Abstract
The microphthalmia-associated transcription factor (Mitf) has emerged as an important model for gene regulation in eukaryotic organisms. In vertebrates, it regulates the development of several cell types including melanocytes and has also been shown to play an important role in melanoma. In vitro, the activity of MITF is regulated by multiple signaling pathways, including the KITL/KIT/B-Raf pathway, which results in phosphorylation of MITF on serine residues 73 and 409. However, the precise role of signaling to MITF in vivo remains largely unknown. Here, we use a BAC transgene rescue approach to introduce specific mutations in MITF to study the importance of specific phospho-acceptor sites and protein domains. We show that mice that carry a BAC transgene where single-amino-acid substitutions have been made in the Mitf gene rescue the phenotype of the loss-of-function mutations in Mitf. This may indicate that signaling from KIT to MITF affects other phospho-acceptor sites in MITF or that alternative sites can be phosphorylated when Ser73 and Ser409 have been mutated. Our results have implications for understanding signaling to transcription factors. Furthermore, as MITF and signaling mechanisms have been shown to play an important role in melanomas, our findings may lead to novel insights into this resilient disease.
THE bHLH-Zip transcription factor MITF (microphthalmia-associated transcription factor) was initially found as the gene mutated at the mouse microphthalmia locus (Hodgkinson et al. 1993). Subsequent work has led to the proposal that MITF acts as a master regulator of melanocyte development by regulating proliferation, survival, and differentiation of this cell type (reviewed by Hou and Pavan 2008). MITF has also been implicated as a regulator of melanocyte stem cell maintenance (Nishimura et al. 2005) as well as a melanoma oncogene (Levy et al. 2006). In addition, the human MITF gene is mutated in individuals with Waardenburg syndrome type 2 and Tietz syndrome (reviewed in Steingrímsson et al. 2004). Clearly, MITF is an important decision maker in melanocytes and may be an important player in melanomas as well.
The many mutations at the mouse Mitf locus have provided key insights into the structure–function relationship of this transcription factor. All of the mouse Mitf mutations affect melanocytes to a varying degree, resulting in animals with coat color phenotypes ranging from white spotting and coat color dilution to a completely white coat due to lack of melanocytes (reviewed in Steingrímsson et al. 2004). Some of the mutations also affect retinal pigment epithelial (RPE) cells, resulting in unpigmented or hypopigmented microphthalmic eyes. Other cell types affected by Mitf mutations include osteoclasts and mast cells as well as natural killer cells, basophils, macrophages, and B cells (reviewed in Steingrímsson et al. 2004). Interestingly, the phenotypes of Mitf mutations overlap partly with the phenotypes of mutations in the Kit and Kitl genes in the mouse. Mutations in these three genes affect melanocyte development, resulting in pigmentation defects (often leading to white coat color), and all have effects on mast cells. However, only mutations in Mitf, and not Kit or Kitl, affect eye development. On the other hand, Kit and Kitl mutations affect hematopoiesis and germ-cell development whereas mutations in Mitf do not, at least not to the same degree.
As Kit and Kitl encode a receptor tyrosine kinase and its ligand, respectively, it was proposed that they form a signaling cascade that culminates in effects on the MITF protein (Dubreuil et al. 1991). Indeed, a number of signaling pathways, including KITL and KIT, have been shown to regulate the activity of the MITF protein. In a landmark series of experiments, David Fisher's group showed that MITF is an important downstream target of the KIT tyrosine kinase receptor. They showed that treating human 501-mel melanoma cells with KIT ligand (KITL) resulted in the phosphorylation of the Ser73 and Ser409 amino acids of MITF (Hemesath et al. 1998; Wu et al. 2000). Signaling to MITF involved the receptor tyrosine kinase KIT, the mitogen-activated protein kinase ERK2, and the serine–threonine kinase p90RSK and resulted in the phosphorylation of Ser73 by ERK2 and of Ser409 by p90RSK (Hemesath et al. 1998; Wu et al. 2000). Phosphorylation of Ser73 increased the transactivation potential of MITF by specifically recruiting the co-activator p300/CBP to the phosphorylated protein (Price et al. 1998). KITL stimulation was also shown to result in the ubiquitination of MITF and its subsequent proteasome-mediated degradation (Wu et al. 2000; Xu et al. 2000). Interestingly, when either Ser73 or Ser409 alone are mutated to alanine in vitro, transcription activation potential is only mildly affected, whereas mutating both sites simultaneously obliterates the transcription activation ability of the MITF protein in in vitro assays (Wu et al. 2000). These findings, together with the fact that mice that carry mutations in Kit and Kitl are white with normal eye development, have suggested that this signaling pathway is essential for MITF function in melanocytes.
Here, we use a BAC transgene rescue strategy to investigate the in vivo role of signaling to MITF in the mouse. Recombineering was used to generate specific mutations in a BAC clone containing the Mitf gene. BAC transgenic animals were made and then crossed to Mitf loss-of-function mutants to determine if the BAC could rescue the Mitf phenotype. Using this method, we show that mutating either Ser73 or Ser409 to alanine leads to rescue of the melanocyte and RPE defects associated with Mitf mutations. Similarly, deletion studies show that the domains encoded by exons 1A, 2A, and 2B do not seem to be essential for MITF function. Our results present a novel and rapid method for analyzing the role of individual amino acids and functional domains in the mouse. The results have implications for our understanding of transcription factor function in general and for melanocyte development in particular. And as MITF and signaling mechanisms have been shown to play an important role in melanoma, our findings may lead to novel insights into this disease.
MATERIALS AND METHODS
Recombineering and generation of transgenic mice:
The BAC-Mi1 (RP23-9A13) and BAC-Mi2 (RP23-21E20) clones were selected from the RPCI-23 BAC library (female C57BL/6J mouse). The BAC-Mi1 clone was introduced into the DY380 strain of Escherichia coli, and the Ser73Ala and Ser409Ala mutations were introduced using the two-step hit-and-fix method as described in Yang and Sharan (2003) and outlined in Figure 1. The deletion mutations were made using the method of Warming et al. (2005). The oligos used for recombineering and BAC screening are listed in Tables 1 and 2. The BACs were screened for the presence of the hit-and-fix sequences using PCR, and the mutated regions were sequenced to verify that the correct changes had been introduced. BAC DNA was prepared for pronuclear injections according to Sharan et al. (2004), and pronuclear injections of supercoiled BAC DNA were performed as described by Hogan et al. (1994).
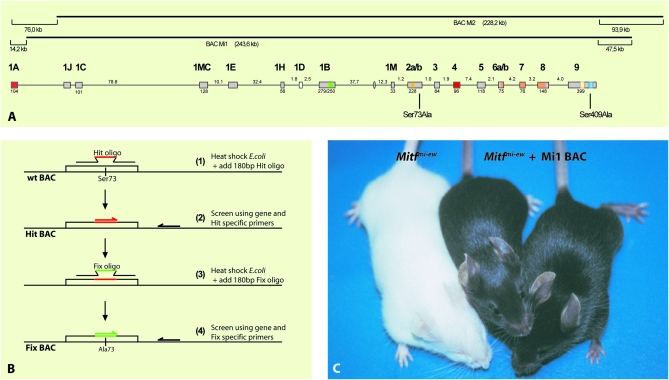
Figure 1.—
The BAC clones and recombineering strategy. (A) The two BAC clones isolated in the study are shown with respect to the structure of the Mitf gene. The size of the two clones are indicated in parentheses. Also indicated is the distance from the ends of the first and last known exons of Mitf to the ends of each clone. BAC Mi1 lacks 14.2 kb of DNA with respect to the first nucleotide of exon 1A whereas it contains an additional 47.5 kb of DNA from the last known nucleotide of exon 9. Similarly, BAC Mi2 lacks 76 kb with respect to the first nucleotide of exon 1A and contains additional 93.9 kb at the 3′-end. The relative sizes of the exons and introns are indicated but are not shown in the correct proportional sizes. The locations of amino acid residues Ser73 and Ser409 are indicated. (B) The two-step recombineering strategy used for generating the Ser73 and Ser409 mutations. The hit oligo was recombineered into the Mitf locus, and successful recombinants were screened using Hit-specific primers. After verifying the recombineered Hit clone, the Fix oligo was recombineered into the positive Hit-BAC and screened by PCR using fix-specific oligos. The resulting mutated BAC clones were further screened for the presence of the mutation and the absence of the Hit sequences before they were used for generating transgenic mice. (C) BAC transgene rescue of the Mitfmi-ew mutation. All three mice are homozygous for the Mitfmi-ew mutation, and therefore all should have the phenotype of the mouse on the left showing a white coat and microphthalmia. However, the two mice on the right also carry the BAC Mi1 clone, which fully rescues both phenotypes.
