Summary
In recent years, informatics studies have predicted several new ways in which the transforming growth factor β (TGFβ) signaling pathway can be post-translationally regulated. Subsequently, many of these predictions were experimentally validated. These approaches include phylogenetic predictions for the phosphorylation, sumoylation and ubiquitylation of pathway components, as well as kinetic models of endocytosis, phosphorylation and nucleo-cytoplasmic shuttling. We review these studies and provide a brief `how to' guide for phylogenetics. Our hope is to stimulate experimental tests of informatics-based predictions for TGFβ signaling, as well as for other signaling pathways, and to expand the number of developmental pathways that are being analyzed computationally.
Introduction
Intercellular signaling is essential for proper pattern formation during development in all multicellular organisms. In metazoan animals, a surprisingly small set of highly conserved developmental signaling proteins, which are encoded as multigene families, such as the transforming growth factor β (TGFβ) family, perform a multitude of tasks. To illustrate the versatility of these signaling molecules, we note just a few of the documented roles that TGFβ family members play in mammals: they maintain the pluripotency of embryonic stem cells, regulate mesenchymal differentiation, coordinate skeletal patterning, mediate epithelial development, synchronize the differentiation of endothelial cells, modulate myeloid cell maturation, influence lineage commitment in neurons and specify male versus female sexual differentiation (reviewed by Derynck and Miyazono, 2008a).
In order to perform these myriad roles, the M. musculus genome encodes 33 TGFβ family members (Derynck and Miyazono, 2008b). By comparison, the D. melanogaster genome encodes seven (Pyrowolakis et al., 2008) and the C. elegans genome encodes five (Savage-Dunn, 2005). Structurally, all family members share several features. Each contains an N-terminal signal sequence that is removed prior to secretion and a large pro-protein region that is also cleaved prior to secretion but that contributes to the dimerization of the C-terminal biologically active ligand. The ligand domain of all family members (∼110 amino acids in length) contains a stereotypical pattern of six cysteines. Most family members have seven cysteines, with the additional residue being centrally located and involved in ligand dimerization via a disulfide bond. A subset of family members has nine cysteines: the common set of seven and two additional residues towards the N-terminus. In addition, strong amino acid similarity exists in the regions between the cysteines of the ligand and, to a lesser extent, in the pro-protein region (reviewed by Derynck and Miyazono, 2008b).
Generating meaningful amino acid alignments, quantifying the level of amino acid similarity between two sequences and then assessing the probability of recent common ancestry are traditional informatics methods that contribute to our understanding of developmental signaling. This approach, known as phylogenetics, is designed to elucidate evolutionary relationships between family members both within and between species. The value of these studies to developmental biologists is that the tree of evolutionary relationships generated by a phylogenetic analysis can be utilized both as a roadmap upon which to interpret experiments conducted by others, and as a hypothesis generator to suggest new functions for poorly understood family members.
Given the universality of multigene families in developmental signaling pathways and the growing application of informatics approaches to understanding pathway regulation, we provide a review of recent advances in TGFβ signaling (also see Box 1 for other reviews on TGFβ signaling published in this issue). First, we briefly summarize TGFβ signal transduction pathways and the phylogenetics of the TGFβ family. Short guides to phylogenetics and its associated computer programs are included. Second, we highlight new applications of phylogenetics that are designed to improve our understanding of TGFβ pathway regulation at the level of the ligand and within the signal transduction pathway. Third, we review the latest reports that describe the kinetic modeling of the TGFβ pathway. We begin with a description of the method and then discuss both biochemical and cell biological models. Lastly, we highlight emerging informatics methods that are currently being applied to the TGFβ pathway, such as Boolean modeling and network reconstruction.
Box 1. Minifocus on TGFβ signaling
This article is part of a Minifocus on TGFβ signaling. For further reading, please see the accompanying articles in this collection: `The extracellular regulation of bone morphogenetic protein signaling' by David Umulis, Michael O'Connor and Seth Blair (Umulis et al., 2009); `The regulation of TGFβ signal transduction' by Aristidis Moustakas and Carl-Henrik Heldin (Moustakas and Heldin, 2009); and `TGFβ family signaling: novel insights in development and disease', a review of a recent FASEB Summer Conference on TGFβ signaling by Kristi Wharton and Rik Derynck (Wharton and Derynck, 2009).
TGFβ signal transduction pathways
Cells respond to instructions from developmental signaling molecules once the information has been transmitted from the cell surface via dedicated signal transduction pathways (Fig. 1). The current model for canonical TGFβ signal transduction begins when a TGFβ ligand (e.g. TGFβ1) binds to a type II transmembrane receptor serine-threonine kinase. Upon ligand binding, this receptor (which contains a constitutively active kinase) recruits a type I receptor and phosphorylates it within a serine/threonine-rich region. Type I receptor phosphorylation, in turn, stimulates this receptor to phosphorylate one or more C-terminal serines in the receptor-associated Smads (R-Smads), Smad2 and Smad3. The phosphorylation of Smad2/Smad3 shifts their subcellular localization towards the nucleus, where they form a heteromeric complex with the common-mediator Smad (Co-Smad), Smad4. This multi-Smad complex then regulates the expression of TGFβ target genes in cooperation with tissue-specific activators and repressors (Massagué, 2008). Mechanisms of TGFβ signal transduction are discussed in more detail in the accompanying review (Moustakas and Heldin, 2009).
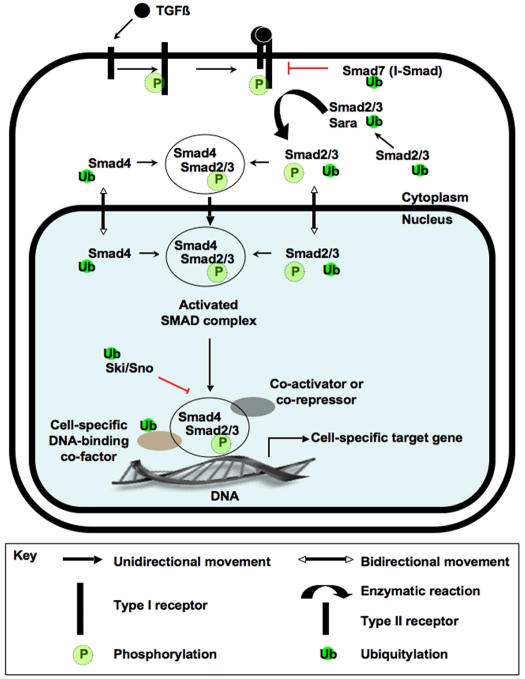
Fig. 1.
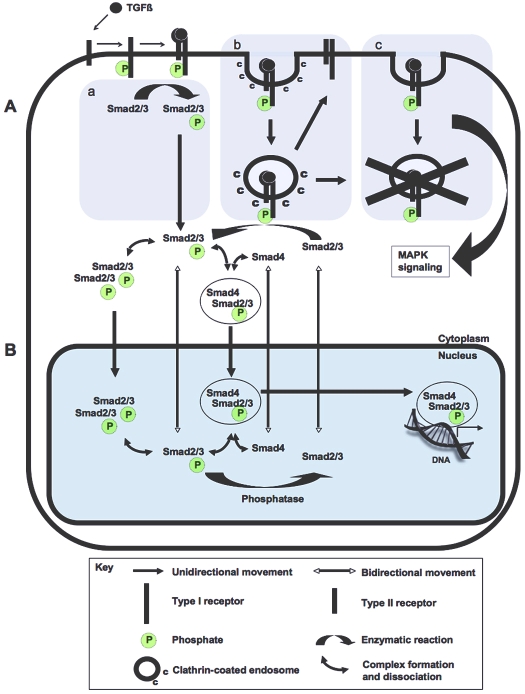
TGFβ signal transduction. Events downstream of ligand-receptor binding in a TGFβ-responsive cell. When a TGFβ ligand binds to a type II transmembrane receptor serine-threonine kinase, the receptor recruits a type I receptor and phosphorylates it (P) within a serine/threonine-rich region. Serine-threonine phosphorylation stimulates the type I receptor to phosphorylate one or more C-terminal serines in a receptor-associated Smad (R-Smad), such as Smad2 and Smad3 (Smad2/3). At the membrane, Smad anchor for receptor activation (Sara) acts as an adapter and facilitates the interaction between R-Smads and their type I receptor. The phosphorylation of Smad2/3 stimulates their translocation into the nucleus, where they form a heteromeric complex with the common-mediator Smad (Co-Smad), Smad4. This multi-Smad complex then regulates the expression of TGFβ target genes in cooperation with tissue-specific activators and repressors. Solid arrows represent the movement of information and protein subcellular localization (see key). Red T-bars represent the activity of proteins that block information transfer, such as I-Smads (Smad7) and members of the Ski/Sno family. The regulation of pathway components by polyubiquitylation (Ub) is also shown.
In addition to Smad-dependent signal transduction pathways, studies in mammalian cells have shown that TGFβ ligands can stimulate TGFβ receptors to activate Smad-independent signal transduction pathways, such as mitogen-activated protein kinases (MAPKs) (Javelaud and Mauveil, 2005), the Rho-like GTPases (Wilkes et al., 2003) and phosphatidylinositol-3-kinase (Bakin et al., 2000). Which signal transduction pathway a TGFβ receptor activates is thought to be influenced by cell type-specific accessory proteins, such as TGFβ-activated kinase 1 (TAK1; MAP3K7 - Mouse Genome Informatics) in glomerular mesangial cells (Kim et al., 2009).
