Abstract
Coordinating homeostasis of multiple metabolites is a major task for living organisms, and complex interconversion pathways contribute to achieving the proper balance of metabolites. AMP deaminase (AMPD) is such an interconversion enzyme that allows IMP synthesis from AMP. In this article, we show that, under specific conditions, lack of AMPD activity impairs growth. Under these conditions, we found that the intracellular guanylic nucleotide pool was severely affected. In vivo studies of two AMPD homologs, Yjl070p and Ybr284p, indicate that these proteins have no detectable AMP, adenosine, or adenine deaminase activity; we show that overexpression of YJL070c instead mimics a loss of AMPD function. Expression of the yeast transcriptome was monitored in a AMPD-deficient mutant in a strain overexpressing YJL070c and in cells treated with the immunosuppressive drug mycophenolic acid, three conditions that lead to severe depletion of the guanylic nucleotide pool. These three conditions resulted in the up- or downregulation of multiple transcripts, 244 of which are common to at least two conditions and 71 to all three conditions. These transcriptome results, combined with specific mutant analysis, point to threonine metabolism as exquisitely sensitive to the purine nucleotide balance.
THE purine nucleotides, ATP and GTP, are involved in almost all aspects of cellular life. In addition to their role as building blocks of nucleic acids, adenylic and guanylic nucleotides have specific roles. For example, GTP is critical for translation and for signaling through GTPases, while ATP is the major energy-providing molecule in the cell. In yeast, intracellular concentrations of ATP and GTP are clearly different (∼5 and 1.5 mm, respectively; Breton et al. 2008; Gauthier et al. 2008), most probably as the result of regulatory processes that maintain homeostasis. In most eukaryotic cells, including yeast, adenylic and guanylic nucleotides either are synthesized from a common precursor (IMP) or are recycled from preformed bases or nucleosides (Figure 1). While most enzymes involved in these processes have been identified, the physiological consequences of purine nucleotide imbalance are far from being understood. Interestingly, drugs specifically inhibiting GTP synthesis, such as mycophenolic acid (MPA), have a strong immunosuppressive effect and are now widely used to limit allograft rejection. We have previously established the effect of MPA on the yeast proteome (Escobar-Henriques et al. 2001) and have identified numerous yeast mutants hypersensitive to this drug (Desmoucelles et al. 2002). MPA effects are due to GTP shortage, since they are reversed by exogenous guanine, allowing replenishment of the GTP pool. However, drugs often have secondary effects and can be detoxified and/or diluted during cell growth. As an alternative, consequences of purine nucleotide imbalance can be investigated using yeast mutants. In two previous studies, we have used specific mutants to increase the GTP pool or decrease the ATP pool (Breton et al. 2008; Gauthier et al. 2008). In this study, we take advantage of a conditional phenotype of an AMP deaminase mutant to revisit GTP shortage consequences in yeast.
Figure 1.—
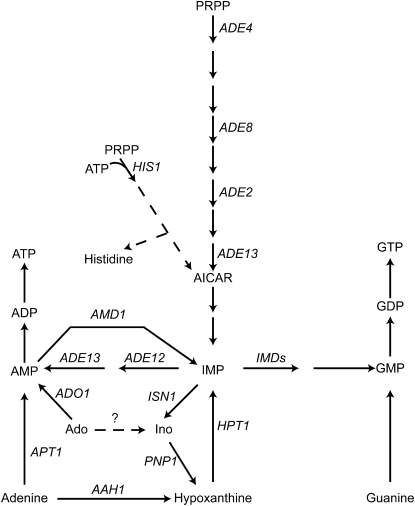
Schematic of purine metabolism in S. cerevisiae. Ado, adenosine; AICAR, 5′-phosphoribosyl-5-amino-4-imidazole carboxamide; Ino, inosine; IMP, inosine 5′-monophosphate; and PRPP, 5-phosphoribosyl-1-pyrophosphate. Gene names are italicized. For simplicity, only the enzymatic steps cited in the text are shown in the figure. Putative adenosine deaminase activity is indicated by a question mark.
AMP deaminase (AMPD; EC 3.5.4.6) is an important enzyme for purine interconversion. During muscle effort, extensive hydrolysis of ATP into ADP results in massive AMP production due to adenylate kinase (myokinase) activity. Under such conditions, AMPD, by draining AMP to IMP, plays a critical role in the stabilization of adenylate energy charge. Consequently, defects in AMPD lead to exercise-induced muscle symptoms such as early fatigue and is the most common muscle enzyme defect in humans (Fishbein et al. 1978). While there are three AMPD human isoforms, in Saccharomyces cerevisiae, a mutation at a single locus (named AMD1; Figure 1) abolishes AMPD activity (Meyer et al. 1989). However, two yeast proteins of unknown function (Ybr284p and Yjl070p) are >30% identical to Amd1p in their C terminus (supporting information, Figure S1) (Duenas et al. 1999; Vandenbol and Portetelle 1999). Strikingly, in most organisms, AMPD isoforms have a highly divergent N terminus and in several cases, including yeast, the N terminus of the protein is not required for activity and is often lost during protein purification due to proteolysis (Merkler et al. 1989; Sabina and Mahnke-Zizelman 2000). Yeast AMP deaminase activity has been characterized in vitro (Meyer et al. 1989) and is presumed to be high under specific growth conditions where massive synthesis of IMP from AMP has been observed (Osorio et al. 2003; Silles et al. 2005; Loret et al. 2007). IMP can then be metabolized through three different pairs of enzymatic reactions. It can give back AMP via two enzymatic steps encoded by the ADE12 and ADE13 genes; it can be transformed to GMP via IMP dehydrogenase and GMP synthetase; and, finally, it can be degraded into inosine and hypoxanthine via the successive action of IMP-specific nucleotidase (Isn1p) and purine nucleoside phosphorylase (Pnp1p) (Figure 1) (Jones and Fink 1982; Lecoq et al. 2001; Itoh et al. 2003). Therefore, yeast AMP deaminase appears as a critical enzyme for both purine interconversion and degradation. While the biochemical properties of yeast AMP deaminase have been well studied (Merkler et al. 1989, 1993; Merkler and Schramm 1990, 1993), the physiological consequences of a amd1 defect have not been investigated.
