and suggested that crossability in Chinese tetraploid and hexaploid wheat may be controlled by different genetic systems. If tetraploid wheat, indeed, exploits a different genetic system, then it brings into question the origin of the hexaploid wheat system. The fact that the Asian wheat pool is highly crossable compared to the European pool suggests that the inhibition barrier is an ancestral state found in the Fertile Crescent where rye was domesticated at the same time as wheat and both have been maintained and cultivated in Europe ever since. In contrast, in Asia where the wheat population structure is clearly different (Balfourier et al. 2007), rye has never been cultivated in high amount in the vicinity of wheat possibly allowing the development of mutation and allelic variants that became crossable with rye. A systematic analysis of crossability with rye in tetraploid and hexaploid wheat core collections (Balfourier et al. 2007) would facilitate the study of the origin of high crossability in the Asian wheat pool. Preliminary data indicate the SSR markers cfb306 and cfb341 can be used also to investigate the tetraploid wheat pool and that cfb306 is highly polymorphic in T. durum accessions (4 different alleles among five varieties from Algeria, data not shown).
High-density mapping using microcolinearity with barley as a vehicle:
Using a SSD recombinant population, we were able to mendelize the crossability gene SKr and localize it in a small (<1 cM) genetic interval with five closely linked molecular markers at the telomeric end of the short arm of chromosome 5B. Our result indicates that the previous selection of the 50 SSD made by D. Lamoureux, (unpublished data) was performed incorrectly because it was based on the assumption that SKr was located between the dl103 and gpw1072 markers. Thus, it is clear that the current estimates of genetic distances around SKr are biased and that the order of the markers very closely linked or completely segregating with SKr remains to be confirmed with a new population. However, the association study performed with the wheat lines used for primary triticale production provides strong evidence for the location of SKr between the gwm234 and GBR0233 markers in close vicinity of the cfb306 and cfb341 markers.
The saturation of the target gene region by molecular markers is an important step in map-based cloning projects. Comparative studies had reported good colinearity between wheat and rice at large scale (Moore et al. 1995; Feuillet and Keller 2002; Sorrells et al. 2003) and more complex relationships at the subcentimorgan or sequence level (Bennetzen and Ramakrishna 2002; Feuillet and Keller 2002) with generally better conservation in the proximal regions (Sanmiguel et al. 2002; Yan et al. 2003) compared to the distal regions (Kilian et al. 1997; Feuillet and Keller 1999; Li and Gill 2002; Linkiewicz et al. 2004). The SKr gene is located in a very distal position on 5BS and low-resolution mapping indicates high disruption of colinearity between wheat and rice in this region making it difficult to use rice as a new source of markers in the first place. Moore et al. (1995) and Sorrells et al. (2003) showed that the wheat homeologous group 5 represents one of the most complex syntenic relationships with rice. Linkiewicz et al. (2004) constructed high-density deletion bin maps of wheat chromosomes 5A, 5B, and 5D, including a total of 2555 loci and revealed that group 5 is, generally, colinear with chromosomes 12, 9, and 3 of rice. Other studies reported that the chromosome arm 5BS is colinear with rice chromosome 11 (Lamoureux et al. 2002). However, in our study the lack of mapped genes near SKr prevented the identification of the rice orthologous region for new marker design in the first place.