The ability of TGFβ family members to elicit powerful responses in target cells via a variety of pathways dictates that their activity is strictly regulated to avoid unintended consequences. Studies in many organisms have shown that negative regulation of the TGFβ pathway can be accomplished by a variety of mechanisms. In an accompanying review article, mechanisms of extracellular antagonism are discussed (Umulis et al., 2009) (see also Box 1). Here, we focus on intracellular antagonism and note that two mechanisms have been widely studied: antagonism by dedicated proteins and the elimination of an essential component of the signaling pathway via polyubiquitin-mediated degradation.
Dedicated intracellular antagonists from two families act at distinct points in the pathway. Inhibitory Smads (I-Smads) antagonize signaling near the top of the pathway by blocking interactions between type I receptors and R-Smads and also by preventing R-Smad-Co-Smad complex formation (Miyazono, 2008). Members of the Sno/Corl/Dac family of nuclear co-repressors bind to promoter-bound Co-Smads and block their ability to activate transcription. These proteins accomplish this by recruiting transcriptional repressors, such as histone deacetylases and Sin3A. Then, upon ligand stimulation, the Sno family proteins are degraded, dissolving the repressor complex and allowing Smad complexes to stimulate transcription (Pot and Bonni, 2008). Alternatively, Sno proteins in M. musculus, D. melanogaster and C. elegans have been shown to facilitate TGFβ signaling. For example, in D. melanogaster, the dSno (Snoo - FlyBase) mutant phenotype mimics the loss of Activin signaling, and biochemical studies have revealed that dSno facilitates Activin signaling by shifting the affinity of the Co-Smad Medea away from Mad (the BMP-dedicated R-Smad) and towards dSmad2 (Smox - FlyBase) (Takaesu et al., 2006). Taken together, these data suggest that the influence of Sno proteins on TGFβ signaling is probably tissue specific.
Ubiquitylation is another frequently employed mechanism for antagonizing TGFβ signaling. The ubiquitin pathway results in protein degradation and is stimulated when a polyubiquitin chain is attached to a lysine in a target protein (Meinnel et al., 2006). Numerous proteins act as ubiquitin E3 ligases (the enzyme that forms the covalent bond between a ubiquitin molecule and a target protein) for TGFβ pathway components, including Ectodermin/Tif1γ (Trim33) (Dupont et al., 2005) and Smurf (Morén et al., 2005). Negative regulation of TGFβ pathway components by polyubiquitylation and their subsequent degradation has been reported for H. sapiens type I receptors, such as ALK5 (TGFβR1 - Human Gene Nomenclature Committee; also known as TβRI), and for Smads, such as SMAD2 (Kuratomi et al., 2005). Interestingly, in several instances, the I-Smad and polyubiquitylation mechanisms are intertwined, as shown by the observation that I-Smads can recruit the Smurf ubiquitin ligase to the TGFβ receptor complex (Kavsak et al., 2000).
By contrast, monoubiquitylation can have positive or negative effects on TGFβ signaling in a tissue-specific fashion. For example, the monoubiquitylation of Lys507 in human SMAD4 (a Co-Smad) can accentuate TGFβ signaling by enhancing its ability to form complexes with R-Smads (Morén et al., 2003). Recently, the E3 ligase Ectodermin was shown to be a deactivating monoubiquitinase, rather than a polyubiquitylating enzyme for Smad4, and this was coupled with the discovery of the first TGFβ pathway deubiquitinase: Dupont et al. demonstrated that reversible monoubiquitylation by Ectodermin/Tif1γ and the deubiquitinase FAM (Fat Facets in Mammals, also known as Usp9x) on Lys519 in human SMAD4, can function as an `on-off' switch for TGFβ signaling in D. melanogaster, X. laevis and M. musculus (Dupont et al., 2009).
Sumoylation, a distinct post-translational modification that targets lysine residues, also plays positive and negative roles in the TGFβ pathway. When mammalian SMAD4 is sumoylated at Lys113 or Lys159, its ability to activate transcription is stimulated in transfected HeLa cells (Lin et al., 2003). In this study, RNAi knockdown of UBC9 (UBE2I) (an E2-conjugating enzyme that participates in the addition of SUMO1 molecules to lysine residues in a target protein) decreased SMAD4 protein levels, whereas overexpression of the SUMO1 protein increased SMAD4 protein levels. A second study showed that sumoylation of the same lysine residues represses the ability of SMAD4 to activate transcription in transfected COS cells (Long et al., 2004). In this study, overexpression of SUMO1 and UBC9 decreased TGFβ-responsive reporter gene expression, whereas overexpression of a SUMO1 protease increased expression from the reporter. Taken together, these studies suggest that the influence of sumoylation on SMAD4 activity is cell-type specific.
To date, numerous questions remain about the role of lysine modification as a mechanism for regulating the activity of the TGFβ pathway. Recently, this topic has been addressed through the use of phylogenetics (Konikoff et al., 2008). Before reviewing this and other phylogenetic studies of TGFβ pathway regulation, we reprise the traditional role of phylogenetics and provide the first description of the evolutionary relationship between TGFβ family members based on full-length protein sequences.
Phylogenetics of TGFβ family members
It is well known that phylogenetic trees are useful for assessing the probability of recent common ancestry between members of a multigene family (reviewed by Whelan, 2008) and that a tree is generated from DNA or protein sequences by computers running unfathomable mathematical equations (reviewed by Giribet, 2007). In Box 2, we provide a practical guide to generating an informative tree. We briefly describe each step (identifying related sequences, creating an alignment, generating a tree and incorporating statistics) and suggest relevant, user-friendly computer programs. In Box 3, for those with an interest in the underlying theory, we provide additional details on four common algorithms for generating trees from alignments and additional computer programs.
Box 2. Practical guide to phylogenetics
Here we provide a practical guide to generating an informative tree. We describe how to identify related sequences, how to create an alignment of the sequences, how to generate a tree from the alignment and how to incorporate statistics.
BLAST is a program available at the NCBI website that identifies related sequences (Johnson et al., 2008) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For many phylogenetic analyses, the most useful BLAST program is BLASTp, which utilizes protein sequences. An important part of a BLAST report is the E-values. These values are a measure of how frequently the alignment between your query and the listed sequence would have occurred by chance. The smaller the E-value (they are usually shown as negative exponents), the less likely the match is due to chance.
Clustal is a program that generates multi-sequence alignments (Larkin et al., 2007) (http://www.clustal.org/). Clustal aligns sequences using a mathematical approach that does not utilize external biological knowledge, such as mutational mechanisms that cause changes in amino acid sequences, transcription factor binding sites or protein structure, as a guide, and therefore the investigator should examine the alignment for deviations from biological `common sense'. If you plan to publish an alignment, several programs such as Boxshade (http://ch.embnet.org/software/BOX_form.html) add easily interpretable black and gray highlights for identical and similar amino acids.
MEGA is a program that generates trees from Clustal alignments (Kumar et al., 2008) (http://www.megasoftware.net/). Within MEGA there are several algorithms available for generating phylogenetic trees, each based on different sets of assumptions (see Box 3 for details on four popular algorithms). In addition to MEGA, there are many programs available for phylogenetic analysis of protein sequences, including PhyML (Guindon and Gascuel, 2003) and Leaphy (Whelan, 2009). An extensive list of programs is available at evolution.genetics.washington.edu/phylip/software.html. Regardless of the method employed in tree construction, it is important to determine how much statistical confidence is present at the node that connects two branches.
Bootstraping (Felsenstein, 1985) is a common statistical technique that can be implemented in MEGA to ascertain the probability that the node connecting two branches or two clusters of branches is significant. This method takes a random sample of the sequences in the alignment and builds a tree from this sample. It does this many times (typically >1000) and then calculates a bootstrap value for each branch in the tree (the percentage of trees in which two branches or clusters are grouped together). Bootstrap values are not strictly equivalent to probabilities, but the higher the bootstrap value the greater the likelihood that a branch has not occurred due to chance. The bootstrap method is considered a conservative estimator and in practice a bootstrap value above 70 is often interpreted as a statistically supported branch (Efron et al., 1996; Sitnikova, 1996).
Box 3. Four popular algorithms for generating trees from alignments
The Neighbor-Joining method (Saitou and Nei, 1987) is a `numerical-based' technique. It first determines the number of amino acid differences between each pair of sequences in an alignment. Then it normalizes this number over the length of each protein to calculate a scalar value known as the `evolutionary distance' between each pair of sequences. A tree is built by joining the two sequences that have the smallest distance between them. Then, the program sequentially adds the next closest sequence or group of similar sequences until all sequences are incorporated. This method is available in MEGA.
The Maximum Parsimony method is a `character-based' technique. It attempts to implement the age-old scientific principle of Occams' Razor (i.e. the best explanation is the simplest one) (Fitch and Farris, 1974). The method is designed to identify the tree that requires the smallest number of evolutionary steps to explain all the observed changes (an amino acid is considered a discrete character) in the alignment. To do this, the program creates all possible trees and then searches for the minimal tree. A drawback is that the number of trees to be analyzed becomes computationally prohibitive even for an alignment of modest size. Computationally tractable variations, such as Branch and Bound (Penny and Hendy, 1987), have been developed to address this issue. The Branch and Bound algorithm is available in MEGA.
