Abstract
Lepidopteran wing scales and Drosophila bristles are considered homologous structures on the basis of the similarities in their cell lineages. However, the molecular mechanisms underlying scale development are essentially unknown as analysis of gene function in Lepidoptera is sorely limited. In this study, we used the Bombyx mori mutant scaleless (sl), which displays a nearly complete loss of wing scales, to explore the mechanism of lepidopteran wing-scale formation. We found that Bm-ASH2, one of four Bombyx achaete-scute homologs, is highly expressed in early pupal wings of wild-type silkworms, but its expression is severely reduced in sl pupal wings. Through molecular characterization of the mutant locus using luciferase and gel shift assays, genetic analysis of recombining populations, and in vivo rescue experiments, we provide evidence that a 26-bp deletion within the Bm-ASH2 promoter is closely linked to the sl locus and leads to loss of Bm-ASH2 expression and the scaleless-wings phenotype. Thus, the Bm-ASH2 appears to play a critical role in scale formation in B. mori. This finding supports the proposed homology of lepidopteran scales and dipteran bristles and provides evidence for conservation of the genetic pathway in scale/bristle development at the level of gene function.
THE wing surface of lepidopteran adults is covered by wing scales, which function in heat preservation, mimesis, touch, etc. (Nijhout 1991). Lepidopteran scales and Drosophila bristles are considered homologous structures on the basis of the similarities in their cell lineages and the expression of a few molecular markers (Overton 1966, 1967; Galant et al. 1998). Development of the Drosophila bristle is regulated by the basic helix-loop-helix (bHLH) transcription factors of the Achaete-Scute Complex (AS-C). Within the clusters of cells expressing achaete (ac) and scute (sc), some are selected to become sensory organ precursors, which then form and innervate the mature bristles (Skeath and Carroll 1991; Jan and Jan 1994). In ac/sc double-mutant flies, the majority of the bristles are lost; on the contrary, ectopic expression of ac and/or sc can induce extra bristles (Garcia-Bellido 1979; Campuzano et al. 1986; Balcells et al. 1988; Rodriguez et al. 1990). In the butterfly Precis coenia, the AS-C homolog B-ASH1 is indeed expressed in scale-forming cells during pupation (Galant et al. 1998), and as we reported, the four AS-C homologs of the silkmoth Bombyx mori are also expressed during wing development, particularly at the early pupal stage when scale formation begins (Zhou et al. 2008). By studying the genomic organization and the evolution of ac/sc genes of multiple distant insect species, Negre and Simpson (2009) have recently argued that the independent evolution of the homologs might have contributed to morphological diversity in Diptera and Lepidoptera.
We show here that a small deletion in the Bm-ASH2 gene underlies the scaleless (sl) mutant phenotype in B. mori. As previously described, this mutant displays a severe reduction in wing scales (Zhou et al. 2004, 2006). We find that Bm-ASH2 expression is strongly reduced in the sl pupal wing. At the molecular level, we identify a 26-bp deletion within the promoter region of Bm-ASH2, which is linked to the mutant phenotype. Using an electrophoresis mobility shift assay, we show that the 26-bp region contains a cis-regulatory element. Finally, we show that targeted expression of Bm-ASH2 in the sl pupal wing can partially rescue the lack of wing scales, confirming the central role of Bm-ASH2 downregulation in the etiology of this mutant. Thus, at least one ASH factor is fundamentally involved in scale formation in B. mori. This finding supports the proposed homology of lepidopteran scales and Drosophila bristles and provides evidence for conservation of the genetic pathway in the formation of these structures.
MATERIALS AND METHODS
Animals:
Silkworms used in the experiments were the wild-type strains 7532 and Furong and the transparent wings mutant strain scaleless (Zhou et al. 2004). The larvae were cultured on mulberry leaves under 25° with 70–80% relative humidity.
In situ hybridization:
DIG-labeled RNA probes were generated by in vitro transcription, using the ORF region of a gene as the template. One-day-old pupal wings were dissected in cold 0.75% NaCl and fixed in fresh 4% formaldehyde in 100 mm HEPES (pH 7.9), 2 mm MgCl2, and 1 mm EDTA for 2–3 hr at room temperature or overnight at 4°. The tissues were washed for 3× for 5 min each time in PBST0.2 (0.2% Tween20) and then digested with Proteinase K (20 μg/ml) for 5 min, followed by treatment with 0.2% glycine in PBS. The wings were refixed in 4% polyformaldehyde for 30 min and washed 2× for 5 min each time in PBST0.2. Prehybridization was processed in hybridization solution (50% formamide, 5× SSC, 2% blocking powder, 10 mg/ml yeast tRNA, 5 mg/ml salmon sperm DNA, and 20 mg/ml heparin) for 2 hr. An antisense RNA probe (100–500 ng) was used for each experiment, and hybridization was done overnight. Then the tissues were washed 4× with PBST0.2, for 10, 20, 30, and 30 min, respectively. Subsequently, they were dipped in 5% goat serum and 2% BSA in PBST0.2 for 2 hr and in anti-DIG-AP Fab fragment for 2 hr. The samples were washed 3× for 10 min each time in PBST0.2 and stained with NBT (nitro blue tetrazolium chloride)/BCIP (5-bromo-4-chloro-3-indolyl phosphate, toluidine salt) in the dark until a signal appeared. The issues were dehydrated successively with 30%, 50%, 70%, and 100% methanol for 15 min each, followed by a successive 1-hr treatment with 50% and 80% glycerol. Sense-strand RNA was used as the control for each experiment.