Here, we show that a defect in AMPD activity impairs yeast cell growth under specific conditions, and we establish that AMPD plays a crucial role in maintaining guanylic nucleotide homeostasis. Study of the Amd1p homologs, Yjl070p and Ybr284p, revealed that overexpression of these proteins cannot suppress phenotypes associated with amd1 deletion. Instead, we show that overexpression of YJL070c mimics a loss of Amd1p function. Transcriptome analyses were performed using these various constructs and MPA-treated cells, thus allowing us to evaluate the consequences of guanylic nucleotide depletion obtained through different means. These new genetic tools were also used to revisit the phenotypes of several MPA hypersensitive mutants.
MATERIALS AND METHODS
Yeast media:
SD minimal medium contains 0.5% ammonium sulfate, 0.17% yeast nitrogen base Difco, and 2% glucose. SC was prepared as described (Sherman et al. 1986). SDcasaW is SD medium supplemented with 0.2% casamino acids (Difco) and tryptophan (200 μm). When indicated, adenine, hypoxanthine, and guanine were added at 300 μm and uracil was added at 180 μm.
Yeast strains:
All strains belong to, or are derived from, a set of disrupted strains isogenic to BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) or BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) purchased from Euroscarf. The following mutant strains were constructed: Y2077 (MATα, amd1∷kanMX4, aah1∷kanMX4, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), Y2362 (MATa, ade8∷kanMX4, amd1∷kanMX4, aah1∷kanMX4, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), and Y2693 (MATa, his1∷kanMX4, ade8∷kanMX4, amd1∷kanMX4, aah1∷kanMX4, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0).
Plasmids:
The IMD2-lacZ (P354, 2μ URA3 and P777, 2μ, LEU2) and the pCM189 (CEN, URA3) plasmids used in this study were previously described (Gari et al. 1997; Escobar-Henriques and Daignan-Fornier 2001). The tet-APT1 (P2091), tet-YJL070c (p2483), tet-HPT1 (P2149), tet-AMD1 (P2479), and tet-YBR284w (P2481) plasmids were obtained by PCR amplification of the corresponding genes using specific oligonucleotides (sequence available upon request) and cloning into pCM189. β-Galactosidase assays were performed as described (Kippert 1995).
Transcriptome analysis:
For DNA microarray analysis, cells were grown overnight in SDcasaWAU medium, diluted in 50 ml of the same medium, and harvested 24 hr later in exponential phase at A600 (absorbance at 600 nm) = 0.6–0.8. RNA were extracted as described in the Gene Expression Omnibus (GEO) entry for this article (GSE9557) and were purified with a RNeasy purification kit (Qiagen) according to the manufacturer's protocol. Normalization was done by the locally weighted scatter plot smoothing (LOWESS) algorithm (Bengtsson et al. 2004). Complete microarray raw data sets and experimental details are available in GEO under accession no. GSE9557. Intracellular nucleotide determination was performed as previously described (Breton et al. 2008; Gauthier et al. 2008).
Growth test:
Yeast cells were resuspended in sterile water to an A600 = 1 and submitted to 1/10 serial dilutions. Drops (5 μl) of each dilution were spotted on freshly prepared plates and were incubated at 30° for 48–72 hr.
RESULTS
Deletion of AMD1 affects growth in the presence of adenine and impairs purine nucleotide balance:
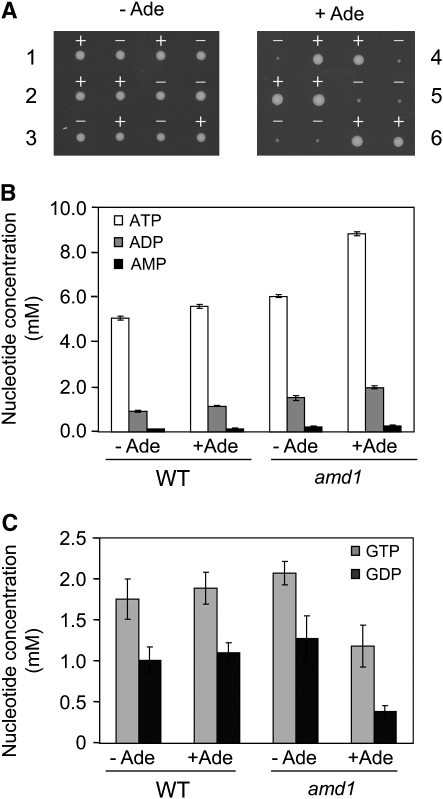
While no major growth defect associated with the lack of AMP deaminase activity has been reported previously (Meyer et al. 1989), we observed a strong defect in germination and/or growth of amd1 spores in the presence of adenine (Figure 2A). During vegetative growth, doubling time of the wild-type strain in SDcasa medium (82 min) was not affected by adenine (83 min) while, for the isogenic amd1 mutant, doubling times were 90 and 140 min in the absence and presence of adenine, respectively. This observation prompted us to measure purine nucleotide pools in these strains. HPLC measurement revealed that adenylic nucleotides were more abundant in the amd1 mutant, which cannot convert AMP to IMP (Figure 2B). This effect was further enhanced by addition of adenine. Second, we observed that both the GDP and the GTP pools were severely diminished (three- and twofold, respectively) in the amd1 mutant grown in the presence of adenine compared to the wild-type strain (Figure 2C). In the absence of adenine, GDP and GTP pools were not significantly affected in the amd1 mutant (Figure 2C). These results establish that Amd1p is critical in maintaining the purine nucleotide balance and suggest that the amd1 growth defect in the presence of adenine could be due to the inability of this mutant to correctly balance adenylic and guanylic nucleotides.