The development of high-density EST maps in barley (Stein et al. 2007) enabled us to circumvent this problem and served as an efficient bridge to reach sufficient resolution and establish finally a close relationship with rice chromosome 12. This illustrates the importance of developing barley resources to support wheat genomics projects and reinforces the fact that barley is the best model for wheat. Barley belongs to the same tribe, shares the same number of chromosomes, and diverged from wheat only 10–14 million years ago, in contrast to Brachypodium (Bossolini et al. 2007) and rice (Wolfe et al. 1989) that diverged from wheat 30–50 million years ago. As a diploid, EST mapping in barley is more efficient than in bread wheat in which genome-specific markers need to be developed for each homeologous copy of a gene. The establishment of high throughput gene mapping platforms in barley (OPA1; http://harvest.ucr.edu/Barley1.htm) together with the construction of anchored physical maps of wheat and barley through international consortia (www.wheatgenome.org; www.barleygenome.org) will greatly support map-based cloning in wheat in the near future. Several map-based cloning projects in wheat have revealed the limits of using rice or Brachypodium as genome models to identify candidate genes in particular for fast evolving pathways such as disease resistance (Keller et al. 2005). It will be interesting to see whether crossability genes are conserved between wheat and rice or Brachypodium. At the moment, we can only speculate about the possible candidates found in the SKr orthologous rice region on chromosome 12L. Genetic and biochemical analyses both suggest that inhibition of crossability is an active mechanism and that the crossability genes may encode a product that stops the penetration of the rye pollen tube between the style base and the embryo sac, i.e., just before fertilization (Lange and Wojciechowska 1976; Jalani and Moss 1980; Jalani and Moss 1981). In dicots, molecular analyses of self-incompatibility and interspecies pollen tube rejection have identified key roles for S-RNAse genes (Franklin-Tong and Franklin 2003) as well as receptor kinases (Escobar-Restrepo et al. 2007). In rice, a number of studies have indicated loci involved in the control of intersubspecific and interspecific reproduction barriers through fertilization prevention, hybrid male sterility, embryo sac abortion, or hybrid breakdown. They were identified on rice chromosomes 1 (S24, Kubo et al. 2008), 2 (S32, Li et al. 2007), 5 (S31, S35, Kubo et al. 2008; Zhao et al. 2007), 6 (S1, S5, esa-1, Liu et al. 2000; Qiu et al. 2005; Chen et al. 2008), and 12 (esa-2 and esa-3, Liu et al. 2000), but not in the orthologous region to SKr. S5 has been cloned recently (Chen et al. 2008) and was shown to encode an aspartic protease conditioning embryo-sac fertility. None of the nine genes colinear between wheat and rice at the SKr locus revealed homology to this type of gene and it will be interesting to see whether SKr corresponds to a new type.
Toward map-based cloning of SKr:
In this study, we mapped new molecular markers and established 2 physical contigs of ∼300 kb and 120 kb at the SKr locus. Because the current SSD population was not selected with flanking markers, we cannot be certain that the physical contigs actually flank the gene. However, the association analysis performed with the cfb306 and cfb341 markers makes us confident that the two markers are extremely close to SKr and that they will be very useful to select recombinants in a new large population mapping. To support our data, we also mapped the gpw1072, gwm234, cfb306, and cfb341 markers using 94 individuals of a Renan × CS F2 population (Saintenac et al. 2009) (data not shown). The results confirmed the distal position of gwm234, cfb306, and cfb341 compared to gpw1072 and indicated recombination between the cfb306 and cfb341 markers. This demonstrates that even though the SSD map underestimated recombination rates and genetic distances, it represents a good basis for further high-resolution mapping of the SKr gene. Together with the identification of a recombination between cfb341 and the GBR0233 marker within a single BAC in the SSD population, these results also suggest high recombination rates in the SKr region, which should greatly support efficient map-based cloning of the gene. We are developing currently a heterogeneous inbred family population (Tuinstra et al. 1997) using a line (SSD254) heterozygous for the molecular markers GBR0233, cfb341, cfb306, gwm234, E41M48, dl103, fbb267, and bcd873 that flank SKr (W. Alfares and C. Feuillet, unpublished data) and carrying the alleles from Courtot for the other loci on 5BS.