Cell culture and dual-luciferase reporter assay:
Cell culture and dual-luciferase reporter assay were processed as described (Zhou et al. 2008). One pupal wing was dissected and lysed with 50 μl of the passive lysis buffer, and 10 μl of the lysate was used for the dual-luciferase reporter assay according to the manufacturer's protocol (Dual-luciferase Reporter Assay System, Promega). Luciferase activity was determined with a 20/20n Luminometer (Turner BioSystems).
Electrophoretic mobility shift assay:
Probes for the electrophoretic mobility shift assay (EMSA) were prepared by annealing two partially overlapping oligos (supporting information, Table S1), synthesized to have two sticky ends to introduce dATP by filling the gaps. The probes were labeled with [α-32P]dATP. Nuclear extracts were prepared from 1-day-old pupal wings as described (Feng et al. 1998; Blough et al. 1999), and the protein concentration was determined by the Bradford Assay (Bradford 1976). Five micrograms of nuclear proteins were incubated with a probe at a concentration of 104 cpm in 0.04 pmol DNA in a final volume of 20 μl containing 50 mm HEPES–KOH (pH 7.9), 250 mm KCl, 20 mm MgCl2, 5 mm DTT, 5 mm EDTA, 0.5 mg/ml BSA, 0.25 μg/μl Poly (deoxyinosinic-deoxycytidylic) (Sigma), and 50% glycerol. After incubation for 30 min at 25°, the binding reactions were analyzed on 7% polyacrylamide gels. The gels were dried and exposed to a phosphoscreen (Bio-Rad) for 3 hr. For competition analysis, 100-fold excess of unlabeled double-stranded oligos was used.
Genomic DNA extraction from silkmoth:
The head and thorax of a moth was cut and homogenized in 2 ml solution A [0.25 m sucrose, 10 mm EDTA, 30 mm Tris–HCl (pH 7.5)] on ice. After centrifugation for 3 min at 1500 × g, the supernate was discarded and the pellet was resuspended in 600 μl solution B [10 mm Tris–HCl (pH 7.5), 10 mm EDTA, 0.15 m NaCl, 1% sarcosyl] and incubated on ice for 15 min. The sample was treated successively with 1 vol phenol/chloroform to denature the proteins. Genomic DNA was precipitated with 2 vol ethanol, and the pellet was washed with 70% ethanol, dried, and dissolved in TE with 20 μg/ml RnaseA and stored at −20° for further use.
Ectopic expression of a foreign gene in the silkworm pupal wing by a transient expression system:
The ORF region of a candidate gene was cloned into a modified pBacPAK8 vector with an ie-1 promoter and a hr3 enhancer (Chen et al. 2004). Ten microliters of the recombinant plasmid (5 μg) with 5 μl of lipofectin were micro-injected into the pupal wing. Injection of 10 μl of double-distilled water with 5 μl of lipofectin was used as the control.
RESULTS AND DISCUSSION
Expression of Bm-ASH2 is severely reduced in scaleless pupal wings:
The scaleless mutant loses nearly all wing scales. Given that AS-C genes play critical roles in the development of Drosophila bristles and that AS-C homologs (ASH) are expressed in the developing wings of butterflies and silkmoths (Jan and Jan 1994; Galant et al. 1998; Zhou et al. 2008), we decided to investigate the potential involvement of these bHLH-type of transcription factors in the etiology of this mutant. We used semiquantitative RT–PCR to assess the expression level of Bm-ASH genes in wild-type and sl wings. The levels of Bm-ASH1, Bm-ASH3, and Bm-ase in sl pupal wings were similar to those from two different wild-type lines, Furong and 7532. On the contrary, the Bm-ASH2 level was severely reduced in sl compared to wild type (Zhou et al. 2006; data not shown).
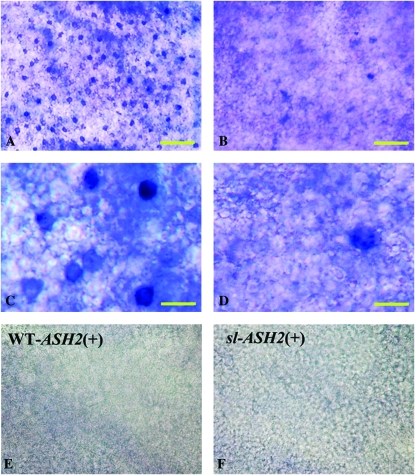
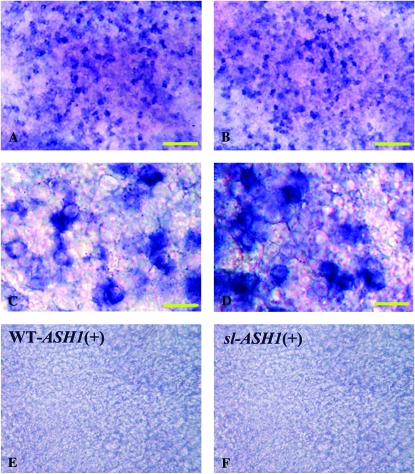
We further used in situ hybridization to confirm these findings and to investigate the pattern of Bm-ASH2 expression. In the wild type, Bm-ASH2 was robustly expressed across 1-day-old pupal wings in regularly spaced clusters of scale mother cells (Figure 1, A and C). Consistent with the reduction in mRNA levels detected by RT–PCR, only a few cells expressing Bm-ASH2 were detected on the sl wing surface (Figure 1, B and D). By contrast, and in agreement with the RT–PCR data, clusters of Bm-ASH1-expressing cells were present across the mutant wings in a pattern essentially indistinguishable from that of wild type (Figure 2, A–D).
Figure 1.—
The expression pattern of Bm-ASH2 is abnormal in the scaleless pupal wing. Cells expressing Bm-ASH2 are regularly located on the wing surface of wild-type pupa (A and C, blue dots). By contrast, in sl pupa, only a very few cells were detected expressing Bm-ASH2 (B and D). C and D are enlargements of a partial area in A and B. No staining was observed in the sense-strand control for wild type [(E)WT-ASH2 (+)] or scaleless [(F)sl-ASH2 (+)]. Distal is to the top. Bars, 100 μm in A, B, E, and F, and 20 μm in C and D.