TABLE 1.
Primers used in the study for BAC recombineering
| Primer | Sequence |
|---|---|
| Ser73 | |
| Ser73HitF | 5′-ACATGCCAGCCAAGTCCTGAGCTCACCATGTCCAAACCAGCCTGGCGACCATGCCATGCCACCAGTGCCGGGGAGCAGCGAAGCTTACTGTCAGCTCGAG-3′ |
| Ser73HitB | 5′-AGTTAATGGAATGTAAGAAACAGAGGAGAAGAGGAGACGTGAATTACCTCTTTTTCACAGTTGGAGTTAAGAGTGAGCATCTCGAGCTGACAGTAAGCTT-3′ |
| Ser73FixF | 5′-ACATGCCAGCCAAGTCCTGAGCTCACCATGTCCAAACCAGCCTGGCGACCATGCCATGCCACCAGTGCCGGGGAGCAGCGCACCCAACGCCCCTATGGCT-3′ |
| Ser73FixB | 5′-AGTTAATGGAATGTAAGAAACAGAGGAGAAGAGGAGACGTGAATTACCTCTTTTTCACAGTTGGAGTTAAGAGTGAGCATAGCCATAGGGGCGTTGGGTG-3′ |
| Ser409 | |
| Ser409HitF | 5′-GGACGATGCCCTCTCACCTGTTGGAGTCACCGACCCACTGCTGTCATCAGTGTCGCCAGGAGCTTCAAAAACAAGCAGCCAAGCTTACTGTCAGCTCGAG-3′ |
| Ser409HitB | 5′-AGTCTCCTGAAGAAGAGAGGGAGCGGTCCGTGCAGAGGCAGAGCAAGGCAGGCTCGCTAACACGCATGCTCCGTTTCTTCCTCGAGCTGACAGTAAGCTT-3′ |
| Ser409FixF | 5′-GGACGATGCCCTCTCACCTGTTGGAGTCACCGACCCACTGCTGTCATCAGTGTCGCCAGGAGCTTCAAAAACAAGCAGCCGGAGGAGCGCTATGAGCGCA-3′ |
| Ser409FixB | 5′-AGTCTCCTGAAGAAGAGAGGGAGCGGTCCGTGCAGAGGCAGAGCAAGGCAGGCTCGCTAACACGCATGCTCCGTTTCTTCTGCGCTCATAGCGCTCCTCC-3′ |
| Del(int1/2A) | |
| Mitf galK F | CTGCCTGAAACCTTGCTATGCTGGAAATGCTAGAATACAGTCACTACCAGCCTGTTGACAATTAATCATCGGCA |
| Mitf galK R | ACTTGGCTGGCATGTTTATTTGCTAAAGTGGTAGAAAGGTACTGCTTTACTCAGCACTGTCCTGCTCCTT |
| galK del(int1/2a)up | CTGCCTGAAACCTTGCTATGCTGGAAATGCTAGAATACAGTCACTACCAGGTAAAGCAGTACCTTTCTACCACTTTAGCAAATAAACATGCCAGCCAAGT |
| galK del(int1/2a)low | ACTTGGCTGGCATGTTTATTTGCTAAAGTGGTAGAAAGGTACTGCTTTACCTGGTAGTGACTGTATTCTAGCATTTCCAGCATAGCAAGGTTTCAGGCAG |
| Del(2B/int2) | |
| Mitf2B galK F | ACCTGGAAAACCCCACCAAGTACCACATACAGCAAGCTCAGAGGCACCAGCCTGTTGACAATTAATCATCGGCA |
| Mitf2B galK R | CCTGGGCACTCACTCTCTGCCCTGCTCTGCTCCTCAAACTTATAAAATGCTCAGCACTGTCCTGCTCCTT |
| Mitf2B S | ACCTGGAAAACCCCACCAAGTACCACATACAGCAAGCTCAGAGGCACCAGGCATTTTATAAGTTTGAGGAGCAGAGCAGGGCAGAGAGTGAGTGCCCAGG |
| Mitf2B AS | CCTGGGCACTCACTCTCTGCCCTGCTCTGCTCCTCAAACTTATAAAATGCCTGGTGCCTCTGAGCTTGCTGTATGTGGTACTTGGTGGGGTTTTCCAGGT |
| Del(int2) | |
| Mitf intr2 galK F | CCAACAGCCCTATGGCTATGCTCACTCTTAACTCCAACTGTGAAAAAGAGCCTGTTGACAATTAATCATCGGCA |
| Mitf2B galK R | CCTGGGCACTCACTCTCTGCCCTGCTCTGCTCCTCAAACTTATAAAATGCTCAGCACTGTCCTGCTCCTT |
| Mitf intr2 S | CCAACAGCCCTATGGCTATGCTCACTCTTAACTCCAACTGTGAAAAAGAGGCATTTTATAAGTTTGAGGAGCAGAGCAGG |
| Mitf intr2 AS | ATGCACGACGCTCGAGAGTGCGTGTTCATACCTGGGCACTCACTCTCTGCCCTGCTCTGCTCCTCAAACTTATAAAATGC |
TABLE 2.
Primers used in this study for BAC and genomic screening
| Primer | Sequence | Function |
|---|---|---|
| Hit ScreenF | 5′-AAGCTTACTGTCAGCTCGAG-3′ | For screening Hit clones |
| S73FixScr | 5′-GGGAGCAGCGCACCCAACGCCCCTATGGCT-3′ | For screening 73 fixed clones |
| S409FixScr | 5′-ACAAGCAGCCGGAGGAGCGCTATGAGCGCA-3′ | For screening 409 fixed clones |
| Ex2/F | 5′-TGTTGGTCAACTGGTCAGTCT-3′ | For sequencing exon 2 |
| Ex2/R | 5′-AGGACAGAGGTTGGTGACAAT-3′ | For sequencing exon 2 and for screening hit-and-fix clones |
| RecEx9Scr5A | 5′-CCTGATCTGGTGAATCGGATCATCAAGC-3′ | For sequencing part of exon 9 |
| RecEx9Scr3A | 5′-CAGATACTGCACGCTGAAGGTACAATAC-3′ | For sequencing part of exon 9 and for screening hit-and-fix clones |
| vga9-for | 5′-CTGCAGCACGGACAATAAACCTCC-3 | Mitf: specific for vga9 mutation |
| mitf-rev | 5′-AAACAAGGCATCCCGAGGCACC-3′ | Mitf: for screening vga9 mutation |
| Mitf-2BtestF | 5′-CAGGGACGTTAAACAGGATAGT-3′ | For screening deletions |
| Mitf-2BtestR | 5′ -AGGTCCTGAAGTCATCAGGA-3′ | For screening deletions |
| CMr-F | 5′-AGCATTCTGCCGACATGGAAGC-3′ | BAC clone forward |
| CMr-R | 5′-CACGACGATTTCCGGCAGTTTC-3′ | BAC clone reverse |
Analysis of mutations:
Genomic DNA was extracted from mouse tail tips according to Jenkins et al. (1982). For detecting the MITFmi-vga9 transgene, we used the primers vga9-for and mitf-rev, and for detecting the CMr sequence of the BAC transgene, we used the primers CMr-F and CMr-R (see primers in Table 1). To verify that the mutations had been correctly introduced into the transgenic mice, RT–PCR was performed on heart tissue using the appropriate Mitf primers, and the resulting product was sequenced.
Analysis of alternative splicing:
Tissues were collected and deposited directly into liquid nitrogen and total RNA extracted using TRIZOL reagent (Invitrogen). RNA quantity was measured on a Nanodrop spectrophotometer, and integrity was determined on an Agilent 2100 Bioanalyzer RNA Nano chips. RNA with an RNA integrity number >7.0 was treated with DNase (Qiagen) to remove genomic contamination and purified on an RNeasy MinElute column (Qiagen). RNA quantity was measured again, and integrity was determined. RNA samples were then reverse transcribed using the Revertaid H first-strand synthesis kit (Fermentas) with both oligo(dT) and random hexamer primers, and the two reactions were combined for subsequent analysis. PCR was performed using the primers indicated in each case and listed in Tables 1–3.
TABLE 3.