The fact that the BAC contigs at the SKr locus span only 54 kb in rice suggests that the physical distance between the two wheat contigs may not be too large and that a single walking step could be sufficient to link both contigs. Currently, we are screening the Chinese Spring BAC library with markers derived from the BAC ends of these contigs to establish a single contig in the region. Finally, one of the potential difficulties that we may face in the final steps of map-based cloning concerns the low polymorphism observed in the SKr/GSP-5B region on 5BS. Only one out of 30 markers originating from the BAC 1793L02 showed polymorphism between CS and Ct and no single sequence difference was found over 1464 bp of sequence including an intronic region for the ATPase1-5B gene. This is very similar to the observations of Chantret et al. (2005, 2008) who found only 58 SNPs and a few short indels between the T. aestivum and the T. turgidum orthologous sequences at the GSP locus on chromosome 5BS. A possible explanation is that deletions in some of the genes present in this region are highly deleterious and have been counter-selected through the domestication and selection processes.
A new and efficient diagnostic marker for crossability:
Until now, the introduction of crossability genes into noncrossable wheat elite lines has been performed mostly by backcrossing without markers. This is laborious, lengthy, and rather inefficient because crossability with rye is recessive and a self-fertilization generation is required to enable the identification of plants carrying the crossability gene before further use. In addition, to avoid losing one generation time in the breeding process, crossability testing in the absence of markers requires that all F2 plants are crossed with rye (12 per population) and with the recurrent wheat parent when only 25% of the F2 plants will be eventually crossable with rye. Thus, markers can greatly improve the efficiency and reliability of the primary triticale backcross process by allowing the identification of crossable progenies without self-fertilization and testcrosses with rye. Our results reveal that cfb306, which is closely linked to SKr, has all of the characteristics of an efficient marker for the introduction of crossability in wheat germplasm through marker-assisted selection. As a SSR marker, it is amenable to high throughput genotyping and its high level of allelic polymorphism will allow its implementation in a large variety of wheat lines. Finally, our results support previous data suggesting that the crossability genes in wheat exhibit high levels of allelic variability. Thus, to ensure the efficient introduction of crossability into wheat varieties, a careful analysis of the allelic composition of the recipient lines at the major crossability loci and, in particular at Kr1 and SKr, will be needed. The markers developed for SKr in this study and for other loci (Kr1, G. Moore, personal communication) will be extremely useful in this regard.
Acknowledgments
We are very grateful to Delphine Boyer, Alain Loussert, Jacqueline Philippon, and Bouzid Charef for the excellent and dedicated technical support they have provided all along this project. We also thank Philippe Leroy, Frederic Choulet, Etienne Paux, Sébastien Faure, and Jérome Salse for their support with the bioinformatics analysis and for fruitful discussions. In addition, we acknowledge Nils Stein (Institut für Pflanzengenetik und Kulturpflanzenforschung, Gatersleben, Germany) for providing us with the barley EST and RFLP probes and Boulos Chalhoub (Unité de Recherche en Génomique Végétale, France) for providing us with BAC clones for the Ha locus in Chinese Spring. The authors thank the GIE Triticale for allowing the use of the crossable wheat progenies for validation of the SSR markers in this study. For editing and critical review of the manuscript, the authors thank Graham Moore, Kellye Eversole, and Pierre Barret. This research has been funded by grants from the Institut National de la Recherche Agronomique (INRA, France) and from the University of Aleppo (Syria).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107706/DC1.