Figure 2.—
The expression patterns of Bm-ASH1 in wild-type and scaleless pupal wings are similar. There are no significant differences in Bm-ASH1 expression patterns between wild type (A and C, blue dots) and sl (B and D). Unlike those expressing Bm-ASH2, cells expressing Bm-ASH1 form special clusters but are not arrayed singly on the wing surface. C and D are enlargements of a partial area in A and B. No staining was observed in the sense-strand control for wild type [(E)WT-ASH1 (+)] or scaleless [(F)sl-ASH1 (+)]. Distal is to the top. Bars, 100 μm in A, B, E, and F, and 20 μm in C and D.
The scaleless wings phenotype is caused by a single locus and correlates with the presence of a 26-bp deletion in the Bm-ASH2 genomic region:
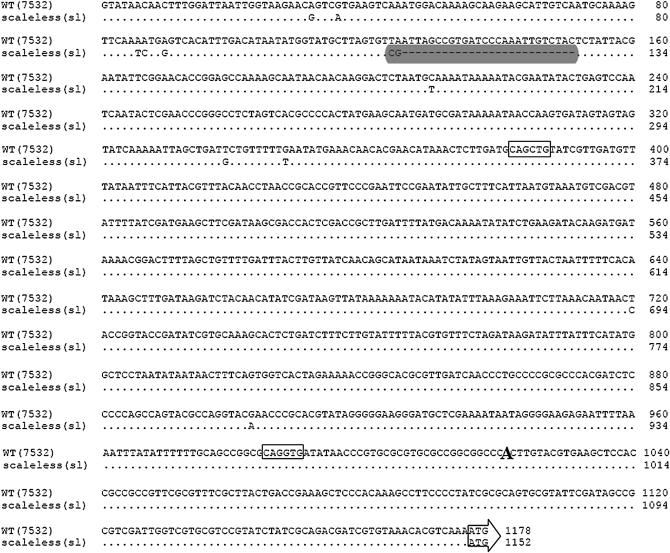
To understand the defect leading to loss of Bm-ASH2 expression in the sl, we decided to characterize the locus at the molecular level. Although in situ analysis (Figure 1) suggested a defect in transcriptional regulation, we nonetheless sequenced the entire Bm-ASH2 coding region. Unsurprisingly, sequences from the mutant and several wild-type strains had no significant differences (data not shown). Since transcription is often controlled through the chromosomal region flanking a gene on the 5′ side, we decided to analyze 1175 bp of genomic DNA just upstream of the Bm-ASH2 translation start site. On the basis of in silico prediction, this fragment spans ∼1010 bp upstream of the start of transcription (Figure 3). A number of isolated single-base-pair changes were identified throughout the region. However, the most striking sequence alteration occurred ∼850 bp upstream of the putative transcription start site, where a 28-bp sequence present in the wild type was replaced by only 2 bp in the sl (Figure 3).
Figure 3.—
Comparison of Bm-ASH2 promoter sequences between scaleless and wild type. There are 11 transversions and one transition in the sl Bm-ASH2 promoter sequence compared to the wild-type one. More importantly, a 26-bp fragment is absent in sl (shaded area). Two deduced E-boxes are boxed. The enlarged “A” at 1021 is the predicted transcription start site.
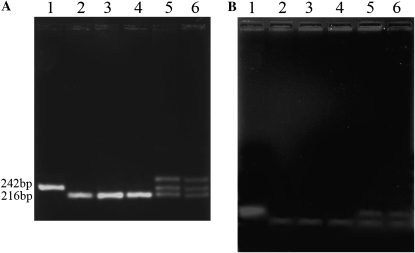
We used this deletion as a molecular marker to investigate whether the sl phenotype segregated with this Bm-ASH2 mutant allele. Two primers flanking the region deleted in sl (Table S1) were used to amplify a 242-bp fragment from the wild-type genomic DNA (Figure 4, lane 1; Figure S1, box 1) and a 216-bp fragment from sl (Figure 4, lane 2; Figure S1, box 1). To ensure that our findings are reasonable, we studied individuals with multiple different genetic backgrounds. In the series of crosses shown in Figure S1, individuals were scored for their wing phenotype and then selected for PCR testing. In all cases, phenotypically sl individuals carried the deletion allele but not the wild-type allele (Figure 4, lanes 3 and 4; Figure S1, boxes 2 and 3). On the contrary, phenotypically wild-type individuals carried either two copies of the wild-type DNA or one copy of the wild type and one copy of the deletion alleles (Figure 4, lane 5; Figure S1, box 3; data not shown).
Figure 4.—
The scaleless wings mutant phenotype is tightly linked to the absence of the 26-bp element in the Bm-ASH2 promoter. PCR was carried out with primers Cash2pF and Cash2pR (Table S1). (A) Native agarose gel electrophoresis. (B) Denaturing alkaline agarose gel electrophoresis. DNA in each lane was from individuals with a specific genotype, and the cross strategies are shown in Figure S1. As expected, a 242-bp fragment was amplified from wild type (7532, lane 1), and a 216-bp fragment was amplified from sl (lane 2). All of the individuals with a mutant phenotype have a sl genetype (lanes 3 and 4). Both 242- and 216-bp fragments were amplified from heterozygous genotype individuals (lanes 5 and 6). In lanes 5 and 6 in A, an extra band moved more slowly than 242 bp. We supposed that this band was produced by the annealing of a 242- and a 216-nt single-stranded DNA. These fragments contained a small loop and had slower migrating speeds in the natural electrophoresis conditions. Consistent with the supposition, there were only two bands under denaturing electrophoresis (B, lanes 5 and 6).