Figure 2.—
External adenine affects growth and nucleotide pools of AMP deaminase-deficient strains. (A) Growth of wild-type and amd1 spores in the absence or the presence of adenine. A heterozygous AMD1/amd1∷KanMX4 diploid strain was sporulated and dissected on SDcasaWU medium containing adenine (+Ade, tetrads 4–6) or not (−Ade, tetrads 1–3). “+” and “−” signs above the colonies refer, respectively, to AMD1 (+) and amd1∷KanMX4 (−) genotypes determined on the basis of sensitivity or resistance to geneticin associated with KanMX4 expression. (B and C). Intracellular nucleotide content is affected in the amd1 mutant grown in the presence of extracellular adenine. Wild-type (WT, BY4742) and amd1 mutant strains were grown in SDcasaW medium supplemented (+Ade) or not (−Ade) with external adenine. Internal adenylic (B) and guanylic (C) nucleotides were measured as previously described (Breton et al. 2008; Gauthier et al. 2008).
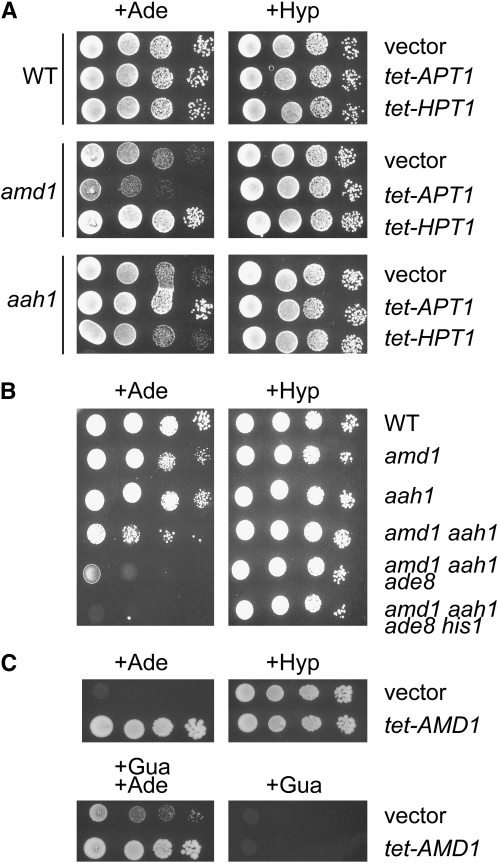
There are three possible routes to synthesize IMP (and in turn GMP) in the presence of adenine. The first route goes through adenine phosphoribosyltransferase (Apt1p) and AMP deaminase (Amd1p), the second route is through adenine deaminase (Aah1p) and hypoxanthine-guanine phosphoribosyltransferase (Hpt1p), and the third route is de novo synthesis from 5-phosphoribosyl-1-pyrophosphate (Figure 1). However, in the presence of adenine, this last route is inhibited due to feedback control on the first enzyme of the pathway (Ade4p) and transcriptional control on all the genes of the de novo pathway (Daignan-Fornier and Fink 1992; Rebora et al. 2001). Thus, in the presence of adenine and in the absence of Amd1p, IMP and GMP synthesis should mostly take place via Aah1p and Hpt1p and could become limiting for growth. Consistently, the amd1 growth defect was suppressed by overexpression of HPT1 (Figure 3A), which drains adenine toward IMP synthesis. On the other hand, the amd1 growth defect was enhanced by overexpression of APT1 (Figure 3A), which metabolizes adenine to AMP and therefore competes with Aah1p for available adenine (Figure 1). As expected, an aah1 mutant, which blocks the second route, also showed a slight growth defect in the presence of adenine (Figure 3A), and consistently, in the case of aah1, the growth defect was suppressed by overexpression of APT1 (Figure 3A), which metabolizes adenine to AMP, the Amd1p substrate (Figure 1).
Figure 3.—
The growth defect of the amd1 mutant is enhanced by mutations affecting IMP synthesis and partially suppressed by guanine addition. (A) Overexpression of APT1 exacerbates the growth defect of the amd1 mutant in the presence of external adenine. Cells were transformed with the pCM189 plasmid (vector), tet-APT1, or tet-HPT1. Transformants were serial diluted and spotted on SDcasaW medium supplemented with adenine (+Ade) or hypoxanthine (+Hyp). (B) Combinations of the amd1 mutation with aah1, ade8, and his1 lead to drastic decrease of growth in the presence of adenine. (C) Growth defect of the amd1 aah1 ade8 his1 mutant in the presence of adenine is due to guanylic nucleotide starvation. The quadruple amd1 aah1 ade8 his1 mutant was transformed with the pCM189 empty plasmid (vector) or the plasmid allowing overexpression of AMD1 (p2479). Cells were spotted on SDcasaW medium supplemented with adenine (+Ade), guanine (+Gua), or hypoxanthine (+Hyp), as indicated.
We then tested the hypothesis that the amd1 phenotype would be exacerbated by combining it with mutations affecting other IMP-supplying enzymes. Indeed, in the presence of adenine, growth of an aah1 amd1 double mutant was much more affected than that of each single mutant (Figure 3B). Residual growth in this mutant was due to IMP synthesis via AICAR from the purine and histidine de novo pathways (Rebora et al. 2005) (Figure 1) and could be totally abolished in the ade8 his1 aah1 amd1 quadruple mutant (Figure 3B). As expected, this quadruple mutant was fully viable when hypoxanthine was provided as a purine source (Figure 3B). Furthermore, growth of the quadruple mutant in the presence of adenine was restored to a certain extent by the addition of guanine, indicating that the growth defect is, at least in part, due to guanylic nucleotide shortage (Figure 3C). Incomplete suppression by guanine could be due to the fact that guanylic nucleotide shortage is not the sole problem faced by this strain. Alternatively, it could be due to poor guanine uptake in the presence of adenine since both compounds are transported by the purine–cytosine permease (Schmidt et al. 1984).