References
- Allouis, S., G. Moore, A. Bellec, R. Sharp, P. Faivre Rampant et al., 2003. Construction and characterisation of a hexaploid wheat (Triticum aestivum L.) BAC library from the reference germplasm ‘Chinese Spring’. Cereal Res. Commun. 31: 331–338. [Google Scholar]
- Backhouse, W. O., 1916. Note on inheritance of crossability. J. Genet. 6: 91–94. [Google Scholar]
- Balfourier, F., V. Roussel, P. Strelchenko, F. Exbrayat-Vinson, P. Sourdille et al., 2007. A worldwide bread wheat core collection arrayed in a 384-well plate. Theor. Appl. Genet. 114: 1265–1275. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and W. Ramakrishna, 2002. Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48: 821–827. [DOI] [PubMed] [Google Scholar]
- Bossolini, E., T. Wicker, P. A. Knobel and B. Keller, 2007. Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J. 49: 704–717. [DOI] [PubMed] [Google Scholar]
- Cadalen, T., C. Boeuf, S. Bernard and M. Bernard, 1997. An intervarietal molecular marker map in Triticum aestivum L. Em. Thell. and comparison with a map from a wide cross. Theor. Appl. Genet. 94: 367–377. [Google Scholar]
- Caldwell, K. S., P. Langridge and W. Powell, 2004. Comparative sequence analysis of the region harboring the hardness locus in barley and its colinear region in rice. Plant Physiol. 136: 3177–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, R. G., and B. J. Reger, 1991. Hybridization barriers between wheat and rye: in vitro pollen assays and electrophoretic survey. Euphytica 52: 147–153. [Google Scholar]
- Chantret, N., A. Cenci, F. Sabot, O. Anderson and J. Dubcovsky, 2004. Sequencing of the Triticum monococcum Hardness locus reveals good microcolinearity with rice. Mol. Genet. Genomics 271: 377–386. [DOI] [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, S. Rahman, A. Bellec et al., 2005. Molecular basis of evolutionary events that shaped the Hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, A. Bellec, B. Laubin et al., 2008. Contrasted microcolinearity and gene evolution within a homoeologous region of wheat and barley species. J. Mol. Evol. 66: 138–150. [DOI] [PubMed] [Google Scholar]
- Chen, J., J. Ding, Y. Ouyang, H. Du, J. Yang et al., 2008. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. USA 105: 11436–11441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo, J. M., N. Huck, S. Kessler, V. Gagliardini, J. Gheyselinck et al., 2007. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Falk, D. E., and K. J. Kasha, 1983. Genetic-studies of the crossability of hexaploid wheat with rye and Hordeum bulbosum. Theor. Appl. Genet. 64: 303–307. [DOI] [PubMed] [Google Scholar]
- Fedak, G., 1985. Alien species sources of physiological traits for wheat improvement. Euphytica 34: 673–680. [Google Scholar]
- Fedak, G., and P. Y. Jui, 1982. Chromosomes of Chinese Spring wheat carrying genes for crossability with Betzes barley. Can. J. Genet. Cytol. 24: 227–233. [Google Scholar]
- Felix, I., J. P. Martinant, M. Bernard, S. Bernard and G. Branlard, 1996. Genetic characterization of storage proteins in a set of F1-derived haploid lines in bread wheat. Theor. Appl. Genet. 92: 340–346. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96: 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 2002. Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann. Bot. (Lond) 89: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., P. Langridge and R. Waugh, 2008. Cereal breeding takes a walk on the wild side. Trends Genet. 24: 24–32. [DOI] [PubMed] [Google Scholar]
- Florell, V. H., 1931. A genetic study of wheat x rye hybrids and back crosses. J Agric Res 42: 315–339. [Google Scholar]
- Franklin-Tong, N., and F. C. H. Franklin, 2003. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 8: 598–605. [DOI] [PubMed] [Google Scholar]
- Fu, Y. B., and D. J. Somers, 2009. Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 49: 161–168. [Google Scholar]