The Maximum Likelihood method is a `probability-based' technique (Felsenstein, 1981). It calculates the likelihood that a randomly generated phylogeny, when evaluated against a specific evolutionary model, will generate the sequence changes seen in an alignment. The evolutionary models employed are matrices of empirically derived amino acid substitution frequencies expressed as mathematical equations. Two frequently employed matrices are those of Dayhoff et al. (Dayhoff et al., 1978) (PAM 30) and Henikoff and Henikoff (Henikoff and Henikoff, 1992) (BLOSUM). The `most likely' phylogeny is the one with the highest probability of generating the observed sequence changes. The Maximum Likelihood algorithm is widely available and can be implemented in PhyML, PAUP* (Swofford, 2001), Leaphy and MEGA. Bootstrapping is an appropriate approach for assessing the degree of confidence in each node, but the Maximum Likelihood approach allows alternative statistical tests for evaluating trees (e.g. the Shimodaira-Hasegawa test and Approximately Unbiased test). These alternative tests are available via the CONSEL software program (Shimodaira and Hasegawa, 2001) (http://www.is.titech.ac.jp/~shimo/prog/consel/).
The Bayesian Inference method is also a `probability-based' technique. It employs Markov chain Monte Carlo simulation (Mau et al., 1999) to estimate the `most likely' tree directly from the data relatively rapidly, rather than identifying it from randomly generated trees as in the Maximum Likelihood approach (but based on the same set of substitution frequency matrices). This technique is a recent innovation in phylogenetic inference and it is available in the program MrBayes (Ronquist and Huelsenbeck, 2003) (http://mrbayes.csit.fsu.edu).
To achieve maximum confidence in phylogenetic analyses, many studies utilize species that belong to distinct phyla. For example, our tree of 45 TGFβ family proteins (33 from M. musculus; seven from D. melanogaster; five from C. elegans) (Fig. 2) focuses on three fully sequenced genetic model organisms. Two of these species are coelomates, animals with three embryonic germ layers and a digestive tract with two openings: mouse (M. musculus is a deuterostome, in which the blastopore becomes the anus) and fruit fly (D. melanogaster is a protostome, in which the blastopore becomes the mouth). Deuterostomes and protostomes are taxonomically equivalent groups of coelomate phyla. The third species is a nematode (C. elegans is a pseudocoelomate, an animal with three germ layers but a digestive tract with one opening). Molecular data suggest that the split between deuterostomes and protostomes occurred ∼990 million years ago and that between coelomates and pseudocoelomates ∼1.2 billion years ago (Hedges and Kumar, 2003).
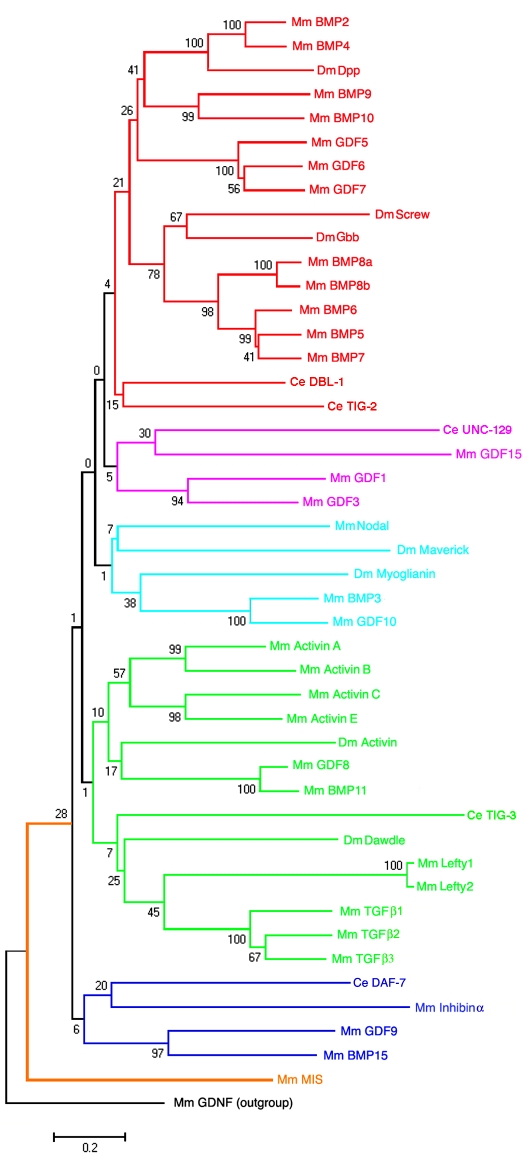
Fig. 2.
TGFβ family tree. The longest isoform of each TGFβ family member from C. elegans (Ce, five) (Savage-Dunn, 2005), D. melanogaster (Dm, seven) (Pyrowolakis et al., 2008) and M. musculus (Mm, 33) (Derynck and Miyazono, 2008b) were aligned in Clustal X. A tree was generated from the alignment by the Neighbor-Joining algorithm with 1000 bootstrap replicates in MEGA as described (Newfeld et al., 1999) (see also Box 3). The length of the alignment was 984 amino acids. This alignment is 871 amino acids longer than the traditional `ligand-only' alignment of 113 amino acids. Mouse GDNF (out-group) provides a root because it shares only the pattern of cysteines with other TGFβ members and uses a novel receptor (Jing et al., 1996). Bootstrap values for each node that joins two or more sequences are shown. Because 88% of the alignment is composed of divergent pro-protein sequences, we consider a bootstrap value (Box 2) of greater than 40 to be biologically meaningful. Branch lengths are drawn to scale, and the scale bar shows the number of amino acid substitutions per site between two connected sequences. Subfamilies with varying levels of statistical support are indicated: bone morphogenetic protein (BMP, red), UNC-129 (purple), Nodal (turquoise), TGFβ/Activin (green), Inhibin (blue) and Mullerian-inhibiting substance (MIS, brown; AMH - Mouse Genome Informatics). For the name, synonym and protein ID of each sequence, see Table S1 in the supplementary material.
Phylogenetic analyses of TGFβ family members have traditionally focused on the ligand domain owing to the easily identifiable amino acid conservation in this region (e.g. Newfeld et al., 1999). In these studies, two large subfamilies, the Decapentaplegic/bone morphogenetic protein (Dpp/BMP) subfamily and the TGFβ/Activin subfamily, were identified based on amino acid similarities. Each subfamily has members in M. musculus, D. melanogaster and C. elegans. In the ligand tree, within these two major subfamilies the level of amino acid conservation between family members from different species (homologs - sequences that are `identical by descent' from a common ancestor) is extremely high. For example, an alignment of D. melanogaster Dpp with its closest relatives (M. musculus BMP2 and BMP4) reveals that these proteins share 75% amino acid identity (Newfeld and Gelbart, 1995). This level of sequence conservation is reflected in the ability of these ligands to function correctly in cross-species experiments. Both human BMP2 and BMP4 rescue dpp mutant phenotypes when expressed in flies (Padgett et al., 1993), and recombinant Dpp induces bone formation in mammalian cell culture (Sampath et al., 1993). These subfamilies and four smaller subfamilies are also present in a phylogeny that we generated from full-length TGFβ proteins (Fig. 2; see Table S1 in the supplementary material).
The experimental demonstration of cross-species functionality for family members that cluster together, such as Dpp and BMP2/4, suggests that clustering can provide clues as to the function of less well-studied proteins. For example, all of the most closely related sequences to D. melanogaster Dpp are present in a supercluster that contains the M. musculus sequences BMP2, BMP4, BMP9 (GDF2 - Mouse Genome Informatics) and BMP10, as well as growth and differentiation factor 5 (GDF5), GDF6 and GDF7 (Fig. 2). A sister supercluster within the BMP subfamily contains the D. melanogaster proteins Glass bottom boat (Gbb) and Screw (Scw), the five M. musculus proteins BMP5/6/7/8a/8b and two C. elegans proteins. The fact that heterodimers composed of Dpp and Gbb (Shimmi et al., 2005a) or Dpp and Scw (Shimmi et al., 2005b) elicit responses distinct from those of their respective homodimers during Drosophila development suggests that each of the seven M. musculus proteins in the Dpp supercluster can potentially dimerize with each of the five M. musculus family members in the Gbb/Scw supercluster (Fig. 2). To date, four of the possible M. musculus intercluster heterodimers have been shown to function in a manner distinct from that of their respective homodimers in cell culture (e.g. Israel et al., 1996). Our tree suggests that these and additional intercluster heterodimers play roles in M. musculus development.
In summary, traditional phylogenetic analyses can still shed new light on the evolutionary relationships between TGFβ family members. Recently, as discussed below, several papers have taken a phylogenetics approach to TGFβ signaling, but with a different purpose.
Phylogenetics of TGFβ pathway regulation
These new applications of phylogenetics are designed to improve our understanding of TGFβ pathway regulation rather than of evolution. The underlying logic of this new approach is the exploitation of evolutionary conservation as a guide to identifying amino acids that are involved in the enzymatic regulation of protein function. Recent reports of new regulatory mechanisms that impact TGFβ ligands, type I receptors and Smads are reviewed.
Ligand cleavage
In order to be effectively bound by their receptors, the pro-region of TGFβ proteins must be cleaved from the receptor-binding ligand domain (Fig. 3). Ligand cleavage is performed by a Furin-type enzyme at a multi-basic amino acid sequence (arginine-X-X-arginine) (Dabovic and Rifkin 2008) prior to secretion of the ligand. Kuunapuu et al. examined the biochemical mechanisms that are employed in the cleavage process for members of the Dpp/BMP2/BMP4 subfamily (Kuunapuu et al., 2009). They began with a phylogenetic analysis of a 60 amino acid region surrounding the cleavage site in 27 species, ranging from the diploblast N. vectensis (an animal even less complex than C. elegans as it has only two germ layers) to H. sapiens - a 1.5 billion year time span (Hayward et al., 2002).