Overexpression of YJL070c phenocopies a amd1 knock-out mutant:
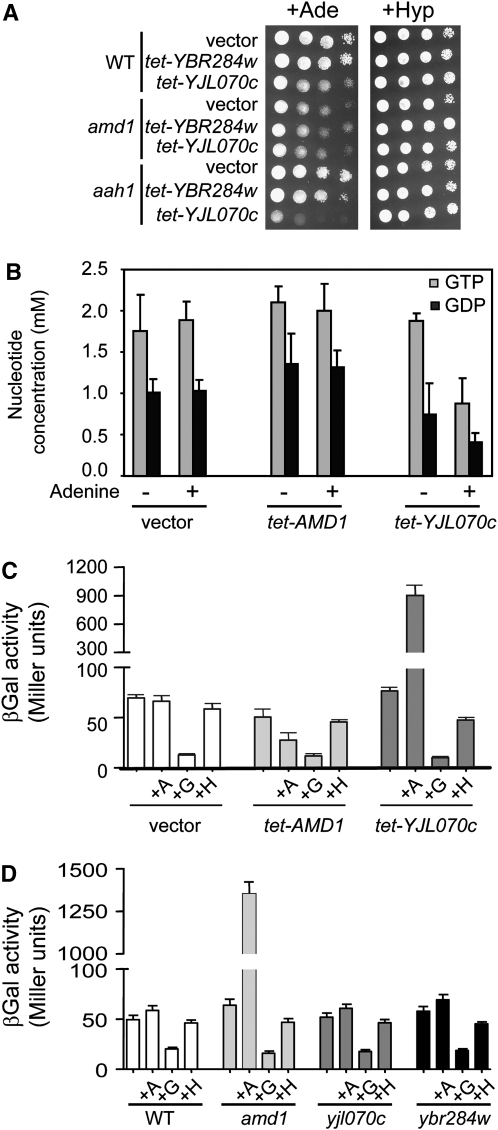
As mentioned above, the function of the two AMD1 homologs, YBR284w and YJL070c, is unknown. Importantly, both proteins lack several residues (Figure S1) conserved in all described purine deaminases (Ribard et al. 2003). Overexpression of these genes could not rescue growth of the quadruple aah1 ade8 amd1 his1 mutant in the presence of adenine (Figure S2A), indicating that these genes do not significantly contribute to AMPD activity. It is notable that these genes do not contribute to adenosine or adenine deaminase activity (Figure S2, A and B). Finally, mutations in these two genes, either alone or combined with amd1, did not result in additional phenotypes (Figure S2C). However, to our surprise, we found that overexpression of YJL070c, but not that of YBR284w, affected the growth of a wild-type strain, specifically in the presence of adenine (Figure 4A). This phenotype is reminiscent of the one found for the amd1 mutant (Figure 2A). Importantly, the effect of YJL070c was dependent on the presence of Amd1p, since YJL070c overexpression had no additional effect in the amd1 deletion mutant (Figure 4A). We thus investigated whether overexpression of YJL070c would mimic other phenotypes of the amd1 knock-out. Indeed, the phenotype, associated with YJL070c overexpression, was strongly enhanced in the aah1 mutant (Figure 4A), as found for the amd1 mutation (Figure 3B). Measurement of guanylic nucleotide pools revealed that overexpression of YJL070c resulted in a strong decrease of both GDP and GTP intracellular concentration (Figure 4B). This effect was observed only when cells were grown in the presence of adenine as for the amd1 mutant (Figure 2C). In yeast, a guanylic nucleotide limitation results in strong induction of IMD2 gene expression (Escobar-Henriques and Daignan-Fornier 2001). Clearly, overexpression of YJL070c as well as the amd1 deletion resulted in strong induction of IMD2-lacZ expression only in the presence of adenine (Figure 4, C and D), while no effect was observed in the ybr284w and yjl070c mutants (Figure 4D). Therefore, according to all tested criteria, overexpression of YJL070c phenocopies a amd1 deletion.
Figure 4.—
Overexpression of YJL070c phenocopies the amd1 deletion mutant. (A) Effect of YJL070c or YBR284w overexpression on growth in the presence of adenine or hypoxanthine. Cells were transformed with the pCM189 empty plasmid (vector) or either the tet-YBR284w or tet-YJL070c plasmids. (B) Intracellular guanylic nucleotide content is specifically decreased in wild-type cells overexpressing YJL070c and grown in the presence of adenine. Wild-type (BY4742) cells were transformed with the pCM189 vector or plasmids allowing overexpression of AMD1, YJL070c, or YBR284w. Transformants were grown in SDcasaW medium supplemented or not with external adenine, and intracellular guanylic nucleotide content was measured as previously described (Breton et al. 2008). (C) Overexpression of YJL070c leads to derepression of the IMD2-LacZ fusion expression. Cells were cotransformed with IMD2-lacZ plasmid and either the vector or the plasmids allowing overexpression of AMD1 or YJL070c. Transformants were grown in SC medium supplemented or not with adenine (+A), guanine (+G), or hypoxanthine (+H). (D) Expression of IMD2-LacZ fusion is drastically increased in a amd1 mutant in the presence of adenine. Cells were transformed with a plasmid carrying a IMD2-lacZ fusion. Transformants were grown in SDcasaW medium supplemented or not with adenine (+A), guanine (+G), or hypoxanthine (+H).
Transcriptome analysis of guanylic nucleotide-depleted yeast cells:
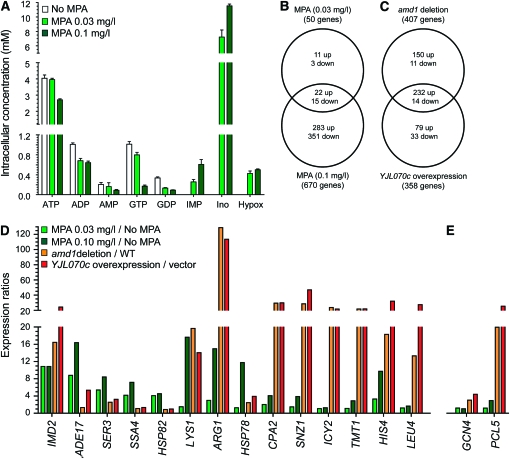
The amd1 mutation and the overexpression of YJL070c provide us with new ways to challenge cells with guanylic nucleotide depletion. We took advantage of these strains to evaluate the effects of GDP and GTP limitation on the yeast transcriptome and to compare them to those of the immunosuppressive drug MPA, which specifically inhibits eukaryotic IMP dehydrogenase (IMPDH). In MPA-treated cells, GDP and GTP concentrations were severely affected in a dose-dependent manner, while adenylic nucleotide pools were only slightly affected (Figure 5A). Importantly, MPA treatment also led to massive accumulation of IMP, the substrate of IMPDH, and its nucleoside (inosine) and base (hypoxanthine) derivatives (Figure 5A). It is notable that the ∼50% decrease in GTP concentration found in both the amd1 mutant and the YJL070c overexpressing strains (Figure 2C and Figure 4B) is intermediary to the 17% and 80% found for the lower and higher MPA concentrations, respectively.