- Gay, G., and M. Bernard, 1994. Production of intervarietal substitution lines with improved interspecific crossability in the wheat cv Courtot. Agronomie 14: 27–32. [Google Scholar]
- Igrejas, G., P. Leroy, G. Charmet, T. Gaborit, D. Marion et al., 2002. Mapping QTLs for grain hardness and puroindoline content in wheat (Triticum aestivum L.). Theor. Appl. Genet. 106: 19–27. [DOI] [PubMed] [Google Scholar]
- Jalani, B. S., and J. P. Moss, 1980. The site of action of the crossability genes (Kr1, Kr2) between Triticum and Secale. I. Pollen germination, pollen tube growth, and number of pollen tubes. Euphytica 29: 571–579. [Google Scholar]
- Jalani, B. S., and J. P. Moss, 1981. The site of action of crossability genes (Kr1, Kr2) between Triticum and Secale. II. Proportion of pistils containing pollen tubes and effects of alternate pollination on seed set. Euphytica 30: 105–112. [Google Scholar]
- Jiang, J. M., B. Friebe and B. S. Gill, 1994. Recent advances in alien gene transfer in wheat. Euphytica 73: 199–212. [Google Scholar]
- Keller, B., C. Feuillet and N. Yahiaoui, 2005. Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize genome. Genet. Res. 85: 93–100. [DOI] [PubMed] [Google Scholar]
- Kilian, A., J. Chen, F. Han, B. Steffenson and A. Kleinhofs, 1997. Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4 using rice as an intergenomic cloning vehicle. Plant Mol. Biol. 35: 187–195. [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann Eugen 12: 172–175. [Google Scholar]
- Krolow, K. D., 1970. Investigations on compatibility between wheat and rye. Z PXanzenzüchtg 64: 44–72. [Google Scholar]
- Kubo, T., Y. Yamagata, M. Eguchi and A. Yoshimura, 2008. A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.). Genes Genet. Syst. 83: 443–453. [DOI] [PubMed] [Google Scholar]
- Lamoureux, D., C. Boeuf, F. Regad, O. Garsmeur, G. Charmet et al., 2002. Comparative mapping of the wheat 5B short chromosome arm distal region with rice, relative to a crossability locus. Theor. Appl. Genet. 105: 759–765. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lange, W., and R. Riley, 1973. The position on chromosome 5B of wheat of the locus determining crossability with rye. Genet. Res. 22: 143–153. [Google Scholar]
- Lange, W., and B. Wojciechowska, 1976. The crossing of common wheat (Triticum aestivum L.) with cultivated rye (Secale cereale L.). I. Crossability, pollen grain germination and pollen tube growth. Euphytica 25: 609–620. [Google Scholar]
- Lein, A., 1943. The genetical basis of the crossability between wheat and rye. Z Indukt Abstamm Vererbungsl 81: 28–59. [Google Scholar]
- Li, D., L. Chen, L. Jiang, S. Zhu, Z. Zhao et al., 2007. Fine mapping of S32(t), a new gene causing hybrid embryo sac sterility in a Chinese landrace rice (Oryza sativa L.). Theor. Appl. Genet. 114: 515–524. [DOI] [PubMed] [Google Scholar]
- Li, W. L., and B. S. Gill, 2002. The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the Triticeae. Genetics 160: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkiewicz, A. M., L. L. Qi, B. S. Gill, A. Ratnasiri, B. Echalier et al., 2004. A 2500-locus bin map of wheat homoeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics 168: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. C., C. Yen, J. L. Yang, Y. L. Zheng and X. J. Lan, 1999. The chromosomal locations of high crossability genes in tetraploid wheat Triticum turgidum L. cv. Ailanmai native to Sichuan, China. Euphytica 108: 79–82. [Google Scholar]
- Liu, Y. S., L. H. Zhu, J. S. Sun and Y. Chen, 2000. Mapping QTLs for defective female gametophyte development in an inter-subspecific cross in Oryza sativa L. Theor. Appl. Genet. 102: 1243–1251. [Google Scholar]
- Moore, G., K. M. Devos, Z. Wang and M. D. Gale, 1995. Cereal genome evolution: grasses, line up and form a circle. Curr. Biol. 5: 737–739. [DOI] [PubMed] [Google Scholar]