Primers used in this study for RT–PCR and qPCR
| Primer | Sequence | Position |
|---|---|---|
| Exon 1bF | 5′-GACACCAGCCATAAACGTCA-3′ | Exon 1b forward |
| Exon 9R | 5′-TGCTTGATGATCCGATTCAC-3′ | Exon 9 reverse |
| Exon 3R | 5′-TGTTCATACCTGGGCACTCA-3′ | Exon 3 reverse |
| Mitf-q3-rev | 5′-CATCAATTACATCATCCATCTGC-3′ | Exon 4 reverse |
| Mitf-1M-e4F | 5′-ACTAAGTGGTCTGCGGTGTCTC-3′ | Exon M forward |
| Mitf 1M-e4R | 5′-TAACTTGATTCCAGGCTGATGA-3′ | Exon 4 reverse |
| Mitf-1H-e9F | 5′-TGAGTCAGACACCAGCCATAA-3′ | Exon 1H forward |
| Mitf-1H-e9-Rev | 5′-TGCTTGATGATCCGATTCAC-3′ | Exon 9 reverse |
| Mitf-e6-8qF | 5′-AGCAAGAGCATTGGCTAAAGA-3′ | Exon 6 forward |
| mMitf-e6-8qR | 5′-GCATGTCTGGATCATTTGACTT-3′ | Exon 8 reverse |
| mHPRT1-for2 | 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ | Hprt control |
| mHPRT1-rev2 | 5′-GATTCAACTTGCGTCATCTTAGGC-3′ | Hprt control |
Relative quantification by quantitative PCR:
RNA was isolated as described above. The presence of inhibitory factors in the RNA samples was tested using the SPUD assay (Nolan et al. 2006). Two micrograms of total RNA were reverse transcribed into cDNA using Superscript III reverse transcriptase and anchored oligo(dT)20 primers (Invitrogen). To find appropriate reference genes, the expression of 12 housekeeping genes (TATAA Biocenter, Göteborg, Sweden) was evaluated in cDNA samples prepared from heart tissues of six wild-type mice, and GeNorm software (Vandesompele et al. 2002) used to identify the most stable reference gene. Quantitative real-time PCR was performed using the SybrGreen I MasterPLUS mix and the LightCycler (Roche Diagnostics) and normalized to the expression of the most stable reference gene (Hprt1). Three mice from each group of BAC transgenic lines were analyzed for Mitf expression. Primers were designed using the Primer3 program (Rozen and Shaletsky 2000). Each sample assay included a cDNA template equivalent to 50 ng of total RNA; each PCR assay was performed in duplicate. Negative controls consisting of non-reverse transcribed samples were included in each set of reactions. Amplification efficiency was determined for the primers using a dilution-curve fivefold series of 6 points) and was 1.03 and 0.978, respectively, for the Mitf (mmitf-e6-8qF and mmitf-e6-8qR) and Hprt1 (Hprt1-F2 and Hprt1-R2) primers. Cycling conditions were the following: denaturation at 95° for 10 min, followed by 45 cycles at 95° for 10 sec, 59° for 10 sec, and 72° for 25 sec. Melting curve analysis and agarose gel electrophoresis were performed to confirm the specificity of the product. Data were analyzed using the Genex program (Vandesompele et al. 2002). Quantitation of Mitf isoforms containing or lacking exon 2B was performed as described in Bharti et al. (2009).
RESULTS AND DISCUSSION
BAC transgene strategy:
To determine the in vivo role of signaling to MITF, we used a BAC rescue strategy. Two BACs that contain the Mitf gene were obtained and characterized with respect to end sequences and the presence of the Mitf gene. The BACs, termed BAC-Mi1 (rpci-23-9a13t7) and BAC-Mi2 (rpci-23-21e20sp6), are shown in Figure 1A. Although they differ at the 5′- and 3′-ends, they contain the entire Mitf gene except exon 1A. We used a two-step recombineering protocol called “hit and fix” (outlined in Figure 1B) for introducing specific mutations into the BAC (Yang and Sharan 2003) and the GalK recombineering strategy (Warming et al. 2005) for introducing deletion mutations.
Phenotypic rescue of wild-type BACs:
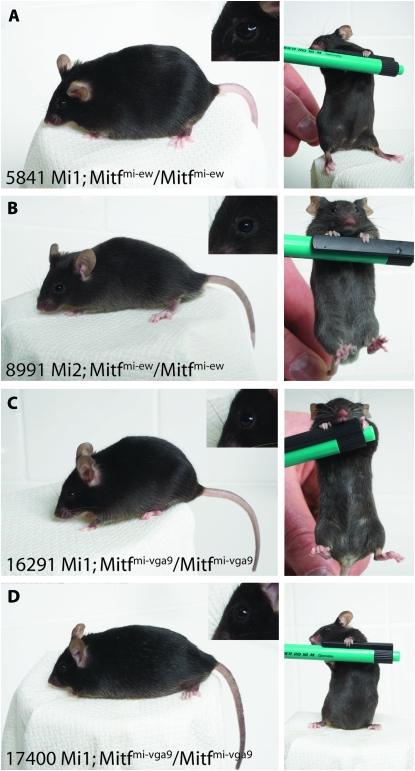
To test if the wild-type BAC clones can rescue the Mitf mutant phenotype, transgenic mice that carry the BAC-Mi1 clone were made. The transgenic mice were made using Mitfmi-ew heterozygous embryos, and the resulting carriers were then crossed to Mitfmi-ew homozygous animals to generate Mitfmi-ew/Mitfmi-ew animals, which also carry the BAC clone. Homozygous Mitfmi-ew/Mitfmi-ew animals are completely white, due to a lack of melanocytes, and have severe microphthalmia due to developmental defects in the retinal pigment epithelium (Figure 1C) (Steingrímsson et al. 2004). However, homozygous Mitfmi-ew mice that are also homozygous for the BAC-Mi1 clone are normally pigmented and have normal eyes (Figure 1C), suggesting that the BAC-Mi1 clone can rescue the phenotype completely, despite lacking exon 1A; they are normally pigmented in every respect, including the belly region (Figure 2A). Transgenic mice were also made using the BAC-Mi2 clone and, again, the BAC can rescue the Mitfmi-ew mutant phenotype with respect to both coat color and eye developmental defects (Figure 2B). The Mitfmi-ew mutation results in a MITF protein that lacks the DNA-binding domain (Steingrímsson et al. 1994; Hallsson et al. 2000), and in vitro studies suggest that the protein acts in a dominant-negative fashion (Hemesath et al. 1994); detailed phenotypic characterization has suggested a mild dominant-negative action of the mutant protein in osteoclasts (Steingrímsson et al. 2002). To avoid possible problems with dominant-negative behavior of the Mitfmi-ew mutation, we also performed rescue studies using the recessive Mitfmi-vga9 mutation, a transgene insertion mutation that obliterates Mitf expression in most tissues (Hodgkinson et al. 1993) and is therefore considered a null mutation. Homozygotes are white and have severe microphthalmia although there is no evidence of a dominant or semidominant phenotype in any tissue, including bone (Hodgkinson et al. 1993; Steingrímsson et al. 2004). Two different transgenic lines were made on the Mitfmi-vga9 mutant background and, as can be seen in Figure 2, C and D, the BAC-Mi1 clone fully rescues the Mitfmi-vga9 mutant phenotype, with respect to both coat color and eye development. Both lines are maintained as homozygotes for both the Mitfmi-vga9 mutation and the BAC transgenes. Thus, we conclude that the BAC clones Mi1 and Mi2 rescue the Mitf mutant phenotype fully and therefore contain all the sequences necessary for normal Mitf expression and function in melanocytes and RPE cells. These results suggest that exon 1A is not necessary for Mitf function in these cell types. This is interesting in light of the relatively high level of conservation of amino acid sequence in this exon between different mammalian species (>95% amino acid conservation between mice and humans) (Hallsson et al. 2007).
Figure 2.—
Wild-type BAC clones rescue Mitf mutant phenotypes. The two wild-type BAC clones rescue Mitf mutant phenotypes completely. (A and B) The BAC Mi1 and Mi2 wild-type BAC clones rescue the phenotype of the Mitfmi-ew mutation fully; no belly spot is visible in the mice (right panels). (C and D) The BAC Mi1 clone also rescues the Mitfmi-vga9 loss-of-function phenotype fully as seen in two independent transgenic lines. Again, not even a belly spot is visible, suggesting full rescue. The inserts show the eyes of the BAC transgenic mice.