Figure 5.—
Global transcriptional response to amd1 deletion, YJL070c overexpression and MPA treatment in the presence of adenine. (A) Intracellular nucleotide content is affected by MPA treatment. Wild-type cells were grown in SDcasaWUA medium supplemented or not with MPA. Internal adenylic and guanylic nucleotides were measured as previously described (Breton et al. 2008; Gauthier et al. 2008). (B and C) Transcriptional response to guanylic nucleotide depletion obtained by MPA treatment, amd1 deletion, and YJL070c overexpression. Transcriptional response was monitored by microarray analyses. Venn diagrams show numbers of genes differentially expressed (by a factor ≥2 compared to untreated wild type) in response to various MPA concentrations (B) in a amd1 mutant and a strain overexpressing YJL070c (C). (D) Expression ratios of the five most upregulated genes for each comparison set used in the transcriptome analysis. (E) Expression of GCN4 and PCL5 in the various conditions.
The effects of guanylic nucleotide shortage on the yeast transcriptome were then evaluated in a wild-type strain treated (or not) with MPA (0.03 and 0.1 mg/liter) in a amd1 mutant and in a wild-type strain overexpressing (or not) YJL070c. The first conclusion was that MPA treatment affected expression of multiple genes that were either up- or downregulated. The number of affected genes was much higher at 0.1 mg/liter MPA compared to 0.03 mg/liter (Figure 5B). While most of the 51 genes affected at low MPA concentration are also affected at the higher concentration, 14 (27%) were not, suggesting that the transcriptional response to the drug is not simple.
Comparison of the transcriptome in YJL070c overexpression and amd1 deletion strains to their cognate control strains revealed an altered expression for 358 and 407 genes, respectively, 246 of which were affected in both conditions (Figure 5C, Table S1). This result strongly supports our genetics and biochemical data, indicating that overexpression of YJL070c phenocopies the amd1 deletion. When looking at the five most affected genes in each condition (Figure 5D), several conclusions can be drawn. First, in all four conditions, IMD2 expression was strongly upregulated (>10-fold), thus confirming the IMD2-lacZ results (Figure 5D). Second, some genes, such as SSA4 or HSP82, were strongly and specifically affected by MPA, even at the lower dose, but did not respond to GTP shortage induced by amd1 or tet-YJL070c. Because these genes are involved in the stress response, this could reflect a side effect of MPA not directly linked to GTP synthesis inhibition. Reciprocally, genes such as ICY2 or LEU4 strongly responded to the amd1 deletion and YJL070c overexpression but not to MPA treatment. Finally, in most cases (LYS1, ARG1, CPA2, SNZ1, TMT1, HIS4), the transcriptional response seems gradual, low at the low MPA dose, higher at 0.1 mg/liter MPA, and even higher when amd1 is mutated or YJL070c overexpressed. This could appear surprising since the high dose of MPA treatment leads to lower GTP concentrations, but could reflect secondary effects of the drug. Interestingly, many of these genes are under the control of the transcriptional factor Gcn4p, and it should be noted that GCN4 itself is transcriptionally induced in the amd1 mutant and under the tet-YJL070c conditions but is unaffected in the MPA-treated cells (Figure 5E). Furthermore, PCL5, a major regulator of Gcn4p stability (Shemer et al. 2002), is also differentially induced (Figure 5E) and could negatively modulate GCN4 overexpression. Global comparison revealed 71 genes similarly affected by amd1 knock-out, YJL070c overexpression, and 0.1 mg/liter MPA treatment (Figure S3), most of which are involved in amino acid metabolism and regulated by Gcn4p (Natarajan et al. 2001). Thus, although GCN4 itself responds differently under the various conditions, GTP shortage is associated with upregulation of multiple GCN4 target genes.
Tet-YJL070c as a tool to revisit MPA hypersensitive mutants:
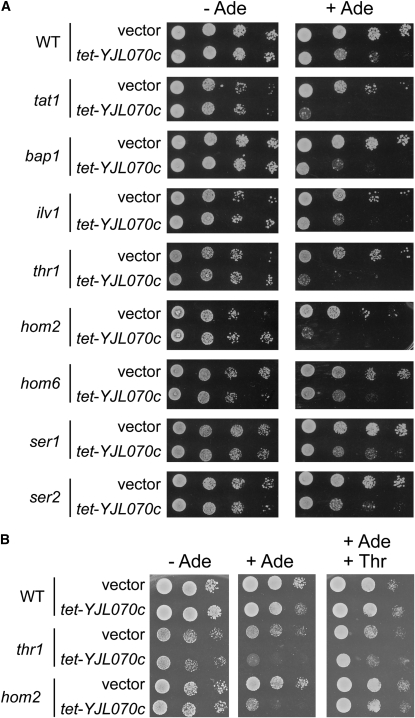
Our earlier search for yeast mutants sensitive to MPA treatment had revealed eight amino acid metabolism mutants (Desmoucelles et al. 2002). Since our transcriptome analysis showed strong induction of several such amino acid metabolism genes when guanylic nucleotides are scarce, we used YJL070c to reinvestigate this phenotype. The tet-YJL070c construct was found to severely affect the growth of three mutants (tat1, thr1, and hom2) on adenine; it had a weaker effect on the ilv1, bap1, ser2, and hom6 mutants, and no major effect on ser1 (Figure 6A). As a control experiment, we showed that a amd1 thr1 double mutant behaved just as the thr1 mutant carrying the tet-YJL070c plasmid did (Figure S4). Importantly, the hom2 and thr1 growth defects, associated with YJL070c overexpression, were suppressed by the addition of threonine in large amounts to the growth medium (Figure 6B). Since both hom2 and thr1 mutations block threonine biosynthesis, this result strongly suggests that threonine uptake is limiting for growth in these mutants when guanylic nucleotides are scarce. Thus, this phenotypic analysis supports the idea that there is a connection between guanylic nucleotide limitation and amino acid metabolism.