Because the original strain in which the mutant was discovered was unknown, a more extensive analysis of linkage was carried out from a cross of sl to another wild-type strain, Furong (Figure 4, lane 6; Figure S1, box 4). Heterozygous F1 progeny were intercrossed to generate a mix of phenotypically sl and wild-type F2 progeny. Among 581 F2 adults, the proportion of phenotypically sl to wild-type individuals was ∼1:3 (Table 1, χ2 = 0.014, 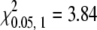 ). We randomly picked some of the moths for molecular analysis. All 40 F2 silkmoths with sl wings carried the Bm-ASH2 deletion allele exclusively. Among 112 F2 individuals with a wild-type phenotype, 34 carried the homozygous wild-type genotype, and the other 78 individuals were heterozygous (Table 1). The ratio of homozygous to heterozygous individuals approximated the predicted 1:2 ratio expected for recessive inheritance at a single locus in a hybrid cross (χ2 = 0.322,
). We randomly picked some of the moths for molecular analysis. All 40 F2 silkmoths with sl wings carried the Bm-ASH2 deletion allele exclusively. Among 112 F2 individuals with a wild-type phenotype, 34 carried the homozygous wild-type genotype, and the other 78 individuals were heterozygous (Table 1). The ratio of homozygous to heterozygous individuals approximated the predicted 1:2 ratio expected for recessive inheritance at a single locus in a hybrid cross (χ2 = 0.322, 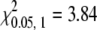 ). The results are consistent with a single gene hypothesis, i.e., that the sl phenotype is caused by a recessive allele at a single locus.
). The results are consistent with a single gene hypothesis, i.e., that the sl phenotype is caused by a recessive allele at a single locus.
TABLE 1.
Genetic analysis of scaleless phenotype and genotype
| Wild type
|
||||
|---|---|---|---|---|
| SS | Ss | sl (ss) | Rate (modified) | |
| Phenotype | 434 | 147 | 3:1 | |
| Genotype | 34 | 78 | 40 | 1:2 (SS:Ss) |
By chi-square test, 434:147 = 3:1 (χ2= 0.014, 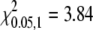 ), 34:78 = 1:2 (χ2 = 0.322,
), 34:78 = 1:2 (χ2 = 0.322, 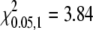 ). “SS” and “Ss” separately indicate the homozygous and heterozygous wild-type genotype, and “ss” indicates the homozygous mutant genotype. The data are from the F2 intercross between F1's produced with the Furong (wild type) and sl strains.
). “SS” and “Ss” separately indicate the homozygous and heterozygous wild-type genotype, and “ss” indicates the homozygous mutant genotype. The data are from the F2 intercross between F1's produced with the Furong (wild type) and sl strains.
These data are consistent with the simple inheritance of sl as a recessive trait and support a close linkage between the 26-bp deletion and the mutant locus. However, this evidence is not sufficient to establish that Bm-ASH2 is the locus responsible for the sl phenotype. Therefore, we proceeded to investigate: (1) whether the 26-bp sequence could be involved in regulating Bm-ASH2 transcription and (2) whether restoring the expression of Bm-ASH2 in the developing wing could rescue the mutant defect.
The 26-bp region contains sequences that contribute to normal transcriptional regulation of Bm-ASH2:
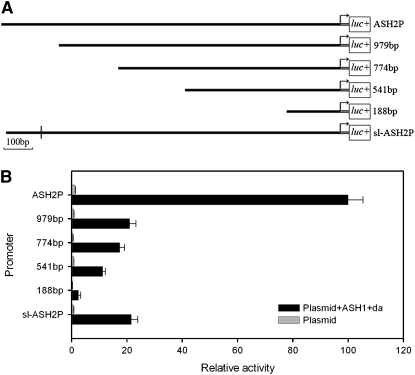
To test whether the 26-bp region was involved in transcriptional regulation, we relied on transient expression assays in cultured cells. We had previously shown that the 1175-bp DNA fragment upstream of the Bm-ASH2 translation start site (Figure 5) is sufficient to drive robust luciferase expression in the presence of the putative upstream regulators Bm-ASH1 and Daughterless and that two E-boxes at positions 194 and 797 likely mediate this regulation (Zhou et al. 2008). As expected, reporters bearing 5′ truncations that eliminate either the 797 E-box or both 797 and 194 E-boxes displayed much lower transcriptional activity, <20% of the full-length ASH2P. However, expression did not rely solely on these regulatory elements because a 979-bp reporter that retained both E-boxes and lacked only 169 bp at the 5′-end was also severely downregulated and expressed at only ∼20% of the ASH2P. Interestingly, this truncated 979 bp lacked the 26-bp region deleted in the sl.
Figure 5.—
Comparison of Bm-ASH2 promoter activities between scaleless and wild type. The transcriptional activity of sl-ASH2P was significantly lower than that of ASH2P (P < 0.01) and similar to that of 979 bp (P > 0.05). ASH2P, the full-length fragment of wild-type Bm-ASH2 promoter; 979 bp, +3 to −976 fragment; 774 bp, +3 to −771 fragment; 541 bp, +3 to −538 fragment; 188 bp, +3 to −185 fragment; sl-ASH2P, the full-length fragment of the Bm-ASH2 promoter in sl. All of the promoter fragments were cloned into the luciferase reporter vector pGL3-Basic. We designated the first base of the start codon as +1, and all ATGs were changed to ATTs for these experiments. “|” in sl-ASH2P shows the missing 26-bp element (A). (B) “Plasmid” shows that only the recombined pGL3-Basic vector containing a particular promoter fragment was transfected into BmN cells, and “Plasmid+ASH1+da” indicates cotransfection of the recombined plasmid together with the transient expression plasmids containing Bm-ASH1 and Dm-da. At least three independent repeats were carried out for each treatment.