Phenotypic rescue of serine-mutant BACs:
To determine the in vivo significance of the Ser73 and Ser409 phosphorylation sites of MITF, we mutated these sites in the BAC-Mi2 clone to alanine using the recombineering strategy described in Figure 1B. We generated three different mutant BAC clones: a Ser73Ala mutation (changing AGC to GCC), a Ser409Ala mutation (changing AGT to GCC), and a clone containing the Ser73Ala and Ser409Ala mutations. After confirming the presence of each mutation, the BACs were used to generate transgenic mice using Mitfmi-vga9/+ heterozygous zygotes, and the resulting transgenic animals were mated to generate animals that were homozygous for the Mitfmi-vga9 mutation and for the BAC transgene.
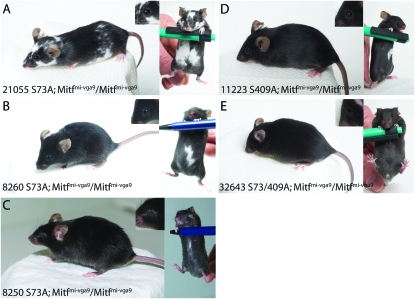
Three independent lines that carry the BAC containing the Ser73Ala mutation were generated (Figure 3, A–C). These lines all rescue the Mitfmi-vga9 phenotype significantly, but to a different extent. The TG21055-Ser73Ala line fully rescues the eye developmental phenotype whereas coat color is partially rescued, resulting in mice with normal eyes, a white belly, and white spots over the rest of the coat (Figure 3A). The TG8260-Ser73Ala line fully rescues the eye defect of the Mitfmi-vga9 mutation, and the coat color is rescued apart from a belly spot (Figure 3B). The TG8250-Ser73Ala line does not rescue the eye developmental defect but rescues the coat color defect nearly fully, leaving a small white belly spot (Figure 3C). Together, the rescue observed with these lines suggests that the Ser73Ala mutant BAC can rescue the eye developmental phenotype and the coat color defects of Mitf mutations, apart from leaving an unpigmented belly spot. Small belly spots are commonly seen in the C57BL/6J strain (Arnheiter 2007), on which the Mitfmi-ew and Mitfmi-vga9 mutations are maintained, suggesting that in fact we observe almost full rescue with the TG8260-Ser73Ala line and full rescue with respect to coat color in the TG8250-Ser73Ala line. The difference in the ability of these three lines to rescue the phenotype probably reflects the integration sites involved.
Figure 3.—
The mutant BAC clones Ser73Ala and Ser409Ala rescue the Mitf mutant phenotype. The BAC clones containing the Ser73Ala and Ser409Ala mutations rescue the phenotype of the Mitfmi-vga9 loss-of-function mutation fully in either single or double-mutant combination. (A–C) Three different transgene lines that contain the Ser73Ala mutation were obtained. The line 21055 fully rescues the microphthalmia associated with the Mitfmi-vga9 mutation whereas the coat color phenotype is rescued only partially (A). These animals lack pigmentation on the belly and have small unpigmented spots over the rest of the body. However, a significant proportion of the coat is pigmented, suggesting significant rescue in this line; the spots on the back are rather small, suggesting failure to rescue during late melanocyte development or differentiation. (B) The line 8260 rescues the phenotype fully, apart from a small belly spot (right). (C) The line 8250 rescues the coat color phenotype fully except for a minor belly streak seen in most of the animals. However, eye development is not rescued. The phenotypic differences in these three lines are most likely due to differences in the integration event of the transgenes. (D) The Ser409Ala mutant BAC clone can rescue the phenotype of the Mitfmi-vga9 mutation completely, although many animals show a significant belly spot. (E) The BAC clone containing the double mutation Ser73Ala; Ser409Ala can also rescue the phenotype of the Mitfmi-vga9 mutation completely, and in this case belly spots are rarely seen. The inserts show the eyes of the BAC transgenic mice.
We obtained one line each of transgenic mice that carry the Ser409Ala single mutant and the Ser73Ala; Ser409Ala double-mutant BAC clones (Figure 3, D and E). Both the TG11223-Ser409 transgenic line and the double-mutant TG32643-Ser73/409Ala transgenic line rescue the eye and coat color phenotype of the Mitfmi-vga9 mutation fully, resulting in normally pigmented mice (Figure 3, D and E). In the TG11223-Ser409 line, some animals show a belly spot (Figure 3D) whereas a belly spot is rarely seen in the double-mutant TG32643-Ser73/409Ala line. Together, these data suggest that the Ser73Ala and Ser409Ala mutations can fully rescue the Mitfmi-vga9 mutation, even in a double-mutant combination. This indicates that these two serines are not essential for MITF function during mouse melanocyte development.
Splicing of Mitf is affected in the Ser73Ala BACs:
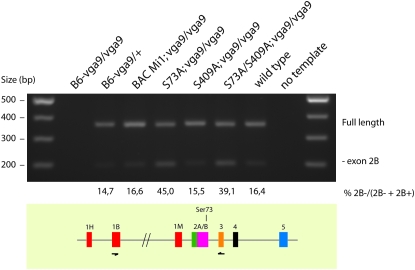
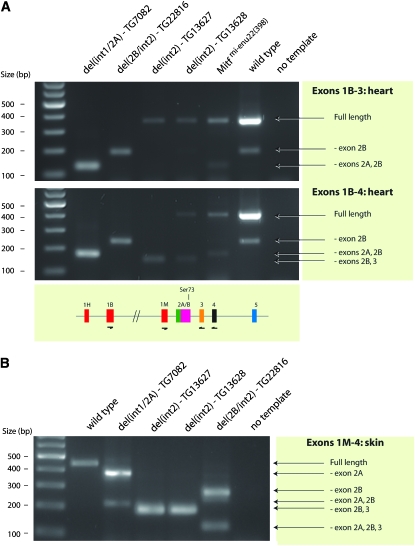
A knock-in mutation has been made in the Mitf gene where Ser73 was replaced with alanine (Bismuth et al. 2008). This knock-in mutation is the very same mutation that we made in the BAC mice, except it also introduced a silent ApaLI restriction site a few base pairs upstream of the Ser73Ala mutation and a LoxP site in the downstream intron (Bismuth et al. 2008). Mice homozygous for this knock-in mutation are normally pigmented and have normal eyes, supporting our conclusions from the BAC rescue experiments. However, the knock-in mutation affected splicing of the Mitf gene such that a normally minor alternative product lacking exon 2B became the major product (Bismuth et al. 2008). We used RT–PCR on heart tissue to determine if the splicing of Mitf was affected in the BAC transgenic mice. Heart tissue expresses the Mitf mRNA at a significant level in wild-type mice as well as in most of the available Mitf mutants but is absent in hearts from animals carrying the Mitfmi-vga9 mutation (Hodgkinson et al. 1993; Steingrímsson et al. 1994). As all the BAC clones are maintained on the Mitfmi-vga9 mutant background, the only Mitf mRNA expressed in the BAC transgenic mice is derived from the BAC clone. The primers used span from exon 1B to exon 3 and resulted in two bands in wild-type mice: a 359-bp full-length band and a 191-bp band that represents a product lacking exon 2B (Figure 4). The same bands are present in all the transgenic lines, and all show the same ratio of the two bands, except in the BAC lines that contain the Ser73Ala mutation, either alone or in the double-mutant combination with Ser409. Quantitative PCR (qPCR) analysis shows that in the two lines containing the Ser73Ala mutation there is approximately three times more of the smaller 198-bp band lacking exon 2B than in either the other BAC lines or wild-type mice (Figure 4). This suggests that the presence of the Ser73Ala mutation alone is sufficient to affect splicing of exon 2B such that the shorter product becomes more abundant than in the other lines. This is consistent with the splicing defect observed in the Ser73Ala knock-in mice and with the idea that the sequence around the Ser73 codon represents an exon splice enhancer sequence (Bismuth et al. 2008). The only difference between the Ser73Ala knock-in and BAC transgene mutations is the ratio of the two bands. In the knock-in mice, the shorter band represents a majority (∼90%) of the resulting Mitf products whereas in the BAC mice it is ∼40–45%. This difference may be due to the more extensive changes introduced in the knock-in mutation than in the BAC as the knock-in mutation generated an ApaLI restriction site (changing a C to a T 10 bp upstream of the Ser73Ala codon) and placed a floxed neo-cassette in a nearby intron, in addition to the Ser73Ala modification.