- Paux, E., D. Roger, E. Badaeva, G. Gay, M. Bernard et al., 2006. Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 48: 463–474. [DOI] [PubMed] [Google Scholar]
- Qiu, S. Q., K. Liu, J. X. Jiang, X. Song, C. G. Xu et al., 2005. Delimitation of the rice wide compatibility gene S5n to a 40-kb DNA fragment. Theor. Appl. Genet. 111: 1080–1086. [DOI] [PubMed] [Google Scholar]
- Riley, R., and V. Chapman, 1967. The inheritance in wheat of crossability with rye. Genet. Res. 9: 259–267. [Google Scholar]
- Saintenac, C., M. Falque, O. C. Martin, E. Paux, C. Feuillet et al., 2009. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181: 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmiguel, P. J., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct. Integr. Genomics 2: 70–80. [DOI] [PubMed] [Google Scholar]
- Sarma, R. N., L. Fish, B. S. Gill and J. W. Snape, 2000. Physical characterization of the homoeologous Group 5 chromosomes of wheat in terms of rice linkage blocks, and physical mapping of some important genes. Genome 43: 191–198. [PubMed] [Google Scholar]
- Sitch, L. A., J. W. Snape and S. J. Firman, 1985. Intrachromosomal mapping of crossability genes in wheat (Triticum aestivum). Theor. Appl. Genet. 70: 309–314. [DOI] [PubMed] [Google Scholar]
- Snape, J. W., V. Chapman, J. P. Moss, C. E. Blanchard and T. E. Miller, 1979. The crossabilities of wheat varieties with Hordeum bulbosum. Heredity 42: 291–298. [Google Scholar]
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdille, P., M. R. Perretant, G. Charmet, P. Leroy, M. F. Gautier et al., 1996. Linkage between RFLP markers and genes affecting kernel hardness in wheat. Theor. Appl. Genet. 93: 580–586. [DOI] [PubMed] [Google Scholar]
- Stein, N., M. Prasad, U. Scholz, T. Thiel, H. Zhang et al., 2007. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor. Appl. Genet. 114: 823–839. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., and S. R. McCouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Thorstenson, Y. R., S. P. Hunicke-Smith, P. J. Oefner and R. W. Davis, 1988. An automated hydrodynamic process for controlled, unbiased DNA shearing. Genome Res. 8: 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier, M. H., P. Sourdille, G. Charmet, G. Gay, C. Jaby et al., 1998. Detection of QTLs for crossability in wheat using a doubled haploid population. Theor. Appl. Genet. 97: 1076–1082. [Google Scholar]
- Tuinstra, M. R., G. Ejeta and P. B. Goldsbrough, 1997. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor. Appl. Genet. 95: 1005–1011. [Google Scholar]
- Turner, M., Y. Mukai, P. Leroy, B. Charef, R. Appels et al., 1999. The Ha locus of wheat: identification of a polymorphic region for tracing grain hardness in crosses. Genome 42: 1242–1250. [PubMed] [Google Scholar]
- Williams, C. E., C. C. Collier, N. Sardesai, H. W. Ohm and S. E. Cambron, 2003. Phenotypic assessment and mapped markers for H31, a new wheat gene conferring resistance to Hessian fly (Diptera: Cecidomyiidae). Theor. Appl. Genet. 107: 1516–1523. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., M. Gouy, Y. W. Yang, P. M. Sharp and W. H. Li, 1989. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86: 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco-Hernandez, J. A., F. Santiveri, A. Michelena and R. Javier Peña, 2005. Durum wheat (Triticum turgidum L.) carrying the 1BL/1RS chromosomal translocation: agronomic performance and quality characteristics under Mediterranean conditions. Eur. J. Agron. 22: 33–43. [Google Scholar]
- Zeven, A. C., 1987. Crossability percentages of some 1400 bread wheat varieties and lines with rye. Euphytica 36: 299–319. [Google Scholar]
- Zhao, Z. G., L. Jiang, W. W. Zhang, C. Y. Yu, S. S. Zhu et al., 2007. Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. L., M. C. Luo, C. Yen and J. L. Yang, 1992. Chromosome location of a new crossability gene in common wheat. Wheat Inf. Serv. 75: 36–40. [Google Scholar]