To investigate whether the lower activity of the 979 bp was due to loss of the 26-bp region (Figure 3, shaded), we assayed the expression of a “full-length ASH2P” reporter generated from the sl genomic DNA (sl-ASH2P) and thus lacking the 26-bp sequence. Notably, the activity of sl-ASH2P was significantly lower than that of wild-type ASH2P and similar to the 979 bp.
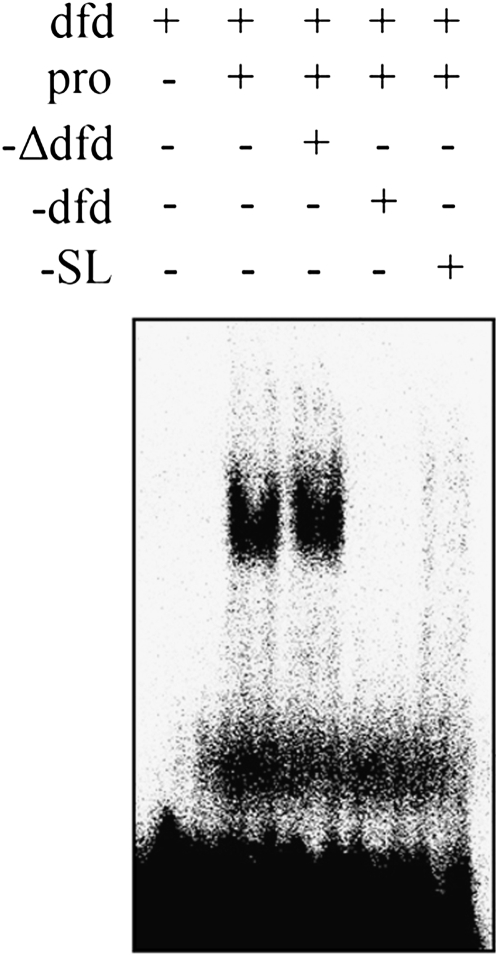
On the basis of these findings, we hypothesized that the 26-bp region contains one or more cis-regulatory elements required for normal expression of Bm-ASH2. Consistent with this proposal, an activator present in nuclear protein extracts from 1-day-old pupal wings bound 49-bp double-strand oligos that contained the 26-bp sequence in EMSA. Binding was competed by either cold 49-bp DNA or a smaller DNA spanning the 26-bp sequence, whereas it was unaffected by a cold 23-bp DNA that excluded the 26 bp (Figure 6). These findings, together with the loss of Bm-ASH2 expression in sl tissue, strongly support a role for the 26-bp region in transcriptional regulation of Bm-ASH2.
Figure 6.—
Fragments containing the 26-bp element can be bound by nuclear protein extracts from a 1-day-old pupal wing in vitro. “+” indicates that the component was included, and “−”indicates that the component was not included in binding reactions. pro, nuclear proteins; dfd, a wild-type 49-bp double-strand DNA containing the 26-bp fragment (“SL”); and Δdfd, the sl 23-bp fragment corresponding to dfd. Minus signs in these terms indicate unlabeled probes.
Ectopic expression of Bm-ASH2 in the pupal wing can rescue the scaleless wing phenotype:
The findings above strongly support a model whereby decreased expression of Bm-ASH2 results in loss of scales in the sl. To test this hypothesis, we attempted to rescue the mutant phenotype by providing exogenous Bm-ASH2 in the wing.
To achieve this goal, we developed a novel method for transient gene expression in vivo. Previous studies have shown that B. mori baculovirus promoters are functional in expression vectors and display expected transcriptional activity when injected into silkworm larvae (Zhou et al. 2002; Chen et al. 2004; Tang et al. 2005). To assess the efficiency of this method in the pupal wing, we generated a constitutive reporter encoding the luciferase gene under the control of the previously described ie-1 promoter and hr3 enhancer (pBacPAK8-ie-1-luc-hr3; Chen et al. 2004). The reporter plasmid was micro-injected into the left forewings of 12-hr-old pupae, and the wings were dissected 36 hr later, lysed, and tested for luciferase activity. Among a total of 14 individuals tested, only 2 (14%) lacked luciferase activity. In the positive samples (n = 12), luciferase activity ranged from 1500 relative light units (RLU; 10 or 71.4%) to >10,000 RLU (3 or 21.4%), the highest reaching 248,064 RLU (Table 2).
TABLE 2.
Luciferase activity after injection of the pBacPAK8-ie-1-luc-hr3 plasmid into the pupal wing
| Luminescence (RLU)
|
|||
|---|---|---|---|
| No. | Left wing | Right wing | Remainder of the right subtracted from the left wings |
| 1 | 248,145 | 81 | 248,064 |
| 2 | 12,017 | 90 | 11,927 |
| 3 | 10,883 | 89 | 10,794 |
| 4 | 8,896 | 93 | 8,803 |
| 5 | 8,439 | 96 | 8,343 |
| 6 | 7,120 | 80 | 7,040 |
| 7 | 4,042 | 76 | 3,966 |
| 8 | 3,054 | 84 | 2,970 |
| 9 | 2,102 | 79 | 2,023 |
| 10 | 1,851 | 88 | 1,763 |
| 11 | 517 | 78 | 439 |
| 12 | 100 | 78 | 22 |
| 13 | 89 | 94 | −5 |
| 14 | 79 | 72 | 7 |
Ten microliters of pBacPAK8-ie-1-luc-hr3 plasmid (5 μg) containing the luc+ gene was micro-injected into the left forewing of 12-hr-old pupa. Thirty-six hours later, the pupal wing was dissected out and lysed with 50 μl of the passive lysis buffer, and then 10 μg of lysate protein was used for the dual-luciferase reporter assay. The right forewing was injected with 10 μl of same dosage lipofectin in double-distilled water as the control.