Figure 4.—
Splicing of exon 2 of Mitf is affected in the BAC transgenic mice containing the Ser73Ala mutation. RT–PCR was used to determine if splicing of exon 2 was affected in the BAC transgenic mice. Primers spanning from exons 1B to exon 3 (bottom) were used to amplify cDNAs isolated from the indicated BAC transgenic mice (top). No products were amplified in the homozygous Mitfmi-vga9 mice whereas two bands were amplified in Mitfmi-vga9 heterozygotes and in wild-type mice as well as in all the other BAC transgenic lines. The 359-bp top band represents the full-length product whereas the 191-bp bottom band represents an alternative product lacking exon 2B. Quantitative PCR using exon-specific primers was used to determine the relative amount of these two products in the different BAC transgenic lines. The results are indicated as the percentage of the product lacking exon 2B over the total amount of products lacking and containing exon 2B. The shorter product is relatively more abundant in the lines that contain the Ser73Ala mutation.
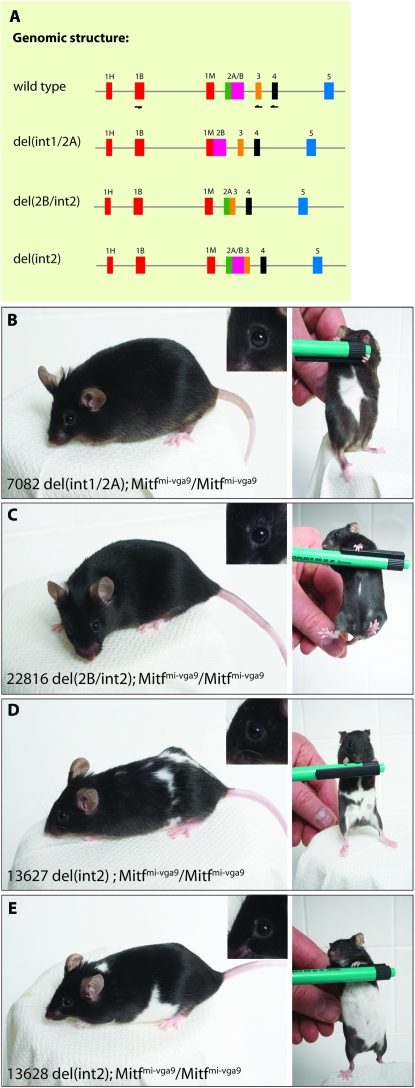
Exon 2 deletion mutations:
Ser73 is located in exon 2B of the Mitf gene. The phenotype of the Ser73Ala knock-in (Bismuth et al. 2008) and BAC transgenic mice suggests that the absence of exon 2B in a majority of the Mitf transcripts produced is not detrimental for the function of the MITF protein. To detail the functional role of exon 2 of Mitf, we created three different deletion mutations in the BAC and generated transgenic mice on the Mitfmi-vga9 background. The mutations represent a deletion of intron 1 and exon 2A, resulting in a fusion of exon 1M with exon 2B [del(int1/2A)]; deletion of exon 2B and intron 2, creating a fusion of exon 2a and exon 3 [del(2B/int2)]; and deletion of only intron 2, creating a fusion of exon 2B and exon 3 [del(int2)] (Figure 5A). For the del(int1/2A) and del(2B/int2) deletions, we obtained one transgenic line each, whereas for del(int2) we obtained two independent lines (Figure 5, B–E). The transgenic line carrying the del(int1/2A) deletion fully rescues the eye developmental defect of the Mitfmi-vga9 mutation as well as the coat color, except for a belly spot (Figure 5B). Similarly, the del(2B/int2) transgene rescues the eye and coat color phenotype, apart from a small belly spot seen in most of the mice (Figure 5C). The two different del(int2) lines fully rescue the eye developmental phenotype of the Mitfmi-vga9 mutation and the coat color phenotype, leaving a large belly spot and occasional white spots on other body regions (Figure 5, D and E). Interestingly, the two independent del(int2) lines have near-identical phenotypes.
Figure 5.—
Generation and analysis of exon 2 deletion mutations. Several different deletion mutations were generated to analyze the role of exons 2A and B in Mitf function. (A) The genomic structure of the deletion mutations that were generated. The exons are indicated as colored boxes. (B–E) The transgenic mice carrying the different deletion mutations. The left panels show a side view of the mice and the right panels show the belly region. The inserts in the upper right corner of the left panels show the eyes of the BAC transgenic mice. (B) The BAC carrying the del(int1/2A) mutation rescues the phenotype of the Mitfmi-vga9 mutation to a large extent, leaving a sizable belly spot. (C) The BAC carrying the del(2B/int2) mutation also rescues the phenotype of the Mitfmi-vga9 mutation; a small belly spot is visible in most of the animals. (D and E) Two different transgenic lines were generated which carry the BAC with the del(int2) mutation. These lines fully rescue the microphthalmia associated with the Mitfmi-vga9 mutation whereas coat color is rescued only partially. These animals have an extensive belly spot in addition to white spots over the rest of the coat.
We used RT–PCR to characterize expression and splicing of the Mitf gene in mice carrying the BAC-deletion mutations. First, we characterized expression of Mitf in heart tissue using primers extending from exon 1B to exon 3 or exon 4 (Figure 6A). In wild-type hearts, both primer sets produced two bands, representing a full-length transcript and a transcript lacking exon 2B, at a ratio of ∼20:1 (Figure 6A). This is consistent with previous results (Hallsson et al. 2000). In the del(int1/2A) transgenic mice, each primer set resulted in the production of only one product. The size of these products suggests that they are missing exons 2A and 2B, and this was further confirmed by sequencing. These results were expected in this line as exons 2A and 2B are not split by an intron; in wild-type mice, the mRNA product lacking exon 2B is produced by the use of an alternative splice donor. As the deletion removes intron 1 and exon 2A, this alternative splice donor is not present and the only possible product in this case fuses exon 1B to exon 3. RT–PCR performed on hearts from the del(2B/int2) deletion mutation also results in only one product for each primer set (Figure 6A). Sequencing shows that, as expected, these products lack exon 2B whereas exon 2A is present.
Figure 6.—
Analysis of splicing in BAC transgenic mice carrying Mitf deletion mutations. RT–PCR analysis was performed on both heart and skin to characterize Mitf transcripts in mice carrying the BAC deletion mutations. (A) Splicing of Mitf in heart tissue was characterized using two primer sets. The first primer set (top) extended from exon 1B to exon 3. In wild-type mice, these primers result in two bands: a full-length 359-bp band and a shorter 191-bp band lacking exon 2B. The full-length band is more prominent, representing 90–95% of the product. In the del(int1/2A) mutation, a single 131-bp PCR product was made, representing a transcript lacking both exons 2A and 2B. Because exon 2A is missing, the only product possible is made by splicing exon 1B directly to exon 3. In the del(2B/int2) mutation, only the 191-bp product lacking exon 2B is generated, as expected. The two del(int2) lines generate only the full-length product. The Mitfmi-enu22(398) mice express both the full-length product and the 131-bp product missing both exons 2A and 2B. The second primer pair (bottom) extended from exon 1B to exon 4. The results are largely the same as above. The only exception is that the two del(int2) lines produce a smaller band in addition to the full-length product observed in line TG13628. The smaller product represents transcripts that lack exons 2B and 3, suggesting that exon 2B is spliced directly to exon 4 in these mice. (B) Splicing of Mitf in skin was analyzed using primers extending from exon 1M to exon 4. In control (B6) animals, this primer set results in the production of a single 440-bp full-length product. These primers result in two products in skin from mice carrying the del(int1/2A) deletion mutation, a 380-bp band representing a transcript lacking exon 2A, and a smaller 212-bp band lacking exons 2A and 2B. This deletion fuses exon 1M directly to exon 2B so none of the products will contain exon 2A. However, the smaller product suggests that the splice donor in exon 1M is still active and that this exon can be fused directly to exon 3. The del(2B/int2) deletion also results in the production of two fragments using this primer set: a 272-bp fragment lacking exon 2B and a fragment lacking exons 2A, 2B, and 3. In these mice, exon 2A is fused directly to exon 3 so all products will lack exon 2B. The small alternative product indicates that exon 1M can also be spliced directly to exon 4. Both del(int2) lines result in only one product, representing a transcript lacking exons 2B and 3. Clearly, the absence of intron 2 leads to the skipping of the two flanking exons, and the only product produced fuses exon 2A directly to exon 4.