Figure 6.—
YJL070c overexpression severely affects growth of threonine biosynthesis mutants in the presence of adenine. Cells transformed with pCM189 (vector) or tet-YJL070c were spotted on SDcasaW medium supplemented (+ Ade) or not (− Ade) with adenine and threonine (+ Thr) as indicated.
In yeast, hypersensitivity to MPA is often used as a criterion to identify transcriptional elongation mutants, the rationale being that GTP shortage (substrate limitation) combined to an elongation default affects transcription efficiency and/or accuracy so severely that it results in growth impairment. However, in several of these MPA-sensitive mutants, such as dst1 (ppr2), expression of IMD2, which encodes the MPA-resistant isoform of IMPDH (Hyle et al. 2003), is drastically decreased (Shaw and Reines 2000). Thus, there are two major and nonexclusive hypotheses to explain why these mutants are hypersensitive to MPA. The first one is that sensitivity to MPA could be due to IMD2 low expression. The second one is that the mutants could be sensitive due to the combination of low GTP concentration and altered transcription elongation. The tet-YJL070c construct, because it allows decreasing GTP concentration independently of MPA, was used to attempt to settle this question.
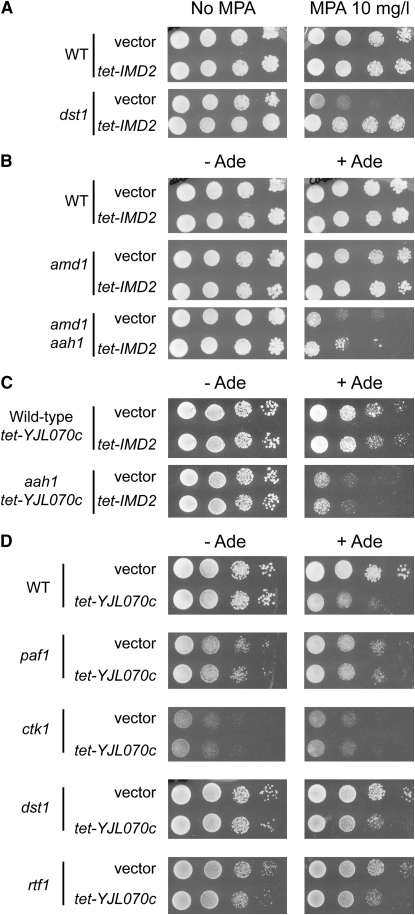
Consistent with the first hypothesis, expression of IMD2 under a heterologous tet promoter reversed MPA hypersensitivity of the dst1 mutant strain (Figure 7A), and, importantly, it did not significantly suppress the adenine-specific growth defect of the amd1 mutant, although it slightly improved growth of all the strains in the presence of adenine (Figure 7B). Thus, although both MPA treatment and amd1 mutation induce IMD2 expression (Figure 4 and Figure S3B), the growth defect associated with the amd1 mutation is independent of IMD2 expression. Similarly, IMD2 overexpression had no effect on the growth defect due to overexpression of YJL070c in the presence of adenine (Figure 7C).
Figure 7.—
Effect of guanylic nucleotide limitation on transcription mutants. Cells were transformed with pCM189 (vector) or the tet-IMD2 plasmid, and transformants were spotted on SDcasaW medium supplemented or not with MPA (10 mg/liter) (A) or with adenine (Ade) as indicated (B). (C) Cells containing the tet-YJL070c overexpression plasmid were transformed with pCM189 (vector) or the tet-IMD2 plasmid, and transformants were spotted either on SC medium lacking uracil and leucine (C) or on SDcasaW (D).
To test the second hypothesis and establish whether a decrease of GTP concentration in various transcription mutants was sufficient to explain their hypersensitivity to MPA, we took advantage of the tet-YJL070c plasmid to decrease GTP pools independently of MPA treatment. We thus overexpressed YJL070c in various MPA hypersensitive mutants proposed to affect transcription elongation and monitored growth and GTP concentration. Strikingly, while overexpression of YJL070c in the mutants and in the isogenic wild-type strain similarly affected GTP levels (Figure S5), it had no drastic effect on growth in the presence of adenine (Figure 7D). It is important to note that the double amd1 dst1 mutant behaved just as the dst1 mutant carrying the tet-YJL070c plasmid did (Figure S4). Together, our results indicate that a significant part of the MPA sensitivity of these mutants is most likely due to poor expression of IMD2 rather than to a synthetic growth defect due to impaired transcription elongation combined with GTP limitation. However, it cannot be excluded that the growth defect of these transcription mutants observed in the presence of MPA could necessitate a more severe GTP limitation than the one caused by YJL070c overexpression in the presence of adenine.
DISCUSSION
A major conclusion from our results is that, in yeast, a defect in AMP deaminase is associated with a severe GDP/GTP pool depletion. This effect could be observed only in the presence of adenine and under conditions where the de novo pathway is turned down. We interpret this result as follows: in the absence of adenine, IMP synthesized from the de novo pathway is sufficient to provide wild-type levels of GDP and GTP. Addition of adenine results in a strong downregulation of the de novo pathway (Daignan-Fornier and Fink 1992) and a subsequent decrease of IMP synthesis, which, in the amd1 mutant, cannot be fully compensated for by IMP synthesis via adenine deaminase. Interestingly, a recent report on Arabidopsis thaliana AMP deaminase inhibition by deaminoformycin showed a synergistic effect of adenine on deaminoformycin toxicity (Xu et al. 2005). This phenotype is highly similar to the inhibitory effect of adenine on growth of the amd1 mutant. However, in A. thaliana, adenylic nucleotide accumulation appears to be the initial cause of growth inhibition; by contrast, in yeast, partial reversion of the adenine effect by guanine (Figure 4B) suggests that guanylic nucleotide shortage is, at least in part, responsible for growth inhibition. In our previous work, we have studied the transcriptional response to ATP limitation (adk1 mutant; Gauthier et al. 2008) and GTP overproduction (constitutive HPT1 mutant; Breton et al. 2008). In this work, we now document the transcriptional response to GTP shortage obtained by different means. Importantly, while all these conditions result in purine nucleotide imbalance, there is no evidence for a common transcriptional response. Furthermore, GTP shortage and overproduction do not result in opposite transcriptional responses.