Having validated this method for the transient induction of exogenous gene expression in the wing, an expression plasmid encoding the Bm-ASH2 was micro-injected into the left forewing of 12-hr sl pupae. Of 50 pupae treated in this fashion, 37 developed into moths, but 16 were excluded because their left forewings were folded and difficult to score. The remaining 21 moths were evaluated for presence/absence of scales. Nine (42.9%) displayed the sl phenotype, whereas 12 (57.1%) displayed significant rescue. In these 12 individuals, the number of scales (as assessed by the presence of scale sockets) on the left forewing was significantly higher than the number of scales on the untreated right one (t-test, P < 0.01; Table 3; Figure 7). Controls subjected to mock injections (33 sl moths) had wings with scale socket totals indistinguishable between treated and untreated sides (P > 0.05; Table 3).
TABLE 3.
Wing-socket cells produced in silkmoths injected with a plasmid (pBacPAK8-ie-1-ASH2-hr3) containing Bm-ASH2
| Sort | Mean of socket cell no./mm2 | Standard deviation | T grouping |
|---|---|---|---|
| Wild type | 462 | 31 | A |
| wild type-left | 464 | 22 | A |
| wild type-right | 464 | 31 | A |
| scaleless | 51 | 8 | B |
| scaleless-CK | 49 | 17 | B |
| scaleless-left | 374 | 134 | C |
| scaleless-right | 53 | 12 | B |
“Wild type” is a wild-type silkworm breed (7532); “wild type-left,” refers to the left wing, and “wild type-right” to the right wing of the wild-type silkmoth whose left wing was injected with pBacPAK8-ie-1-ASH2-hr3; “scaleless,” the scaleless wing mutant; “scaleless-CK,” the scaleless wing injected with the empty vector pBacPAK8-ie-1-hr3; “scaleless-left,” the left wing; and “scaleless-right,” the right wing of the scaleless silkmoth whose left wing was injected with the pBacPAK8-ie-1-ASH2-hr3 plasmid. There are no significant differences among wild type, wild type-left, and wild type-right, and also no significant differences among scaleless, scaleless-CK, and scaleless-right. However, the number of cells per square millimeter of wing portion of scaleless-left was significantly greater than those of scaleless, scaleless-CK, and scaleless-right, although the standard deviation was much larger for scaleless-left. Different letters in the “T grouping” column indicate a statistically significant difference (ANOVA, P < 0.01). At least 10 wings were used for each treatment.
Figure 7.—
Wing scales on the scaleless left forewing injected with the Bm-ASH2 expression plasmid are increased significantly. An expression plasmid containing the Bm-ASH2 gene was injected into the left forewing at the early pupal stage. (A) The enlargement of a partial left wing of the moth. (C) The enlargement of the same area of the right wing. The presence of more wing scales on the left forewing than on the right forewing is evident (B). Bars, 100 μm in A and C.
Interestingly, expression of exogenous Bm-ASH1 did not rescue the mutant phenotype, indicating that loss of scales in the mutant was not due to a reduction of AS-C-type factors in general, but to the specific loss of Bm-ASH2 protein. We cannot exclude, however, the possibility that Bm-ASH2 cooperates with Bm-ASH1 in the selection of scale mother cells. Moreover, we did not observe formation of supernumerary scales in wild-type wings injected with either Bm-ASH2 or Bm-ASH1 expression vectors (Table 3 and Table 4). This is in contrast to the induction of ectopic bristles in Drosophila by targeted expression of ac/sc (Rodriguez et al. 1990). This may be due to methodological differences in the introduction of exogenous gene expression, or, alternatively, it may reflect a significant difference in the genetic control of scale vs. bristle development. Nonetheless, that transient expression of exogenous Bm-ASH2 was sufficient to rescue the loss of wing scales in the mutant supports the identity of the sl locus and the Bm-ASH2 gene.
TABLE 4.
Wing-socket cells produced in silkmoths injected with a plasmid (pBacPAK8-ie-1-ASH1-hr3) containing Bm-ASH1
| Sort | Mean of socket cell no./mm2 | Standard deviation | T grouping |
|---|---|---|---|
| Wild type | 462 | 31 | A |
| Wild type-left | 457 | 27 | A |
| Wild type-right | 460 | 41 | A |
| scaleless | 51 | 8 | B |
| scaleless-left | 55 | 19 | B |
| scaleless-right | 50 | 39 | B |
Column headings are the same as described in Table 3. There are no significant differences among wild type, wild type-left, and wild type-right, and no significant differences among scaleless, scaleless-left, and scaleless-right. Different letters in the “T grouping” column indicate a statistically significant difference (ANOVA, P < 0.01). At least 10 wings were used for each treatment.
Conclusions
The proneural gene Bm-ASH2 plays a key role in the formation of silkmoth wing scales:
Formation of dipteran bristles and lepidopteran scales is controlled mainly by achaete-scute genes and homologs (Galant et al. 1998). Independent duplication of achaete-scute homologs between different insect lineages may have contributed to morphological diversity in insects (Negre and Simpson 2009). During evolution, the emergence of new phenotypes may have been due to changes in the spatial/temporal expression of particular genes, usually caused by mutations in relative cis-regulatory elements and/or in trans-acting factors (Mannervik et al. 1999; Bonifer 2000). Generally, mutations in cis-regulatory elements occur more frequently (Stern 2000), and have been shown at genetic and molecular levels (Belting et al. 1998; Stern 1998; Sucena and Stern 2000; Gompel et al. 2005; Wittkopp 2006).