The two del(int2) transgenic lines (13627 and 13628) affect mRNA splicing similarly, with some minor differences. Using primers from exon 1B to exon 3, only one product is produced, representing a full-length Mitf transcript (Figure 6A, top). This transcript is produced from the transgene by the splicing of exon 1B to exon 2A/B, which is then fused directly to exon 3 due to the deletion of intron 2. No alternative product is detected lacking exon 2B using this primer combination because such a product is not possible when intron 2 has been removed. RT–PCR performed using primers from exon 1B to exon 4 results in two products in the transgenic line TG13628, representing a full-length product and a truncated product lacking exons 2B and 3 (Figure 6A, bottom). This smaller PCR product is produced by alternative splicing from exon 2A to exon 4 using the alternative splice donor at the juncture of exons 2A and B, which is then spliced to the acceptor in exon 4 rather than the acceptor in exon 3 as it is lacking due to the deletion. In transgenic line TG13627, only the smaller product lacking exons 2B and 3 is detected using the primer set from exon 1B to exon 4, whereas in line TG13628, the full-length product is also detected. Clearly, the full-length product is expressed in both transgenic lines at some level since a product is observed using the primers extending from exon 1B to exon 3. However, the absence of such a product in the TG13627 line using the exon 1B to exon 4 primer set suggests that this is a minor product and that the majority of transcripts are lacking exons 2B and 3. Nevertheless, the phenotypes of both transgenic lines are nearly identical (Figure 5, D and E), suggesting that, although a MITF protein lacking exons 2B and 3 is able to rescue eye development fully, it is not quite able to fully rescue the coat color phenotype of the Mitfmi-vga9 mutation. Because the transgene deleting only exon 2B can rescue the phenotype, this would indicate that the simultaneous absence of both domains encoded by exons 2B and 3 is detrimental to function. Alternatively, it is possible that exon 3 plays a more important functional role than exon 2B. Sequence comparison of representative species of the mammalian, avian, amphibian, fish, and echinoderm lineages reveals that exon 3 is found only in mammals and birds. Although this means that exon 3 is not evolutionarily conserved in all species, it is possible that the recruitment and retention of this sequence in mammalian and avian species indicates a functional role. At this point, however, this function is not known.
The deletion del(int1/2A) fuses exons 1M and 2B of Mitf, and as exon 1M is melanocyte specific and thus not expressed in the heart, we also performed RT–PCR on skin tissue from all deletion strains using primers extending from exon 1M to exon 4. As seen in Figure 6B, the RT–PCR results indicate the presence of the expected Mitf transcripts in skin. Using primers extending from exon 1M to exon 4 results in a 440-bp normal band whereas in the transgenic strain carrying the del(int1/2A) deletion this results in two bands: one major 380-bp band missing exon 2A and one minor 212-bp band missing both exons 2A and 2B. This suggests that the splice donor in exon 1M is functional when fused to exon 2B. This is consistent with the fact that the splice donor sequence remains the same (ACCAG/GT). In the del(2B/int2) strain, the primer pair resulted in a major 272-bp band missing exon 2B and a minor band lacking also exons 2A and 3. This smaller product represents a rare alternative Mitf transcript, where exon 1M has been spliced directly to exon 4. A similar Mitf transcript, extending from exon 1A to exon 4, has been shown to exist in heart (Hallsson et al. 2000). In the two del(int2) transgenic strains, the only product present is a 188-bp band lacking exons 2B and 3. This is the major band seen in skin from the mutants, suggesting that the full-length transcript observed in heart is rarely, if at all, expressed in skin.
Our analysis of Mitf expression and splicing in skin is consistent with the results obtained using heart tissue. Because the del(int1/2A) and del(2B/int2) BAC transgenes rescue the Mitfmi-vga9 phenotype (Figure 5, B and C), we conclude that neither exon 2A nor exon 2B is essential for MITF function in eye or melanocyte development in the mouse. Exon 2B contains the codon for amino acid Ser73, so these results are consistent with results obtained by the Ser73Ala knock-in mutation (Bismuth et al. 2008) and the Ser73Ala BAC transgenes described above. These results are rather surprising because a section of exon 2A (amino acids 11–39) is highly conserved among different species (Hallsson et al. 2007).
The Mitfmi-enu22(398) mutation:
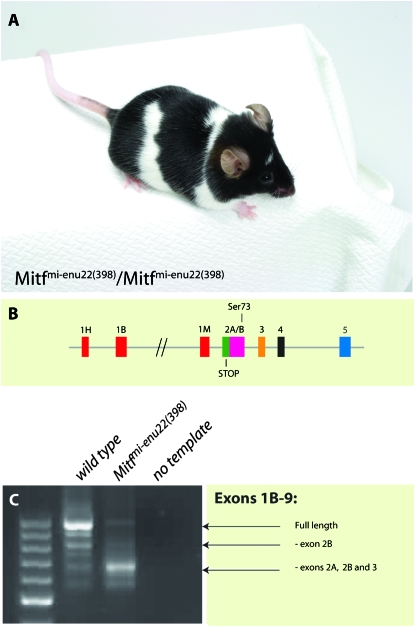
Analysis of a novel Mitf mutation called Mitfmi-enu22(398) provides independent support for the molecular and phenotypic characteristics of the deletion mutations. This mutation was induced by treatment with ethylnitrosourea as part of a genetic screen performed at the Novartis Institute for Functional Genomics. Animals homozygous for this mutation have normally developed eyes, a white belly, and large unpigmented spots over the rest of the coat (Figure 7A). The phenotype is remarkably similar to the phenotype of the del(int2) transgenic lines on the Mitfmi-vga9 background (compare Figure 7A to Figure 5, D and E), except Mitfmi-enu22(398) has a bit more extensive white spotting. Sequence analysis shows that the mutation is a C-to-T change at nucleotide position 205 of the cDNA, which introduces a stop codon in exon 2A, changing glutamine at position 26 of the MITF protein to STOP (Figure 7B). This would terminate the MITF protein prematurely, producing only a short peptide. RT–PCR analysis using primers that extend from exon 1B to exon 9 results in multiple bands in wild-type hearts, indicating the multiple alternative splice products made; sequencing confirms that the full-length product is the most prominent product and that another prominent product lacks exon 2B (Figure 7C). However, in hearts from Mitfmi-enu22(398) homozygous animals, the major Mitf product lacks exons 2A, 2B, and 3, resulting in a product in which exon 1B is fused to exon 4 (Figure 7C). A functional protein can be made although it lacks the domains encoded by these three exons. The molecular characteristics of this mutation are similar to the del(int2) transgenic lines on the Mitfmi-vga9 background in that all are lacking exons 2B and 3. They are different, however, in that the del(int2) lines retain exon 2A and express the full-length wild-type transcript at a low level, at least in heart, whereas in Mitfmi-enu22(398) hearts, exon 2A is largely missing. The full-length RNA product is also made in Mitfmi-enu22(398) animals, as well as a product lacking exons 2A and 2B (Figure 6A). However, the full-length transcript can be translated only into a truncated peptide. The phenotypic similarity of animals carrying the del(int2) BACs and the Mitfmi-enu22(398) mutations suggests that the transgenic approach accurately reflects the functional aspect of Mitf during melanocyte and eye development.
Figure 7.—
Molecular and phenotypic characterization of the Mitfmi-enu22(398) mutation. (A) The phenotype of the Mitfmi-enu22(398) mutation. The mice have normal eye size. However, although they have extensive white belly spots and white spots over the rest of the body, a significant portion of their coat is normally pigmented. (B) The mutation associated with this phenotype is a stop codon in exon 2A of the Mitf gene. The position of the Ser73 amino acid is also indicated. (C) Characterization of splicing of the Mitf gene in normal and Mitfmi-enu22(398) mutant mice using primers extending from exon 1B to exon 9. In normal mice, this results in the production of a major 997-bp full-length band and several alternative splice products, including the 829-bp product lacking exon 2B. In the Mitfmi-enu22(398) mutation, the major 685-bp product is lacking exons 2A, 2B, and 3.
The Mitfmi-black-and-white spot (Mitfmi-bws) mutation also results in black and white animals (Hallsson et al. 2000). The molecular defect described in this mutation is an alteration in a splice acceptor site 12 bp upstream of exon 2A, which results in partial exon skipping such that exon 2B is missing in a significant portion of the transcripts (Hallsson et al. 2000). This is therefore not consistent with our BAC results or with our analysis of the Mitfmi-enu22(398) mutation. Thus, we used quantitative PCR to characterize the expression of the Mitf gene in the Mitfmi-bws mutation. This shows that ∼40% of Mitf transcripts in the hearts of Mitfmi-bws animals lack exon 2B. However, the total expression of the Mitf transcript is only 9.5% in Mitfmi-bws animals as compared to wild type when analyzed using primers from exon 6 to exon 7 or from exon 7 to exon 8. Thus, the level of Mitf expression is significantly affected in Mitfmi-bws animals. At present, it is not well understood how the intron mutation found in Mitfmi-bws results in the alternative splicing of a downstream exon or how it affects expression of the gene.