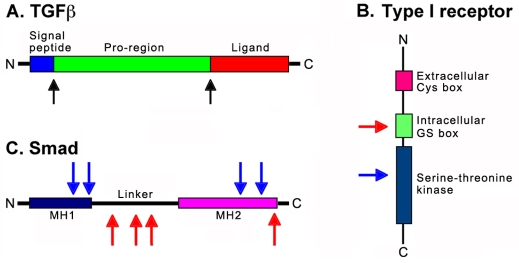
Fig. 3.
Enzymatic regulation of TGFβ signaling. (A) TGFβ proteins are processed by two enzymatic cleavage events (black arrows). The first removes the N-terminal signal peptide and the second separates the pro-region from the C-terminal receptor-binding ligand. (B) Type I receptors are activated by type II receptor phosphorylation on serine in the intracellular glycine-serine (GS) box (red arrow). The H. sapiens type I receptor ALK5 is deactivated by sumoylation on lysine in the kinase domain (blue arrow). The extracellular Cys box is shown for reference. (C) Smad proteins are activated by phosphorylation on serine at their C-terminus (single red arrow). X. laevis Smad1 is deactivated by tyrosine phosphorylation by MAPK and by serine phosphorylation by GSK3β in the linker region (red arrows). H. sapiens SMAD4 is regulated by ubiquitylation and sumoylation of lysine in a tissue-specific manner (blue arrows).
They found that pro-domain cleavage from the ligand is not accomplished at a single site, but that one to three sites are involved. They also discovered that the number of cleavage sites, their amino acid sequence and their location (the number of amino acids from the first cysteine of the ligand domain) are highly variable across species. This hypervariability contrasts with the astounding conservation of the immediately adjacent ligand domain (∼80% amino acid similarity across this 1.5 billion year time span). Further, they found that within deuterostomes (the phylum that includes vertebrates), groups of sequences with similar cleavage site organization do not match the established relationships of their respective species. This phenomenon is not uncommon in the field of molecular evolution, in which it is known as `incongruity between the gene tree and the species tree'. The authors found that vertebrates and echinoderms (echinoderms are a sister phylum to hemichordates and only distantly related to vertebrates) share the same cleavage site organization, whereas cephalochordates and urochordates (both sister phyla to vertebrates and only distantly related to hemichordates) share the same cleavage system as hemichordates.
Kuunapuu et al. then conducted a series of biochemical studies of ligand cleavage in this subfamily. They showed that cleavage site sequence diversity leads to differences in the enzymatic mechanics of cleavage. For example, H. sapiens BMP2 and BMP4 have two Furin sites, whereas their homolog in D. melanogaster, Dpp, has three. In both species, the Furin sites are cut sequentially to produce the ligand. In H. sapiens, the site adjacent to the ligand is cleaved first, but in D. melanogaster the site furthest from the ligand is cut first.
Kuunapuu et al. concluded that the diversity of cleavage site organization seen in different species results from compensatory changes that occurred in response to mutations arising independently in each lineage (Kuunapuu et al., 2009). As the TGFβ ligand must be cleaved from the pro-domain to bind receptors, compensatory changes that create new Furin sites ensure the survival of the organism. We agree with this conclusion and would like to suggest that the phylogenetic data also have implications for understanding TGFβ regulation. In our view, the existence of `gene tree-species tree incongruity' for cleavage sites implies that regulatory functions associated with these sites are likely to be implemented differently in each species.
Type I receptor sumoylation
The phosphorylation of R-Smads by TGFβ type I receptors is regulated by a variety of antagonistic mechanisms, including I-Smads and ubiquitylation (e.g. Wicks et al., 2005; Yamaguchi et al., 2006). In the case of TGFβ type I receptor ubiquitylation, the specific lysines targeted by E3 ubiquitin ligases are unknown. To aid in the identification of lysines that become ubiquitylated in these receptors, Konikoff et al. conducted a phylogenetic analysis of lysine conservation in type I receptors using the three-species strategy (Konikoff et al., 2008) (Fig. 4; see Table S2 in the supplementary material). The authors identified five lysines that are conserved in all type I receptors. However, they noted that in the crystal structure of H. sapiens ALK5 (a receptor for TGFβ/Activin subfamily ligands), only two lysines (which correspond to Lys326 and Lys490 in M. musculus ALK5) are exposed on the molecule's surface and could therefore be considered as candidates for ubiquitylation.
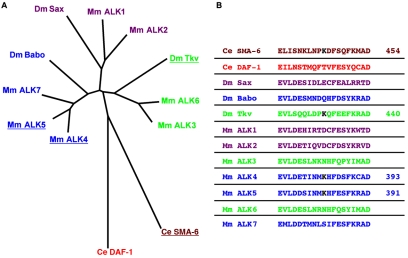
Fig. 4.
Conservation of a TGFβ type I receptor sumoylation site. An alignment was generated from the longest isoform of each protein as described by Konikoff et al. (Konikoff et al., 2008). The length of the alignment was 796 amino acids. (A) A star tree was generated in MEGA as described (Newfeld et al., 1999). The tree is unrooted with branch lengths drawn to scale. Clusters of sequences with significant statistical support are indicated: TGFβ/Activin receptors (purple and blue); BMP receptors (green); DAF-1 (red) and SMA-6 (brown). H. sapiens ALK5 is sumoylated at Lys393, which regulates its activity (Kang et al., 2008). Four receptors with a conserved lysine residue in the same context (I/L-X-X-K) as Lys393 in H. sapiens ALK5 are underlined. (B) Color-coded amino acid sequences from each receptor corresponding to the region of Lys393 in H. sapiens ALK5. The conserved lysine is shown in black in ALK4, ALK5, Tkv and SMA-6, with its amino acid number to the right. For the name, synonym and protein ID of each sequence, see Table S2 in the supplementary material.
The sumoylation of H. sapiens ALK5 within its kinase domain (Fig. 3) was recently shown to regulate the activity of this receptor (Kang et al., 2008). Konikoff et al. reported that the sumoylated lysine (Lys393 in M. musculus ALK5) is also present in three other receptors: M. musculus ALK4 (ACVR1B - Mouse Genome Informatics), D. melanogaster Thickveins (Tkv, a Dpp receptor) and C. elegans SMA-6 (Fig. 4). This pattern of sequence conservation and its presence in C. elegans, D. melanogaster and M. musculus suggest that the sumoylation of this lysine is an ancient mechanism for regulating TGFβ type I receptor activity and that D. melanogaster Tkv and C. elegans SMA-6 might also be regulated by sumoylation.
Conservation of this sumoylated lysine is, however, inconsistent with the evolutionary relationships between the full-length receptors. M. musculus ALK5, which has the conserved lysine, is more closely related to the D. melanogaster receptors Baboon (Babo; an Activin receptor) and Saxophone (Sax; a receptor for both Dpp and Gbb), neither of which has the conserved lysine, than it is to D. melanogaster Tkv (Fig. 4). This pattern of sequence conservation suggests that the sumoylated lysine was lost independently three times: from Babo, from the M. musculus ALK3/ALK6 (BMPR1A/BMPR1B - Mouse Genome Informatics) pair and from the cluster that contains Sax and M. musculus ALK1/ALK2 (ACVRL1/ACVR1). From the analysis, Konikoff et al. concluded that the phylogenetic analysis of lysine conservation, when coupled with experimental data, can be fruitfully employed to identify changes in pathway regulation by ubiquitylation or sumoylation (Konikoff et al., 2008).
R-Smad linker phosphorylation
The Smad family of multifunctional proteins comprises four major subfamilies: two subfamilies of R-Smads, one of Co-Smads and one of I-Smads (Fig. 5). All human and fly Smad proteins cluster together in these four subfamilies, and there are three subfamilies composed solely of nematode family members. Like the Dpp/BMP2/BMP4 ligands, H. sapiens and D. melanogaster Smads in the same subfamily function similarly in transgenic experiments (Marquez et al., 2001; Takaesu et al., 2005). The functions of Smad proteins are effected via three highly conserved regions (Fig. 3). There is an N-terminal MH1 domain that mediates transcription factor interactions and DNA binding, a near C-terminal MH2 domain that modulates a wide variety of protein-protein interactions, and a C-terminal receptor phosphorylation region that contains the serine residues targeted by the type I receptor (Lin et al., 2008). As noted above, when a C-terminal serine is phosphorylated, an R-Smad translocates from the cytoplasm to the nucleus (Fig. 1).
Fig. 5.