The silkworm scaleless mutant exemplifies a potential molecular mechanism for “morphological divergence” resulting from an alteration of gene expression (Figure 1, B and D). As shown by analysis of cultured cells and gel shift assays, the change in wing phenotype is most likely due to the loss of an essential cis-regulatory site in the control region of the proneural gene Bm-ASH2 (Figures 5 and 6). This hypothesis is strengthened by the genetic analysis of the mutant (Table 1; Figure 4). Indeed, the sl trait displays a “single-gene recessive” inheritance pattern and shows genetic linkage to chromosome 13 (J. Qin, personal communication) on which the Bm-ASH2 gene is located (SilkDB).
To test this hypothesis, we chose a gain-of-function approach. Foreign gene expression in B. mori has been achieved by transgenesis (Nikolaev et al. 1993; Zhang et al. 1999; Uhlirova et al. 2002; Imamura et al. 2003; Uchino et al. 2007) or by using modified baculovirus (Mori et al. 1995; Guo et al. 2005; Xia et al. 2006). Here, we have extended our earlier method to induce transient expression of a foreign gene in silkworm tissues. Using this method, we were able to drive Bm-ASH2 gene expression in cells of the sl pupal wing and significantly rescue the mutant phenotype (Figure 7; Table 3). These findings support our hypothesis that loss of expression of the Bm-ASH2 underlies the sl phenotype and that this bHLH factor plays a critical role in the formation of silkworm wing scales.
Developmental mechanisms for the formation of insect wing scales and bristles are most likely conserved:
The formation of fly notum bristles and butterfly wing scales is controlled by the expression of AS-C family genes (Skeath and Carroll 1991; Galant et al. 1998). There are two types of bristles on the thorax of Drosophila: macrochaetes and microchaetes. Development of both kinds of bristles is controlled mainly by the proneural genes ac and sc. Lower Diptera have only one or two AS-C-type proneural genes, with the single one or the scute-like one being responsible for bristle formation (Wülbeck and Simpson 2000, 2002; Pistillo et al. 2002). In the butterfly Precis coenia, only one ac/sc homolog, B-ASH1, has been identified, and it is indeed expressed in a pattern that is consistent with a role in wing-scale formation (Galant et al. 1998). Largely on the basis of this observation, it has been suggested that lepidopteran wing scales and dipteran sensory bristles are homologous structures. We have recently reported that four AS-C homologs are found in another lepidopteran insect, the silkworm B. mori, and that these genes were all broadly expressed in different tissues (Zhou et al. 2008). In particular, all four genes are expressed in early pupal wings. Notably, Bm-ASH2 protein, which in this study is found to be important in the formation of silkworm wing scales, is highly conserved with the B-ASH1 protein of P. coenia, and the two proteins bear identical bHLH domains (Zhou et al. 2008). In this work, we show that loss of Bm-ASH2 gene expression results in the scaleless mutant defect. This finding directly supports the proposed evolutionary relationship between the scales of Lepidoptera and the bristles of Diptera.
Acknowledgments
We thank F. Pignoni, T. Zhang, and D. Yang-Zhou for helpful discussion of the data and critical reading of the manuscript and H. Bellen for the Drosophila daughterless cDNA. This work was supported by grants from the National Natural Sciences Foundation of China (30770279) and the National High Technology Research and Development Program of China (“863” Project no. 2006AA10A119).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.102848/DC1.
References
- Balcells, L., J. Modolell and M. Ruiz-Gómez, 1988. A unitary basis for different Hairy-wing mutations of Drosophila melanogaster. EMBO J. 7: 3899–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belting, H. G., C. S. Shashikant and F. H. Ruddle, 1998. Modification of expression and cis-regulation of Hoxc8 in the evolution of diverged axial morphology. Proc. Natl. Acad. Sci. USA 95: 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blough, E., B. Dineen and K. Esser, 1999. Extraction of nuclear proteins from striated muscle tissue. BioTechniques 26: 202–204, 206. [DOI] [PubMed] [Google Scholar]
- Bonifer, C., 2000. Developmental regulation of eukaryotic gene loci: Which cis-regulatory information is required? Trends Genet. 16: 310–315. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 7: 248–254. [DOI] [PubMed] [Google Scholar]
- Campuzano, S., L. Balcells, R. Villares, L. Carramolino, L. García-Alonso et al., 1986. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell 44: 303–312. [DOI] [PubMed] [Google Scholar]
- Chen, Y., B. Yao, Z. Zhu, Y. Yi, X. Lin et al., 2004. A constitutive super-enhancer: homologous region 3 of Bombyx mori nucleopolyhedrovirus. Biochem. Biophys. Res. Commun. 318: 1039–1044. [DOI] [PubMed] [Google Scholar]
- Feng, Z., A. Z. Wu, Z. Zhang and C. C. Chen, 1998. GATA-1 and GATA-4 transactivate inhibin/activin b-B-subunit gene transcription in testicular cells. Mol. Endocrinol. 14: 1820–1835. [DOI] [PubMed] [Google Scholar]
- Galant, R., J. B. Skeath, S. Paddock, D. L. Lewis and S. B. Carroll, 1998. Expression pattern of a butterfly achaete-scute homolog reveals the homology of butterfly wing scales and insect sensory bristles. Curr. Biol. 8: 807–813. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido, A., 1979. Genetic analysis of the achaete-scute system of Drosophila melanogaster. Genetics 91: 491–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel, N., B. Prudhomme, P. J. Wittkopp, V. A. Kassner and S. B. Carroll, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Guo, T., J. Wang, X. Guo, S. Wang and C. Lu, 2005. Transient in vivo gene delivery to the silkworm Bombyx mori by EGT-null recombinant AcNPV using EGFP as a reporter. Arch. Virol. 150: 93–105. [DOI] [PubMed] [Google Scholar]
- Imamura, M., J. Nakai, S. Inoue, G. Quan, T. Kanda et al., 2003. Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics 165: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, Y. N., and L. Y. Jan, 1994. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu. Rev. Genet. 28: 373–393. [DOI] [PubMed] [Google Scholar]
- Mannervik, M., Y. Nibu, H. Zhang and M. Levine, 1999. Transcriptional coregulators in development. Science 284: 606–609. [DOI] [PubMed] [Google Scholar]
- Mori, H., M. Yamao, H. Nakazawa, Y. Sugahara, N. Shirai et al., 1995. Transovarian transmission of a foreign gene in the silkworm, Bombyx mori, by Autographa californica nuclear polyhedrosis virus. Biotechnology 13: 1005–1007. [DOI] [PubMed] [Google Scholar]
- Negre, B., and P. Simpson, 2009. Evolution of the achaete-scute complex in insects: convergent duplication of proneural genes. Trends Genet. 25: 147–152. [DOI] [PubMed] [Google Scholar]
- Nijhout, H. F., 1991. The Development and Evolution of Butterfly Wing Patterns. Random House (Smithsonian Institution Press), Washington, D.C./London.