In this study, important data were collected on the yeast AMP deaminase gene family. On the basis of phenotypic analysis, we established that none of the two AMD1 homologs, YBR284w and YJL070c, encodes AMP, adenosine, or adenine deaminase activity (Figure S2). These results are in agreement with the fact that both proteins lack important residues (Figure S1) conserved in all described purine deaminases (Ribard et al. 2003). However, because these genes are syntenic with genes in other fungi (S. paradoxus, S. bayanus, Ashbya gossypii, etc.), and because the Yjl070p protein has been detected by mass spectrometry (Ghaemmaghami et al. 2003), it seems unlikely that these genes could be pseudogenes. Concerning Yjl070p, we favor the hypothesis that it could have a noncatalytic regulatory function. Indeed, we found that overexpression of Yjl070p, in the presence of adenine, has a strong effect on guanylic nucleotide concentration, and growth was impaired. These results are highly similar to those obtained with the amd1 deletion, indicating that overexpression of YJL070c mimics the amd1 knock-out. Importantly, Amd1p and Yjl070p copurified in a global proteome interaction analysis (Krogan et al. 2006), thus suggesting a possible direct interaction that could affect AMP deaminase activity. An attractive working hypothesis would be that Yjl070p could form inactive heterodimers with Amd1p; however, our attempts to document a direct inhibitory effect in vitro were unsuccessful (C. Saint-Marc and B. Daignan-Fornier, unpublished data). The fact that YJL070c overexpression mimicked the amd1 deletion was confirmed by our transcriptome analysis showing that a large set of common genes were induced in both conditions. The transcriptional response to MPA was more divergent. This could be due to the fact that MPA treatment, by inhibiting IMPDH, leads to massive accumulation of inosine (≤12 mm) as well as IMP and hypoxanthine, although to a lesser extent. No accumulation of these compounds was observed in the amd1 mutant or YJL070c overexpression strains.
We also took advantage of GTP limitation induced by YJL070c overexpression to revisit the proposed phenotypical link between GTP shortage and transcription elongation mutants, which are hypersensitive to MPA. We found that all tested mutants (paf1, dst1, rtf1, and ctk1) were not more affected than the wild-type control by YJL070c overexpression. Interestingly, a study of transcription elongation in vivo on a specific gene indicated that dst1, rtf1, and ctk1 mutations, on their own, do not affect transcription elongation or processivity (Mason and Struhl 2005). We therefore favor the hypothesis that MPA hypersensitivity of these mutants is due to the need for these factors in MPA-induced IMD2 upregulation and is not a secondary effect of low GTP concentrations upon transcription per se. These results cast a doubt upon the use of MPA sensitivity as a criterion in identifying transcription elongation mutants.
Our transcriptome data revealed 71 genes affected at least twofold in all three conditions leading to GDP/GTP shortage (Figure 6B). Strikingly, many of these genes are involved in amino acid metabolism, and the vast majority of them are under the control of the Gcn4p transcription factor. This result is in good agreement with previous reports showing induction of GCN4 target genes in response to severe purine limitation (Rolfes and Hinnebusch 1993). An important result is that some amino acid metabolism mutants appear exquisitely sensitive to guanylic nucleotide limitation. We found that the threonine biosynthesis mutants, thr1 or hom2, are strongly affected by YJL070c overexpression and that this growth defect could be alleviated by increasing external threonine concentration. Interestingly, thr1 and hom2 mutants are hypersensitive to hydroxyurea (Hartman and Tippery 2004), and threonine metabolism has been shown to be important for dNTP pool homeostasis in yeast (Hartman 2007); however, the precise mechanism leading to this cross-pathway effect is not known. In addition, both thr1 and hom2 mutants showed reduced fitness under various growth conditions (Giaever et al. 2002), suggesting that threonine availability could be central in responding to different cellular perturbations. Together, our results point to amino acid uptake as limiting under guanylic nucleotide shortage, thus revealing a new metabolic crosstalk that could be relevant for understanding the immunosuppressive and antiproliferative effects of mycophenolate derivatives.
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique, Université Bordeaux 2, and the Conseil Régional d'Aquitaine. O.L. was supported by a North Atlantic Treaty Organization fellowship.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.105858/DC1.