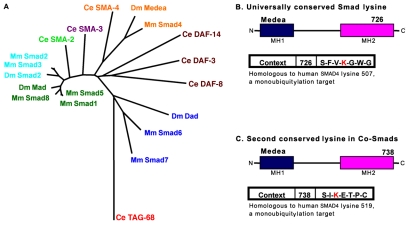
Conserved lysine residues in Smads correspond to ubiquitylation targets. An alignment was generated from the longest isoform of each protein as described (Konikoff et al., 2008). The length of the alignment was 892 amino acids. (A) A star tree was generated in MEGA as described (Newfeld and Wisotzkey, 2006). The tree is unrooted with branch lengths drawn to scale. Clusters of sequences with significant statistical support are indicated: BMP R-Smads (green), TGFβ/Activin R-Smads (turquoise), SMA-2 (red), SMA-3 (purple), Co-Smads (gold), DAF pathway Smads (brown) and I-Smads (blue). (B) These protein alignments revealed that a single lysine is conserved in all Smads. The location, amino acid number and immediate context for this lysine (Lys726) are shown in a schematic of D. melanogaster Medea. The homologous lysine in human SMAD4 (Lys507) is a target of monoubiquitylation (Morén et al., 2003). (C) A second lysine, 12 amino acids downstream from the universally conserved lysine, is conserved only in Co-Smads. The location, amino acid number and immediate context for this lysine (Lys738) are shown in a schematic of Medea. The homologous lysine (Lys519) was subsequently shown to be a target of monoubiquitylation in human SMAD4 (Dupont et al., 2009). For the name, synonym and protein ID of each sequence, see Table S3 in the supplementary material.
The linker region between the conserved MH1 and MH2 domains was initially thought to function largely as a spacer. This was owing to an apparent lack of conserved amino acids in this region in R-Smads from different phyla (e.g. C. elegans SMA-2 and SMA-3 and D. melanogaster Mad) (Sekelsky et al., 1995). Subsequently, sets of conserved extracellular signal-regulated kinase (ERK) serine-threonine phosphorylation sites were identified in the linker domains of mammalian R-Smads. These ERK sites are phosphorylated in vivo (Kretzschmar et al., 1997), where they promote the recruitment of Smurf ubiquitin ligases via a conserved PY motif also present in the linker region (Chong et al., 2006; Sapkota et al., 2007). Genetic analyses revealed that ERK phosphorylation of M. musculus SMAD1 contributes to embryonic events, such as primordial germ cell formation (Aubin et al., 2004).
In 2006, Newfeld and Wisotzkey extended the analysis of linker conservation to D. melanogaster and C. elegans and reported that one of the mammalian ERK sites is present in all Mad/Smad1 subfamily members (Fig. 3), but that none of the mammalian ERK sites in the Smad2/Smad3 subfamily is conserved in D. melanogaster or C. elegans (Newfeld and Wisotzkey, 2006). This linker conservation study also revealed that two consensus sites for phosphorylation by the glycogen synthase kinase 3 β (GSK3β) serine-threonine kinase are conserved in all members of the Mad/Smad1 subfamily and that these sites are not present in any other family member (Fig. 3). Based on the subfamily-wide conservation of a GSK3β site, the authors predicted that linker phosphorylation by GSK3β represented a subfamily-specific function. This prediction was experimentally validated by Fuentealba et al., who demonstrated that the X. laevis Smad1 linker region was phosphorylated by GSK3β (Fuentealba et al., 2007). They showed that GSK3β phosphorylation regulated TGFβ signal duration by stimulating the polyubiquitylation and degradation of Smad1.
This was the first of three phylogenetic predictions for new mechanisms of Smad regulation to be experimentally validated. Subsequent studies employing the same strategy, the identification of conserved amino acids that are associated with enzymatic activity, shifted from phosphorylation targets (serine and threonine) to the ubiquitylation and sumoylation target (lysine).
R-Smad and Co-Smad ubiquitylation and sumoylation
It is known that R-Smads in D. melanogaster and mammals are negatively regulated by polyubiquitin-mediated degradation (e.g. Kuratomi et al., 2005). Nevertheless, the identity of the lysines that are targeted for ubiquitylation remains largely unknown. By contrast, the monoubiquitylation of the H. sapiens Co-Smad (SMAD4) has a positive influence on its ability to activate gene expression and the target of monoubiquitylation has been identified (Lys507) (Morén et al., 2003). To aid in the process of identifying ubiquitylated lysines in Smad family members, Konikoff et al. conducted a phylogenetic analysis of lysine conservation in which they incorporated information from Smad crystal structures (e.g. Shi et al., 1998; Wu et al., 2001) to eliminate lysines that are conserved for structural reasons (Konikoff et al., 2008). They found that every Smad contains a single conserved lysine in the MH2 domain (Fig. 5; see Table S3 in the supplementary material) that is homologous to H. sapiens SMAD4 Lys507. This finding led the authors to predict that this universally conserved lysine is a strong candidate for regulation by ubiquitylation in all Smads.
Konikoff et al. then analyzed lysine conservation within each Smad subfamily. They found that all Co-Smads contain two conserved lysines, including the universal Smad lysine in the MH2 domain (Fig. 5). The conservation of the context for Lys738 in D. melanogaster Medea (homologous to H. sapiens SMAD4 Lys519) is stronger than that of the universal Smad lysine (H. sapiens SMAD4 Lys507), leading the authors to predict that Lys738 is also targeted for ubiquitylation. Several months later, experiments validating this prediction were published. Dupont et al. demonstrated that Lys519 in H. sapiens SMAD4 is the target of reversible monoubiquitylation by the E3 ligase Ectodermin/Tif1γ and the deubiquitinase FAM (Dupont et al., 2009). Further, the ubiquitylation state of Lys519 acted as an `on-off' switch for TGFβ signaling. As a result, we now predict that Co-Smads in all species are subject to regulative monoubiquitylation at the homologous lysine.
Additional analyses of the conserved lysines in Co-Smads revealed that neither lies within a sumoylation consensus site (Konikoff et al., 2008). However, Lys185 in D. melanogaster Medea is within a consensus site (Yang et al., 2006), and is as conserved as Lys159 in H. sapiens SMAD4, which is sumoylated (Long et al., 2004). Lys113 in H. sapiens SMAD4 is also sumoylated (Long et al., 2004), and is as conserved as Lys141 (also within a sumoylation consensus site) in Medea. Given that both lysines, and their contexts, are conserved between H. sapiens SMAD4 and D. melanogaster Medea, Konikoff et al. predicted that both lysines are sumoylated in Medea (Konikoff et al., 2008). In the same month, Miles et al. demonstrated that Lys141 and Lys185 in Medea are sumoylated and that sumoylation reduces Medea activity in embryonic dorsal/ventral patterning (Miles et al., 2008), validating this prediction. Note that the lysine numbering used in this review and in Konikoff et al. derives from GenBank NP 524610 (Wisotzkey et al., 1998; Das et al., 1998), whereas that employed by Miles et al., who report sumoylation of Lys115 and Lys159, derives from GenBank AF039232 (Hudson et al., 1998).
In summary, the frequent convergence of phylogenetic and experimental data for TGFβ signaling suggests that the analysis of lysine conservation in receptors and signal transducers from other pathways will be similarly informative. A strength of the phylogenetic approach for understanding signaling pathway regulation is that it employs publicly available sequences and computer programs. A weakness of this approach is that distinguishing between structural conservation and regulative conservation can be difficult. As such, we are currently developing computational methods to address this issue.
Kinetic modeling of TGFβ pathway regulation
Computational modeling presents information derived from a biological system in mathematical form. Models often aim to identify emergent properties of a system that are not detectable from observations of the underlying interactions. Models typically depict a network of molecular components (genes, proteins, metabolites) and their functional interactions but they can also include features such as phenotypes. Over the last decade, mathematical modeling has been successfully applied to a number of biological problems. For example, models of signaling pathways have revealed unsuspected features, such as negative feedback or bistability (e.g. Ferrell, 2002). More complex models integrate multiple aspects of cellular biology. For example, a model incorporating signaling, gene expression and metabolism has demonstrated the contribution of these mechanisms to the stress response of the yeast cell (Klipp et al., 2005).
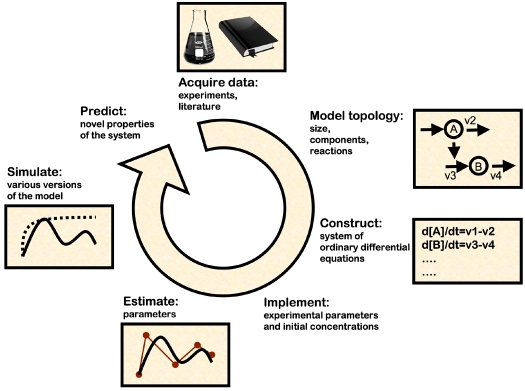
Systems of ordinary differential equations (equations that describe how a function changes in response to a single independent variable over time) are frequently used to model dynamic processes in biological networks. Models built on differential equations are known as kinetic models (Klipp, 2007). The differential equations used in kinetic models describe changes in the concentration of components over time and express molecular events (e.g. phosphorylation) according to empirical biochemical data. Kinetic equations were originally established to represent classical enzymatic reactions (e.g. oxidation-reduction) (Cope, 1963), but are now applied to describe protein-protein or protein-DNA interactions in cellular settings [e.g. transcription (Chen et al., 1999)]. The accuracy of the predictions that arise from computational models of a given biological question can vary according to the quality of the initial biochemical data, the model's topology and its assumptions. Therefore, the systematic experimental assessment of predictions is required to validate a model. A schematic of the steps necessary for kinetic model construction and application are shown in Fig. 6.
Fig. 6.
Constructing a kinetic model of a biological system. The steps required to create a kinetic model of a biological system based on published experimental data. Modeling a signaling pathway requires both qualitative and quantitative data. Molecular and biochemical knowledge of protein-protein interactions shape the overall topology of the model. Details of protein amounts, transport and subcellular localization, phosphorylation states, kinetics of synthesis and degradation and gene expression levels provide the parameters and parameter values that constrain the model to biological reality. The relationships between the model components are then defined mathematically in the form of ordinary differential equations. Quantitative parameter values are implemented in the model and missing parameter values (e.g. an experiment that has yet to be done) are computationally estimated to fit the quantitative dataset. The model gains in explanatory power with the number of datasets incorporated. Once the model is set, simulations using modified forms of the model can predict what the crucial components are and rule out regulatory roles for other components. Next, experimental testing of new hypotheses for biochemical mechanisms that underlie model predictions validate or invalidate the model, potentially providing new insights into the pathway. Then the model begins a new cycle.