Relative quantification of MITF expression in BAC transgenic mice:
To determine the expression level of the Mitf gene in BAC transgenic mice, we performed qPCR analysis on tissues from the transgenic animals as well as from controls. We followed the protocols and recommendations of Nolan et al. (2006) and tested for inhibitory factors using the SPUD assay, tested 12 different housekeeping genes to identify the most stable reference gene (we used Hprt1), and determined amplification efficiency for each primer pair used. The primers used for quantifying Mitf extend from exon 6 to exon 8 and should quantify all currently know splice forms of the gene. We measured expression in hearts from three animals and skin from two animals of each genotype, and each reaction was performed in duplicate using identical amounts of total RNA in each case. Expression of Mitf was first determined in heart, a tissue that expresses significant levels of Mitf in wild-type mice, as well as in mutants that lack melanocytes and show severe microphthalmia (Steingrímsson et al. 1994). In the Mitfmi-vga9 mutants, however, we detected no Mitf expression because no significant difference was found in Mitf expression between cDNA samples prepared from Mitfmi-vga9 homozygotes and from no-template controls (average cycle threshold values were 32.5 ± 0.95 and 33.2 ± 1.34, respectively). Thus, the Mitfmi-vga9 mutants do not express the Mitf gene in heart, and the only transcripts detected in the BAC transgenic lines must therefore be expressed from the transgenes and not from the Mitfmi-vga9 locus.
The results of the qPCR analysis in heart are presented in Table 4 as the fold change of Mitf expression as compared to the control after normalization. The data show that expression of the Mitf transcript in heart from the BAC transgenic lines ranges from −1.15-fold to 0.55-fold as compared to controls. Interestingly, expression of Mitf in Mitfmi-vga9 heterozygotes is −0.9-fold less than in controls, indicating that they have half the amount of the Mitf transcript as compared to controls. This is exactly as expected, given that these animals express Mitf from only one copy of the gene. Expression of Mitf in the BAC-Mi1 transgene, which contains the wild-type BAC, is very similar to controls or −0.17-fold. This suggests that the BAC construct truly replicates expression of the wild-type Mitf gene in heart. These data confirm both the robustness of our qPCR analysis and the BAC transgene rescue method. Analysis of Mitf expression in the remaining BAC transgenic lines shows that expression in heart is in most cases similar to wild type, or lower [the Ser73Ala, Ser73Ala; Ser409Ala double mutant, del(2B/int2), Ser409Ala, del(int2)13628, and del(int1/2A) transgenic lines]. In only one case is Mitf expressed at a level higher than seen in the controls [the BAC transgene line del(int2)13627]. This line, however, shows a phenotype identical to the phenotype of the del(int2)13628 line, suggesting that expression levels are less important for phenotypic expression than the molecular defect involved. The results suggest that the BAC transgenes more or less replicate the wild-type expression of the Mitf gene in heart.
TABLE 4.
Relative expression of Mitf in heart and skin
| Fold changea
|
||||
|---|---|---|---|---|
| Heart | Skin | |||
| B6 | 0 | ±0,62 | 0 | ±1.51 |
| B6-vga9/+ | −0.90 | ±0,26 | −1.05 | ±0.33 |
| BAC-Mi1 | −0.17 | ±0,62 | −0.22 | ±1.70 |
| Ser73Ala | −0.50 | ±0,32 | −1.31 | ±1.53 |
| Ser409Ala | −0.82 | ±0,19 | −2.04 | ±2.55 |
| Ser73/409Ala | −0.13 | ±0,60 | −4.80 | ±0.25 |
| del(int2)13627 | 0.55 | ±0,25 | −0.19 | ±1.73 |
| del(int2)13628 | −0.88 | ±1,40 | −1.27 | ±1.68 |
| del(int1/2A) | −1.15 | ±0,53 | −1.70 | ±3.45 |
| del(2B/int2) | −0.52 | ±0,60 | −2.18 | ±2.00 |
Relative to the reference gene Hprt1.
Melanocytes are the cells responsible for pigmentation. To determine Mitf expression levels in melanocytes of the BAC transgenic mice, skin of transgenic 5-day-old mice was prepared for RNA isolation, cDNAs were prepared, and qPCR analysis was performed as described above. The results are shown in Table 4 and show that in all the lines but one, Mitf is expressed at lower levels than seen in controls, ranging from 0.19- to 4.80-fold lower expression. Again, the expression of Mitf in Mitfmi-vga9 heterozygotes is at half the level seen in the controls, suggesting that our method accurately measures expression of the gene in skin. In conclusion, the gene expression data in both heart and skin show that most of the transgenes express Mitf at a level similar to or lower than what is seen in wild-type mice. Thus, we conclude that the rescue observed is not due to overexpression of the Mitf gene. The level of transcripts is of course only one parameter in determining the levels of functional protein, and it is possible that the cells can compensate for the lack of transcripts through regulating translation, activity, or other aspects of producing a transcriptionally active MITF protein.
Conclusions:
We have used a BAC transgene rescue method to determine the in vivo functional role of two phospho-acceptor sites in the MITF protein, serine 73 and serine 409, as well as of two domains encoded by exons 2A and 2B. These two phospho-acceptor sites had previously been suggested to be important for MITF function, although the effects observed differed depending on the cell type used. Hemesath et al. (1998) showed that in baby hamster kidney (BHK) cells the wild-type and Ser73Ala mutant MITF proteins can activate expression of the tyrosinase promoter at equal levels. However, in BHK cells harboring constitutively active Raf, the wild-type protein was a 2.5 times more potent activator of tyrosinase than the Ser73Ala mutant protein, suggesting that the Raf signaling pathway affects transactivation specifically through this phospho-acceptor site. Price et al. (1998) showed that the increased transcription activity was due to the recruitment of the co-activator p300 to the phosphorylated protein; a protein containing the Ser73Ala mutation failed to activate expression. Wu et al. (2000) then showed that in NIH3T3 cells the Ser73Ala mutant protein is significantly less able to activate transcription than the wild-type protein whereas the Ser409Ala mutation is similar to wild type. In these cells, the Ser73Ala; Ser409Ala double-mutant protein is not able to activate transcription at all. In 501mel melanoma cells, the wild type, Ser73Ala, and Ser409Ala proteins are similar in their transcription activation potential whereas the double mutant is transcriptionally inactive (Wu et al. 2000). From these data and the fact that the phenotypes of Mitf, Kitl, and Kit mutations all affect coat color, we would have expected that mutations in these phospho-acceptor sites in mice would result in mice with a white coat color. Our results, however, indicate that, in vivo, neither of the two phospho-acceptor sites is essential for MITF function.
We believe that our results accurately depict the in vivo roles of these amino acids and domains. First, the BAC transgene rescue strategy clearly works because wild-type BACs rescue the Mitfmi-vga9 phenotype fully and replicate the endogenous expression of the Mitf gene. It should be emphasized that this strategy calls for the rescue of a severe phenotype, where melanocytes are missing and where RPE cells have lost their normal identity. The strategy therefore represents a “positive” test of Mitf function in vivo. Second, expression of the Mitf gene in heart and skin from the BAC transgenic mice is in the range (or less) observed in wild-type and Mitfmi-vga9 heterozygous mice, which have normal coat color and eye development. This suggests that the transgenes have not resulted in overexpression of the Mitf gene, which might have confounded the results. Third, our results are consistent with a previous characterization of the role of Ser73 using a knock-in strategy (Bismuth et al. 2008). Fourth, the phenotype and molecular defect of the Mitfmi-enu22(398) mutation is consistent with the phenotype of the del(int2) mutations, suggesting that the BAC constructs can replicate the phenotype of Mitf mutations.