- Nikolaev, A., T. Tchkonia, C. Kafiani-Eristavi and V. Tarantul, 1993. Preferential extrachromosomal localization of exogenous DNA in transgenic silkworm Bombyx mori L. Mol. Gen. Genet. 236: 326–330. [DOI] [PubMed] [Google Scholar]
- Overton, J., 1966. Microtubules and microfibrils in morphogenesis of the scale cells of Ephestia kuhniella. J. Cell Biol. 29: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton, J., 1967. The fine structure of developing bristles in wild type and mutant Drosophila melanogaster. J. Morphol. 122: 367–380. [DOI] [PubMed] [Google Scholar]
- Pistillo, D., N. Skaer and P. Simpson, 2002. scute expression in Calliphora vicina reveals an ancestral pattern of longitudinal stripes on the thorax of higher Diptera. Development 129: 563–572. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., R. Hernandez, J. Modolell and M. Ruiz-Gomez, 1990. Competence to develop sensory organs is temporally and spatially regulated in Drosophila epidermal primordia. EMBO J. 9: 3583–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath, J., and S. B. Carroll, 1991. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev. 5: 984–995. [DOI] [PubMed] [Google Scholar]
- Stern, D. L., 1998. A role of Ultrabithorax in morphological differences between Drosophila species. Nature 396: 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D. L., 2000. Evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Sucena, E., and D. L. Stern, 2000. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA 97: 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., Q. Zhao, Y. Yi, Z. Zhang and Y. Li, 2005. Homologous region 3 from Bombyx mori nucleopolyhedrovirus enhancing the transcriptional activity of heat shock cognate 70–4 promoter from Bombyx mori and Bombyx mandarina in vitro and in vivo. Biosci. Biotechnol. Biochem. 69: 1014–1017. [DOI] [PubMed] [Google Scholar]
- Uchino, K., M. Imamura, K. Shimizu, T. Kanda and T. Tamura, 2007. Germ line transformation of the silkworm, Bombyx mori, using the transposable element Minos. Mol. Genet. Genomics 277: 213–220. [DOI] [PubMed] [Google Scholar]
- Uhlirova, M., M. Asahina, L. Riddiford and M. Jindra, 2002. Heat-inducible transgenic expression in the silkmoth Bombyx mori. Dev. Genes Evol. 212: 145–151. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., 2006. Evolution of cis-regulatory sequence and function in Diptera. Heredity 97: 139–147. [DOI] [PubMed] [Google Scholar]
- Wülbeck, C., and P. Simpson, 2000. Expression of achaete-scute homologues in discrete proneural clusters on the developing notum of the medfly Ceratitis capitata, suggests a common origin for the stereotyped bristle patterns of higher Diptera. Development 127: 1411–1420. [DOI] [PubMed] [Google Scholar]
- Wülbeck, C., and P. Simpson, 2002. The expression of pannier and achaete-scute homologues in a mosquito suggests an ancient role of pannier as a selector gene in the regulation of the dorsal body pattern. Development 129: 3861–3871. [DOI] [PubMed] [Google Scholar]
- Xia, A., Q. Zhou, L. Yu, W. Li, Y. Yi et al., 2006. Identification and analysis of YELLOW protein family genes in the silkworm, Bombyx mori. BMC Genomics 7: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., X. Chen, Y. Zhao, G. Qi, J. Huang et al., 1999. Transgenic silkworm of anti-NPV ribozyme. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 31: 331–333. [PubMed] [Google Scholar]
- Zhou, Q., S. Tang, Y. Chen, Y. Yi, Z. Zhang et al., 2004. A scaleless wings mutant associated with tracheal system developmental deficiency in wing discs in the silkworm, Bombyx mori. Int. J. Dev. Biol. 48: 1113–1117. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., Y. Li, X. Shen, Y. Yi, Y. Zhang et al., 2006. The scaleless wings mutant in Bombyx mori is associated with a lack of scale precursor cell differentiation followed by excessive apoptosis. Dev. Genes Evol. 216: 721–726. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., T. Zhang, W. Xu, L. Yu, Y. Yi et al., 2008. Analysis of four achaete-scute homologs in Bombyx mori reveals new viewpoints of the evolution and functions of this gene family. BMC Genet. 9: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Q. Xiao, Z. Zhang, J. He and Y. Zhang, 2002. Foreign insect hormones stimulating the transcription of the ie-1 promoter of Bombyx mori nuclear polyhedrosis virus in vivo and in vitro. Biosci. Biotechnol. Biochem. 66: 1488–1494. [DOI] [PubMed] [Google Scholar]