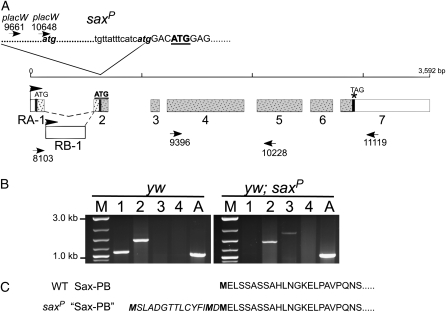
Figure 6.—
P-insertion site of saxP does not disrupt transcription. (A) Genomic structure of sax locus shown with exon (numbered) distribution for the two splice forms of sax mRNAs (RA and RB). The locations of two ATG initiation codons and the TAG termination codon (vertical thick solid lines) indicate the two overlapping open reading frames (speckles) giving rise to the putative protein products PA and PB (shaded speckles). Positions of PCR primers are indicated by arrowheads. The site and junctional sequence of the placW insertion giving rise to saxP is shown at the top. The endogenous second ATG within the sax transcription unit is bold and underlined. (B) RT–PCR products from different primer pairs generated from RNA isolated from control (yw) (left) and saxP homozygous mutant flies (right). Lane 1, primers 8103 + 10228; lane 2, primers 9396 + 11119; lane 3, primers placW10648 + 11119; lane 4, primers placW9661 + 10228. Note the insertion of placW disrupts the wild-type transcription unit initiating at RA-1 (lane 1) but allows transcription to initiate within the placW element between primers placW9661 and placW10648 (presence of PCR product in lane 3 of saxP animals and not in wild type, or in lane 4 of yw or saxP). Transcription in both genotypes extends through the expected translational termination site (PCR product present in lane 2 of both genotypes). M, marker lane. A, actin control. (C) The predicted amino acid sequence of SaxPB produced by open reading frame initiating at second endogenous ATG (bold and underlined in A) is shown in normal type with potential additional amino acids (italics) if translation initiated at an atg within the 3′ P-element of placW (shown in A).