Kinetic models of TGFβ signaling offer the ability to conduct real-time simulations of biochemical interactions as a means of understanding pathway regulation. The difficulty in obtaining suitable, fine-scale quantitative biochemical data on all aspects of the pathway has led modelers to focus on selected parts of the pathway to test specific hypotheses. For example, recent studies of the TGFβ pathway have analyzed receptor endocytosis and nuclear accumulation of phosphorylated Smads, as we discuss in more detail below.
Receptor endocytosis
Although TGFβ receptor activation occurs at the plasma membrane, studies with labeled type II receptors have suggested that downstream signaling through the SMADs requires receptor internalization. For example, Di Guglielmo et al. studied the trafficking of labeled TGFβ type II receptors into clathrin-coated endosomes and into lipid-raft caveolae (Di Guglielmo et al., 2003). The authors associated receptor internalization by clathrin-coated endosomes with the activation of SMAD2 and receptor recycling, whereas internalization into lipid-raft caveolae was associated with receptor degradation. Alternatively, Mitchell et al., who also studied TGFβ type II receptors, demonstrated that internalization of receptors in clathrin-coated endosomes led to receptor recycling and eventually to receptor degradation, but concluded that lipid-raft caveolae have no significant role in TGFβ signaling (Mitchell et al., 2004). A third view of the role of receptor internalization was recently reported by Zuo and Chen, who noted that TGFβ type I receptors localized in lipid-raft caveolae do not activate Smads but instead activate the MAPK pathway (Zuo and Chen, 2009).
To advance our understanding of the relationship between type I and type II receptor activity and their subcellular location, Vilar et al. generated a kinetic model of receptor trafficking (Vilar et al., 2006). Their model included the following components: ligands, both types of receptor and two receptor locations - at the cell surface in the plasma membrane or internalized within endosomes. The model was initiated with biochemical and cell biological data from Di Guglielmo et al. (Di Guglielmo et al., 2003) and Mitchell et al. (Mitchell et al., 2004). Model simulations suggested that receptor location influenced a number of features of receptor activity, such as type I-type II complex formation, ligand binding and receptor degradation.
Subsequently, Zi and Klipp generated an expanded kinetic model that included both the role of receptor compartmentalization and Smad activity (Zi and Klipp, 2007). Model parameters included: ligand-receptor binding, receptor endocytosis by clathrin-coated endosomes and by lipid-raft caveolae, receptor recycling and degradation, Smad nuclear import and export, and R-Smad-Co-Smad complex formation. The model was initiated with biochemical and cell biological data from Di Guglielmo et al. (Di Guglielmo et al., 2003). Model simulation over a range of conditions for each parameter suggested that TGFβ signaling via Smads is modulated by a balance between receptor endocytosis into clathrin-dependent endosomes versus lipid-raft caveolae (Fig. 7A, top). Thus, the model suggests an explanation for the variation in the kinetics of receptor activity observed by Di Guglielmo et al. and Mitchell et al. in that each cell type utilizes a distinct ratio of the available endocytic pathways. Looking ahead, it will be interesting to see whether the endocytosis balance hypothesis holds when the model is expanded with additional parameters, such as Smad activation in the absence of receptor internalization (e.g. Lu et al., 2002).
Fig. 7.
Models of TGFβ receptor endocytosis and Smad nucleo-cytoplasmic shuttling. (A) Three receptor endocytosis models. (a) No internalization accompanies the direct phosphorylation of Smad2 and Smad3 by activated receptors at the membrane. (b) Internalization of activated receptors into clathrin-coated endosomes leads to the activation of Smad2 and Smad3 and to Rab11-dependent receptor recycling. (c) Internalization of the receptors into lipid-raft caveolae is followed by receptor degradation and the activation of MAPK signal transduction pathways. (B) Smad nucleo-cytoplasmic shuttling model. The movement of TGFβ-activated R-Smads (Smad2 and Smad3, with P representing phosphorylation on C-terminal serines) and the Co-Smad (Smad4) between the cytoplasm and nucleus is shown. Arrows indicate the activities of Smad proteins (see key). Enzymatic reactions such as Smad2 and Smad3 phosphorylation by the type I receptor and dephosphorylation of Smad2 and Smad3 by a nuclear phosphatase are indicated with broad arrows.
Smad nuclear accumulation
A key step in transducing the TGFβ signal is Smad nuclear accumulation. Several mechanisms have been proposed to regulate this step in TGFβ signaling, including regulative monoubiquitylation (Dupont et al., 2009), orchestrated nucleo-cytoplasmic shuttling via the TAZ transcription factor (Varelas et al., 2008), nucleo-cytoplasmic shuttling kinetics [in which different forms of the Smads have different kinetics of nuclear import and export, such that the phosphorylated Smads accumulate in the nucleus (Inman et al., 2002)] and retention factors [proteins in the cytoplasm that have a higher affinity for unphosphorylated Smads versus proteins in the nucleus that have a higher affinity for phosphorylated Smads (Xu et al., 2002)].
To distinguish between the shuttling kinetics and retention factor mechanisms, Clarke et al. developed a phosphorylation model of canonical Smad signaling that incorporated R-Smad phosphorylation, R-Smad heterodimerization with Smad4 and the nuclear accumulation of R-Smad-Smad4 complexes (Clarke et al., 2006). The authors also considered the absence of export of activated Smad complexes from the nucleus, as suggested by Schmierer and Hill (Schmierer and Hill, 2005). The model relied on data from both the literature and their own experimental measurements. By systematically altering model features and statistically analyzing sets of parameter values, the simulation showed that perturbing the rate constants for R-Smad phosphorylation, R-Smad dephosphorylation and R-Smad-Smad4 complex dissociation in the nucleus led to the largest changes in Smad nuclear accumulation. This simple model reproduced the major features of Smad signaling and demonstrated that the imbalance between R-Smad phosphorylation and dephosphorylation rates is likely to be a significant mechanism in governing Smad nuclear accumulation during TGFβ signaling. In addition to supporting the shuttling model, the results from repeated simulations falsified the retention hypothesis as no nuclear Smad-binding proteins were needed to recreate the experimental data. Their findings led the authors to generate the new hypothesis that Smad oligomerization protects phospho-R-Smads from dephosphorylation, thereby promoting Smad nuclear accumulation.
Schmierer et al. then generated a model of Smad nucleo-cytoplasmic shuttling that incorporated a much larger base of biochemical data (Schmierer et al., 2008) (Fig. 7B). Simulations generated by this model were entirely consistent with the results of the Smad phosphorylation model of Clarke et al. (Clarke et al., 2006). The new study agreed that the imbalance between R-Smad phosphorylation and dephosphorylation rates is a fundamental contributing factor to Smad nuclear accumulation and that Smad oligomerization protects phosphorylated Smad from a phosphatase in the nucleus. A new insight gained from the broader scope of this model is that the inhibition of nuclear export of activated Smad is not itself sufficient to fit the experimental data. For the model to match the observations, the inhibition of export of activated Smad must be accompanied by the faster import of activated versus monomeric Smads into the nucleus.
In summary, the strengths of kinetic modeling are its ability to incorporate the `real-time' dynamics of biochemical processes within a biological system and its ability to simulate the effects of parameter changes `in silico'. Then, comparisons of simulations with biochemical data allow the investigators to identify parameters or parameter values that `break' the model. Hypotheses regarding the biochemical mechanisms underlying the incongruity can then be tested experimentally. One weakness of this approach is that large quantities of detailed biochemical data are required to establish the initial conditions and rates of change for each component. A second weakness is that models cannot predict activities for which there are no pre-existing data. For example, models of Smad activity equate dephosphorylation with inactivation, but we now know that inactivation of Smads is also achieved by monoubiquitylation. New models that incorporate Smad ubiquitylation are currently being developed.
Conclusions
One of the most satisfying moments in science occurs when two independent lines of research arrive at the same conclusion. The convergence of phylogenetic and molecular studies in identifying new regulatory features of TGFβ signaling is a prime example. There is nothing that prevents other developmental pathways from enjoying similar synergies. An analysis of the Wnt pathway showed that all Frizzled receptors have a single conserved lysine and that Dishevelled signal transducers share six conserved lysines (Konikof et al., 2008). The authors predict that ubiquitylation and sumoylation are as important in regulating the Wnt pathway as they are in the TGFβ pathway. In addition, we have identified a previously unrecognized and conserved lysine in Cubitus interruptus (a Hedgehog pathway signal transducer in D. melanogaster) that might influence its regulation.
The dexterity with which we apply phylogenetics to additional pathways is based upon two principles. First, that all the protein sequence data are publicly available. Second, that the necessary user-friendly computer programs are also freely available (Boxes 2 and 3). Thus, the extension of the analysis of lysine conservation to receptors and signal transducers in other pathways is straightforward. From a broader perspective, the concept of exploiting amino acid conservation to understanding pathway regulation can be applied to any post-translational modification that targets a specific amino acid.