So how can we then explain the discrepancy between the in vitro and the in vivo observations? One possible explanation is that in vivo, alternative phospho-acceptor sites take over when Ser73 and Ser409 have been mutated to alanine, thus leading to total rescue of the phenotype. Due to differences in the environment (culture conditions vs. in vivo development) this compensation may not take place in the in vitro assays, leading to the observed effects on transcription activation and protein stability. This would imply that there is a redundancy in the signaling mechanisms to ensure that the protein receives the signal. Detailed analysis of MITF phosphorylation would need to be performed to test this. Another possibility is that KITL and KIT signal to other phospho-acceptor sites in the MITF protein. Several other phospho-acceptor sites are known in the MITF protein, including Ser307, which has been shown to be phosphorylated by p38 upon RANKL/RANK signaling in osteoclasts (Mansky et al. 2002), and Ser298, which was shown to be phosphorylated by GSK3β (Takeda et al. 2000). In contrast to KIT signaling to MITF in melanocytes, the RANKL/RANK signaling to Ser307 of MITF in osteoclasts appears to be persistent and not transient (Mansky et al. 2002). Little is known about the biochemical consequences or in vivo significance of this signaling event, and currently it is not known if it is also active in melanocytes. Studies seem to indicate that Ser298 is crucial for normal DNA binding and transcription activation of the MITF protein and that this phosphorylation is an important regulatory event. At present it is not clear whether Ser298 is constitutively phosphorylated or regulated in a cell-specific or developmental manner or whether it is a bona fide GSK3β phospho-acceptor site (Frame and Cohen 2001). In addition to Ser307 and Ser298, additional, yet uncharacterized phosphorylation sites of functional importance may exist in the MITF protein. A third explanation for the differences observed between the in vivo and in vitro results might be that the in vitro assays used to determine protein stability and transcription activation (Wu et al. 2000; Xu et al. 2000) are more sensitive to subtle differences between the different mutations than the in vivo assay. This higher sensitivity would lead to significant differences in cell assays, whereas no such differences would be detected in the organism. The mice do not appear to be sensitive to Mitf dose as the phenotype of some of the mutant BAC-transgenic mice is normal although the Mitf gene is expressed at lower levels as compared to wild-type controls. In some cases, only ∼30% of the Mitf transcript is sufficient for a normal phenotype, and in Mitfmi-bws mutant mice the Mitf gene is expressed at only 9.5% level in heart as compared to wild-type controls. Reduced expression would presumably enhance the sensitivity of Mitf to mutations that affect the function of the gene. Finally, it is possible that the stability of the MITF protein is affected in the Ser73Ala and Ser409Ala BAC mutant mice such that the protein is more stable and therefore present for a longer time in the BAC mice than in wild-type mice. This would be consistent with the observation that the Ser73Ala; Ser409Ala double-mutant protein is more stable in cell culture than the wild-type protein (Wu et al. 2000; Xu et al. 2000). Despite being a poor transcription activator in vivo (due to the Ser73 and Ser409 mutations), a more stable protein might be able to rescue the phenotype as it persists for longer in the cells. If this were the case, one would not necessarily expect Kitl and Kit mutant mice to share a phenotype with Mitf mutations as the lack of signaling would presumably lead to a more stable MITF protein. This requires further studies on protein stability in the transgenic mice.
Our studies have also addressed the roles of several different exons in the Mitf gene. The lack of exon 1A from both BAC constructs used in these studies indicates that this exon is dispensable for Mitf function in melanocytes as well as during eye development. Like exon 1A, exons 2A and 2B seem to be dispensable for Mitf function. This is consistent with the normal phenotype of Ser73Ala mutant mice. The del(int2) BACs, together with the del(int1/2A) and del(2B/int2) mutations, suggest that exon 3 may play a more important role than exon 2. However, the role of exon 3 is currently not known.
In summary, our studies imply that signaling to Mitf may be more complex in vivo than the in vitro experiments would suggest. It may involve alternative phospho-acceptor sites or even alternative signaling mechanisms downstream of the Kit and Kitl signaling pathway. Further studies on this fascinating transcription factor are needed to characterize the role of signaling in melanocyte development.
Acknowledgments
We thank Pétur Henry Petersen, Benedikta St. Haflidadóttir, Merle Fleischer, Helga Eyja Hrafnkelsdóttir, Gunnhildur A. Traustadottir, Helgi Már Jónsson, and Gudrún Valdimarsdóttir for expert assistance. We also thank Gudmundur Ingólfsson for assistance with photography. This work was supported by grants from The Icelandic Research Fund and from the Science Fund of the University of Iceland and by an award from the Sigurdur Jónsson and Helga Sigurdardóttir Memorial Fund (E.S.). This work was supported in part by the intramural research program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke.
References
- Arnheiter, H., 2007. Mammalian paramutation: A tail's tale? Pigment Cell Res. 20: 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti, K., J. Debbache, J. Wang and H. Arnheiter, 2009. The basic helix-loop-helix leucine-zipper gene Mitf: analysis of alternative promoter choice and splicing in Methods in Molecular Biology: Transcription Factors, edited by Paul Higgins. Humana Press, Totowa, NJ (in press). [DOI] [PubMed]
- Bismuth, K., S. Skuntz, J. H. Hallsson, E. Pak, A. S. Dutra et al., 2008. An unstable targeted allele of the mouse Mitf gene with a high somatic and germline reversion rate. Genetics 178: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, P., L. Forrester, R. Rottapel, M. Reedijk, J. Fujita et al., 1991. The c-fms gene complements the mitogenic defect in mast cells derived from mutant W mice but not mi (microphthalmia) mice. Proc. Natl. Acad. Sci. USA 88: 2341–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, S., and P. Cohen, 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux et al., 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsson, J. H., B. S. Haflidadottir, A. Schepsky, H. Arnheiter and E. Steingrímsson, 2007. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res. 20: 185–200. [DOI] [PubMed] [Google Scholar]
- Hemesath, T. J., E. Steingrímsson, G. McGill, M. J. Hansen, J. Vaught et al., 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8: 2770–2780. [DOI] [PubMed] [Google Scholar]
- Hemesath, T. J., E. R. Price, C. Takemoto, T. Badalian and D. E. Fisher, 1998. MAP kinase links the transcription factor microphthalmia to c-Kit signalling in melanocytes. Nature 391: 298–301. [DOI] [PubMed] [Google Scholar]
- Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrímsson, N. G. Copeland et al., 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74: 395–404. [DOI] [PubMed] [Google Scholar]
- Hogan, B., R. Beddington, F. Costantini and E. Lacy, 1994. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hou, L., and W. J. Pavan, 2008. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res. 18: 1163–1176. [DOI] [PubMed] [Google Scholar]
- Jenkins, N. A., N. G. Copeland, B. A. Taylor and B. K. Lee, 1982. Organization, distribution and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 43: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, C., M. Khaled and D. E. Fisher, 2006. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12: 406–414. [DOI] [PubMed] [Google Scholar]
- Mansky, K. C., U. Sankar, J. Han and M. C. Ostrowski, 2002. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J. Biol. Chem. 277: 11077–11083. [DOI] [PubMed] [Google Scholar]
- Nishimura, E. K., S. R. Granter and D. E. Fisher, 2005. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307: 720–724. [DOI] [PubMed] [Google Scholar]
- Nolan, T., R. E. Hands, W. Ogunkolade and S. A. Bustin, 2006. SPUD: a quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal. Biochem. 351: 308–310. [DOI] [PubMed] [Google Scholar]
- Price, E. R., H. F. Ding, T. Badalian, S. Bhattacharya, C. Takemoto et al., 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J. Biol. Chem. 273: 17983–17986. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Shaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers., pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Sharan, S. K., A. Pyle, V. Coppola, J. Babus, S. Swaminathan et al., 2004. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142. [DOI] [PubMed] [Google Scholar]
- Steingrímsson, E., K. J. Moore, M. L. Lamoreux, A. R. Ferre-D'Amare, S. K. Burley et al., 1994. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 8: 256–263. [DOI] [PubMed] [Google Scholar]
- Steingrímsson, E., L. Tessarollo, B. Pathak, L. Hou, H. Arnheiter et al., 2002. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. USA 99: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson, E., N. G. Copeland and N. A. Jenkins, 2004. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38: 365–411. [DOI] [PubMed] [Google Scholar]
- Takeda, K., C. Takemoto, I. Kobayashi, A. Watanabe, Y. Nobukuni et al., 2000. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum. Mol. Genet. 9: 125–132. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy et al., 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming, S., N. Costantino, D. L. Court, N. A. Jenkins and N. G. Copeland, 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., T. J. Hemesath, C. M. Takemoto, M. A. Horstmann, A. G. Wells et al., 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14: 301–312. [PMC free article] [PubMed] [Google Scholar]
- Xu, W., L. Gong, M. M. Haddad, O. Bischof, J. Campisi et al., 2000. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp. Cell Res. 255: 135–143. [DOI] [PubMed] [Google Scholar]
- Yang, Y., and S. K. Sharan, 2003. A simple two-step, ‘hit and fix’ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 31: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]