References
- Bengtsson, H., G. Jonsson and J. Vallon-Christersson, 2004. Calibration and assessment of channel-specific biases in microarray data with extended dynamical range. BMC Bioinformatics 5: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, A., B. Pinson, F. Coulpier, M. F. Giraud, A. Dautant et al., 2008. Lethal accumulation of guanylic nucleotides in Saccharomyces cerevisiae HPT1-deregulated mutants. Genetics 178: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier, B., and G. R. Fink, 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89: 6746–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoucelles, C., B. Pinson, C. Saint-Marc and B. Daignan-Fornier, 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277: 27036–27044. [DOI] [PubMed] [Google Scholar]
- Duenas, E., C. R. Vazquez de Aldana, T. de Cos, C. Castro and M. Henar Valdivieso, 1999. Generation of null alleles for the functional analysis of six genes from the right arm of Saccharomyces cerevisiae chromosome II. Yeast 15: 615–623. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques, M., and B. Daignan-Fornier, 2001. Transcriptional regulation of the yeast gmp synthesis pathway by its end products. J. Biol. Chem. 276: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques, M., A. Balguerie, C. Monribot, H. Boucherie and B. Daignan-Fornier, 2001. Proteome analysis and morphological studies reveal multiple effects of the immunosuppressive drug mycophenolic acid specifically resulting from guanylic nucleotide depletion. J. Biol. Chem. 276: 46237–46242. [DOI] [PubMed] [Google Scholar]
- Fishbein, W. N., V. W. Armbrustmacher and J. L. Griffin, 1978. Myoadenylate deaminase deficiency: a new disease of muscle. Science 200: 545–548. [DOI] [PubMed] [Google Scholar]
- Gari, E., L. Piedrafita, M. Aldea and E. Herrero, 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848. [DOI] [PubMed] [Google Scholar]
- Gauthier, S., F. Coulpier, L. Jourdren, M. Merle, S. Beck et al., 2008. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol. Microbiol. 68: 1583–1594. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle et al., 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Hartman, J. L., IV, 2007. Buffering of deoxyribonucleotide pool homeostasis by threonine metabolism. Proc. Natl. Acad. Sci. USA 104: 11700–11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, J. L., IV, and N. P. Tippery, 2004. Systematic quantification of gene interactions by phenotypic array analysis. Genome Biol. 5: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyle, J. W., R. J. Shaw and D. Reines, 2003. Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 278: 28470–28478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R., C. Saint-Marc, S. Chaignepain, R. Katahira, J. M. Schmitter et al., 2003. The yeast ISN1 (YOR155c) gene encodes a new type of IMP-specific 5′-nucleotidase. BMC Biochem. 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E. W., and G. R. Fink, 1982. Regulation of amino acid and nucleotide biosynthesis in yeast, pp. 181–299 in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, edited by J. N. Strathern, E. W. Jones and J. R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kippert, F., 1995. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128: 201–206. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo et al., 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- Lecoq, K., I. Belloc, C. Desgranges, M. Konrad and B. Daignan-Fornier, 2001. YLR209c encodes Saccharomyces cerevisiae purine nucleoside phosphorylase. J. Bacteriol. 183: 4910–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret, M. O., L. Pedersen and J. Francois, 2007. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 24: 47–60. [DOI] [PubMed] [Google Scholar]
- Mason, P. B., and K. Struhl, 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17: 831–840. [DOI] [PubMed] [Google Scholar]
- Merkler, D. J., and V. L. Schramm, 1990. Catalytic and regulatory site composition of yeast AMP deaminase by comparative binding and rate studies. Resolution of the cooperative mechanism. J. Biol. Chem. 265: 4420–4426. [PubMed] [Google Scholar]
- Merkler, D. J., and V. L. Schramm, 1993. Catalytic mechanism of yeast adenosine 5′-monophosphate deaminase. Zinc content, substrate specificity, pH studies, and solvent isotope effects. Biochemistry 32: 5792–5799. [DOI] [PubMed] [Google Scholar]
- Merkler, D. J., A. S. Wali, J. Taylor and V. L. Schramm, 1989. AMP deaminase from yeast. Role in AMP degradation, large scale purification, and properties of the native and proteolyzed enzyme. J. Biol. Chem. 264: 21422–21430. [PubMed] [Google Scholar]
- Merkler, D. J., P. C. Kline, P. Weiss and V. L. Schramm, 1993. Transition-state analysis of AMP deaminase. Biochemistry 32: 12993–13001. [DOI] [PubMed] [Google Scholar]
- Meyer, S. L., K. L. Kvalnes-Krick and V. L. Schramm, 1989. Characterization of AMD, the AMP deaminase gene in yeast. Production of amd strain, cloning, nucleotide sequence, and properties of the protein. Biochemistry 28: 8734–8743. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts et al., 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio, H., E. Carvalho, M. del Valle, M. A. Gunther Sillero, P. Moradas-Ferreira et al., 2003. H2O2, but not menadione, provokes a decrease in the ATP and an increase in the inosine levels in Saccharomyces cerevisiae. An experimental and theoretical approach. Eur. J. Biochem. 270: 1578–1589. [DOI] [PubMed] [Google Scholar]
- Rebora, K., C. Desmoucelles, F. Borne, B. Pinson and B. Daignan-Fornier, 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol. Cell. Biol. 21: 7901–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora, K., B. Laloo and B. Daignan-Fornier, 2005. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribard, C., M. Rochet, B. Labedan, B. Daignan-Fornier, P. Alzari et al., 2003. Sub-families of alpha/beta barrel enzymes: a new adenine deaminase family. J. Mol. Biol. 334: 1117–1131. [DOI] [PubMed] [Google Scholar]
- Rolfes, R. J., and A. G. Hinnebusch, 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina, R. L., and D. K. Mahnke-Zizelman, 2000. Towards an understanding of the functional significance of N-terminal domain divergence in human AMP deaminase isoforms. Pharmacol. Ther. 87: 279–283. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., M. F. Manolson and M. R. Chevallier, 1984. Photoaffinity labeling and characterization of the cloned purine-cytosine transport system in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 81: 6276–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. J., and D. Reines, 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20: 7427–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer, R., A. Meimoun, T. Holtzman and D. Kornitzer, 2002. Regulation of the transcription factor Gcn4 by Pho85 cyclin PCL5. Mol. Cell. Biol. 22: 5395–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Silles, E., H. Osorio, R. Maia, M. A. Gunther Sillero and A. Sillero, 2005. Micromolar HgCl2 concentrations transitorily duplicate the ATP level in Saccharomyces cerevisiae cells. FEBS Lett. 579: 4044–4048. [DOI] [PubMed] [Google Scholar]
- Vandenbol, M., and D. Portetelle, 1999. Disruption of six ORFs on Saccharomyces cerevisiae chromosome X: the YJL069c gene of unknown function is essential to cell viability. Yeast 15: 1411–1417. [DOI] [PubMed] [Google Scholar]
- Xu, J., H. Y. Zhang, C. H. Xie, H. W. Xue, P. Dijkhuis et al., 2005. EMBRYONIC FACTOR 1 encodes an AMP deaminase and is essential for the zygote to embryo transition in Arabidopsis. Plant J. 42: 743–756. [DOI] [PubMed] [Google Scholar]