The same philosophy applies to kinetic modeling as this approach can be employed to analyze any pathway for which quantitative biochemical data are available. The amount of published biochemical data for any particular pathway is likely to be sufficient, but a greater understanding of mathematics is required to generate the appropriate equations for kinetic modeling as compared with the mathematical skills required to utilize phylogenetic computer programs. To productively employ kinetic modeling in studies of signal transduction, we suggest that developmental biologists collaborate with computer scientists. Such collaborations have been successful in characterizing embryonic morphogen gradients in D. melanogaster, as noted in the accompanying review (Umulis et al., 2009).
Looking beyond the fine-scale methods of phylogenetics and kinetic modeling, two studies of TGFβ signaling utilizing network-scale modeling are in progress. First, a strategy for applying Boolean logic to signaling pathways has been developed (Mendoza and Xenarios, 2006). In this method, quantitative parameters are ignored and the focus is solely on the topology of the pathway (e.g. the direction of information transfer and the factors that affect this process). The authors are currently analyzing the tumor necrosis factor α and TGFβ interaction network in dendritic cells. Second, a strategy for signaling network reconstruction from microarray data has been developed (Adler et al., 2009). In this method, a matrix is constructed of genes, the expression of which correlates with a particular experimental regime. Matrices derived from different experiments employing variations to the same regime are then compared statistically to reveal robust correlations that are visible across multiple experiments. The authors are currently analyzing TGFβ signaling utilizing publicly available microarray data from ArrayExpress (www.ebi.ac.uk/microarray-as/ae). Their initial results suggest 108 new candidates for inclusion in the pathway.
In summary, we have reviewed the successful application of both fine-scale and network-scale informatics approaches to developmental signaling pathways, utilizing TGFβ as our model. In our view, these efforts suggest that the application of informatics to improve our understanding of developmental signaling pathways is limited only by the investigator's imagination.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/22/3729/DC1
Supplementary Material
We are grateful to Charlotte Konikoff, Simon Whelan and Robert Wisotzkey for sharing their knowledge of phylogenetics. We thank Caroline Hill, Marty Kreitman, Xuedong Liu, Hedi Peterson, Osamu Shimmi, Ioannis Xenarios and Zhike Zi for valuable discussions. P.K. is supported by ENFIN, a Network of Excellence funded by the European Commission. Research in the laboratory of S.J.N. is supported by the NIH (CA095875; CA109552 HG002516) and by the Science Foundation Arizona. Deposited in PMC for release after 12 months.
References
- Adler, P., Peterson, H., Agius, P., Reimand, J. and Vilo, J. (2009). Ranking genes by their co-expression to subsets of pathway members. Ann. NY Acad. Sci. 1158, 1-13. [DOI] [PubMed] [Google Scholar]
- Aubin, J., Davy, A. and Soriano, P. (2004). In vivo convergence of BMP and MAPK signaling pathways: impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev. 18, 1482-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin, A., Tomlinson, A., Bhowmick, N., Moses, H. and Arteaga, C. (2000). Phosphatidylinositol 3-kinase function is required for TGF-β-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803-36810. [DOI] [PubMed] [Google Scholar]
- Chen, T., He, H. and Church, G. (1999). Modeling gene expression with differential equations. Pac. Symp. Biocomput. 1999, 29-40. [PubMed] [Google Scholar]
- Chong, P., Lin, H., Wrana, J. and Forman-Kay, J. (2006). An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J. Biol. Chem. 281, 17069-17075. [DOI] [PubMed] [Google Scholar]
- Clarke, D., Betterton, M. and Liu, X. (2006). Systems theory of Smad signaling. Syst. Biol. 153, 412-424. [DOI] [PubMed] [Google Scholar]
- Cope, F. (1963). A kinetic theory of enzymatic oxidation-reduction reactions based on a postulate of electron conduction in a macromolecular enzyme with application to active transport of small ions across biological membranes. Bull. Math. Biophys. 25, 165-176. [DOI] [PubMed] [Google Scholar]
- Dabovic, B. and Rifkin, D. (2008). TGF-β bioavailability: latency, targeting and activation. In The TGF-β Family (ed. R. Derynck and K. Miyazono), pp. 179-202. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Das, P., Maduzia, L., Wang, H., Finelli, A., Cho, S., Smith, M. and Padgett, R. (1998). The Drosophila gene Medea demonstrates the requirement for different classes of Smads in Dpp signaling. Development 125, 1519-1528. [DOI] [PubMed] [Google Scholar]
- Dayhoff, M., Schwartz, R. and Orcutt, B. (1978). A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure (ed. M. Dayhoff), pp. 345-352. Washington, DC: National Biomedical Research Foundation.
- Derynck, R. and Miyazono, K. (2008a). The TGF-β Family. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Derynck, R. and Miyazono, K. (2008b). TGF-β and the TGF-β family. In The TGF- β Family, pp. 29-43. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Di Guglielmo, G., Le Roy, C., Goodfellow, A. and Wrana, J. (2003). Distinct endocytic pathways regulate TGF-β receptor signaling and turnover. Nat. Cell Biol. 5, 410-421. [DOI] [PubMed] [Google Scholar]
- Dupont, S., Zacchigna, L., Cordenonsi, M., Soligo, S., Adorno, M., Rugge, M. and Piccolo, S. (2005). Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121, 87-99. [DOI] [PubMed] [Google Scholar]
- Dupont, S., Mamidi, A., Cordenonsi, M., Montagner, M., Zacchigna, L., Adorno, M., Martello, G., Stinchfield, M., Soligo, S., Morsut, L. et al. (2009). FAM/USP9x a deubiquitinating enzyme essential for TGF-β signaling controls Smad4 monoubiquitination. Cell 136, 123-135. [DOI] [PubMed] [Google Scholar]
- Efron, B., Halloran, E. and Holmes, S. (1996). Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 93, 13429-13434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368-376. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783-791. [DOI] [PubMed] [Google Scholar]
- Ferrell, J. (2002). Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140-148. [DOI] [PubMed] [Google Scholar]
- Fitch, W. and Farris, J. (1974). Evolutionary trees with minimum nucleotide replacements from amino acid sequences. J. Mol. Evol. 3, 263-278. [DOI] [PubMed] [Google Scholar]
- Fuentealba, L., Eivers, E., Ikeda, A., Hurtado, C., Kuroda, H., Pera, E. and De Robertis, E. (2007). Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131, 980-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet, G. (2007). Efficient tree searches with available algorithms. Evol. Bioinform. Online 12, 341-356. [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696-704. [DOI] [PubMed] [Google Scholar]
- Hayward, D., Samuel, G., Pontynen, P., Catmull, J., Saint, R., Miller, D. and Ball, E. (2002). Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc. Natl. Acad. Sci. USA 99, 8106-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, S. and Kumar, S. (2003). Genomic clocks and timescales. Trends Genet. 19, 200-206. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Henikoff, J. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89, 10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, J., Podos, S., Keith, K., Simpson, S. and Ferguson, E. (1998). The Drosophila Medea gene is required downstream of Dpp and encodes a functional homolog of human Smad4. Development 125, 1407-1420. [DOI] [PubMed] [Google Scholar]
- Inman, G., Nicolas, F. and Hill, C. (2002). Nucleocytoplasmic shuttling of Smads2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell 10, 283-294. [DOI] [PubMed] [Google Scholar]
- Israel, D., Nove, J., Kerns, K., Kaufman, R., Rosen, V., Cox, K. and Wozney, J. (1996). Heterodimeric BMPs show enhanced activity in vitro and in vivo. Growth Factors 13, 291-300. [DOI] [PubMed] [Google Scholar]
- Javelaud, D. and Mauviel, A. (2005). Crosstalk mechanisms between the MAPK pathways and Smad signaling downstream of TGF-β: implications for carcinogenesis. Oncogene 24, 5742-5750. [DOI] [PubMed] [Google Scholar]
- Jing, S., Wen, D., Yu, Y., Holst, P., Luo, Y., Fang, M., Tamir, R., Antonio, L., Hu, Z., Cupples, R. et al. (1996). GDNF-induced activation of the ret protein tyrosine kinase is mediated by a novel receptor for GDNF. Cell 85, 1113-1124. [DOI] [PubMed] [Google Scholar]
- Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S. and Madden, T. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., Saunier, E., Akhurst, R. and Derynck, R. (2008). The type I TGF-β receptor is covalently modified and regulated by sumoylation. Nat. Cell Biol. 10, 654-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak, P., Rasmussen, R., Causing, C., Bonni, S., Zhu, H., Thomsen, G. and Wrana, J. (2000). Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF-β receptor for degradation. Mol. Cell 6, 1365-1375. [DOI] [PubMed] [Google Scholar]
- Kim, S., Kwak, J., Na, H., Kim, J., Ding, Y. and Choi, M. (2009). TGF-β1 activates TAK1 via TAB1-mediated autophosphorylation, independent of TGF-β receptor kinase activity in mesangial cells. J. Biol. Chem. 284, 22285-22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp, E. (2007). Modeling dynamic processes in yeast. Yeast 24, 943-959. [DOI] [PubMed] [Google Scholar]
- Klipp, E., Herwig, R., Kowald, A., Wierling, C. and Lehrach, H. (2005). Systems Biology in Practice: Concepts, Implementation and Application. Weinheim, Germany: Wiley-VCH.
- Konikoff, C., Wisotzkey, R. and Newfeld, S. (2008). Lysine conservation and context in TGF-β and Wnt signaling suggests new targets and general themes for post-translational modification. J. Mol. Evol. 67, 322-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar, M., Doody, J. and Massagué, J. (1997). Opposing BMP and EGF signaling pathways converge on the TGF-β family mediator Smad1. Nature 389, 618-622. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Dudley, J., Nei, M. and Tamura, K. (2008). MEGA5: biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnapuu, J., Bjorkgren, I. and Shimmi, O. (2009). The Drosophila Dpp signal is produced by the cleavage of its proprotein at evolutionarily diversified furin recognition sites. Proc. Natl. Acad. Sci. USA 106, 8501-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratomi, G., Komuro, A., Goto, K., Shinozaki, M., Miyazawa, K., Miyazono, K. and Imamura, T. (2005). NEDD4-2 negatively regulates TGF-β signaling by inducing ubiquitin-mediated degradation of Smad2 and Type I receptor. Biochem. J. 386, 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M., Blackshields, G., Brown, N., Chenna, R., McGettigan, P., McWilliam, H., Valentin, F., Wallace, I., Wilm, A., Lopez, R. et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948. [DOI] [PubMed] [Google Scholar]
- Lin, X., Lang, M., Liang, Y., Brunicardi, F. and Feng, X. (2003). Sumo-1/Ubc-9 promotes nuclear accumulation and metabolic stability of Smad4. J. Biol. Chem. 278, 31043-31048. [DOI] [PubMed] [Google Scholar]
- Lin, X., Chen, Y. and Feng, X. (2008).Transcriptional control via Smads. In The TGF-β Family (ed. R. Derynck and K. Miyazono), pp. 287-332. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Long, J., Wang, G., He, D. and Liu, F. (2004). Repression of Smad4 transcriptional activity by Sumo modification. Biochem. J. 379, 23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., Murray, J., Luo, W., Li, H., Wu, X., Xu, H., Backer, J. and Chen, Y. (2002). TGF-β activates Smad2 in the absence of receptor endocytosis. J. Biol. Chem. 277, 29363-29368. [DOI] [PubMed] [Google Scholar]
- Marquez, R., Singer, M., Takaesu, N., Waldrip, W., Kraytsberg, Y. and Newfeld, S. (2001). Transgenic analysis of the Smad family of TGF-β signal transducers in Drosophila suggests new roles and interactions between family members. Genetics 157, 1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué, J. (2008). TGF-β in cancer. Cell 134, 215-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau, B., Newton, M. and Larget, B. (1999). Bayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics 55, 1-12. [DOI] [PubMed] [Google Scholar]
- Meinnel, T., Serero, A. and Giglione, C. (2006). Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 387, 839-851. [DOI] [PubMed] [Google Scholar]
- Mendoza, L. and Xenarios, I. (2006). A method for the generation of standardized qualitative dynamical systems of regulatory networks. Theor. Biol. Med. Model. 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, W., Jaffray, E., Campbell, S., Takeda, S., Bayston, L., Basu, S., Li, M., Raftery, L., Ashe, M., Hay, R. et al. (2008). Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 22, 2578-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, H., Choudhury, A., Pagano, R. and Leof, E. (2004). Ligand-dependent and -independent TGF-β receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol. Biol. Cell 15, 4166-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono, K. (2008). Regulation of TGF-β family signaling by inhibitory Smads. In The TGF-β Family (ed. R. Derynck and K. Miyazono), pp. 363-387. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PMC free article] [PubMed]
- Morén, A., Hellman, U., Inada, Y., Imamura, T. and Heldin, C. H. (2003). Differential ubiquitination defines the functional status of Smad4. J. Biol. Chem. 278, 33571-33582. [DOI] [PubMed] [Google Scholar]
- Morén, A., Imamura, T., Miyazono, K., Heldin, C. H. and Moustakas, A. (2005). Degradation of the tumor suppressor Smad4 by WW and HECT ubiquitin ligases. J. Biol. Chem. 280, 22115-22123. [DOI] [PubMed] [Google Scholar]
- Moustakas, A. and Heldin, C.-H. (2009). The regulation of TGFβ signal transduction. Development 136, 3699-3714. [DOI] [PubMed] [Google Scholar]
- Newfeld, S. and Gelbart, W. (1995). Identification of two Drosophila TGF-β family members in the grasshopper Schistocerca americana. J. Mol. Evol. 41, 155-160. [DOI] [PubMed] [Google Scholar]
- Newfeld, S. and Wisotzkey, R. (2006). Molecular evolution of Smad proteins. In Smad Signal Transduction (ed. C. Heldin and P. ten Dijke), pp. 15-35. Dordrecht, The Netherlands: Springer.
- Newfeld, S., Wisotzkey, R. and Kumar, S. (1999). Molecular evolution of a developmental pathway: phylogenetic analyses of TGF-β family ligands, receptors and Smad signal transducers. Genetics 152, 783-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, R., Wozney, J. and Gelbart, W. (1993). Human BMP sequences can confer normal dorsal-ventral patterning in Drosophila. Proc. Natl. Acad. Sci. USA 90, 2905-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny, D. and Hendy, M. (1987). TurboTree: a fast algorithm for minimal trees. Comput. Appl. Biosci. 3, 183-187. [PubMed] [Google Scholar]
- Pot, I. and Bonni, S. (2008). SnoN in TGF-β signaling and cancer biology. Curr. Mol. Med. 8, 319-328. [DOI] [PubMed] [Google Scholar]
- Pyrowolakis. G., Hartmann, B. and Affolter, M. (2008). TGF-β family signaling in Drosophila, in The TGF-β Family (ed. R. Derynck and K. Miyazono), pp. 493-526. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Ronquist, F. and Huelsenbeck, J. (2003). MrBayes 3, bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- Sampath, T., Rashka, K., Doctor, J., Tucker, R. and Hoffmann, F. M. (1993). Drosophila TGF-β superfamily proteins induce endochondral bone formation in mammals. Proc. Natl. Acad. Sci. USA 90, 6004-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota, G., Alarcón, C., Spagnoli, F., Brivanlou, A. and Massagué, J. (2007). Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell 25, 441-454. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn, C. (2005). TGF-β signaling. WormBook 9, 1-12. www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer, B. and Hill, C. (2005). Kinetic analysis of Smad nucleo-cytoplasmic shuttling reveals a mechanism for TGFβ-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 25, 9845-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer, B., Tournier, A., Bates, P. and Hill, C. (2008). Mathematical modeling identifies Smad nucleo-cytoplasmic shuttling as a dynamic signal-interpreting system. Proc. Natl. Acad. Sci. USA 105, 6608-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J., Newfeld, S., Raftery, L., Chartoff, E. and Gelbart, W. (1995). Genetic characterization and cloning of Mothers against dpp: a gene required for decapentaplegic function in Drosophila. Genetics 139, 1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Wang, Y., Jayaraman, L., Yang, H., Massagué, J. and Pavletich, N. (1998). Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94, 585-594. [DOI] [PubMed] [Google Scholar]
- Shimmi, O., Ralston, A., Blair, S. and O'Connor, M. (2005a). The crossveinless gene encodes a new member of the Twisted gastrulation family of BMP-binding proteins which, with Short gastrulation, promotes BMP signaling in the crossveins of the Drosophila wing. Dev. Biol. 282, 70-83. [DOI] [PubMed] [Google Scholar]
- Shimmi, O., Umulis, D., Othmer, H. and O'Connor, M. (2005b). Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120, 873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira, H. and Hasegawa, M. (2001).CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246-1247. [DOI] [PubMed] [Google Scholar]
- Sitnikova, T. (1996). Bootstrap method of interior-branch test for phylogenetic trees. Mol. Biol. Evol. 13, 605-611. [DOI] [PubMed] [Google Scholar]
- Swofford, D. (2001). PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4. Sunderland, MA: Sinauer Associates.
- Takaesu, N., Herbig, E., Zhitomersky, D., O'Connor, M. and Newfeld, S. (2005). DNA-binding domain mutations in Smad genes yield dominant negative or neomorphic proteins that can activate Wg target genes in Drosophila. Development 132, 4883-4894. [DOI] [PubMed] [Google Scholar]
- Takaesu, N., Hyman-Walsh, C., Ye, S., Wisotzkey, R., Stinchfield, M., O'Connor, M., Wotton, D. and Newfeld, S. (2006). dSno facilitates Baboon signaling in the Drosophila brain by switching the affinity of Medea away from Mad and toward dSmad2. Genetics 174, 1299-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umulis, D., O'Connor, M. B. and Blair, S. S. (2009). The extracellular regulation of bone morphogenetic protein signaling. Development 136, 3715-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas, X., Sakuma, R., Samavarchi-Tehrani, P., Peerani, R., Rao, B., Dembowy, J., Yaffe, M., Zandstra, P. and Wrana, J. (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837-848. [DOI] [PubMed] [Google Scholar]
- Vilar, J., Jansen, R. and Sander, C. (2006). Signal processing in the TGF-β superfamily ligand-receptor network. PLoS Comput. Biol. 2, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton, K. and Derynck, R. (2009). TGFβ family signaling: novel insights in development and disease. Development 136, 3691-3697. [DOI] [PubMed] [Google Scholar]
- Whelan, S. (2008). Inferring trees. Methods Mol. Biol. 452, 287-309. [DOI] [PubMed] [Google Scholar]
- Whelan, S. (2009). New approaches to phylogenetic tree search and their application to large numbers of protein alignments. Syst. Biol. 56, 727-740. [DOI] [PubMed] [Google Scholar]
- Wicks, S., Haros, K., Maillard, M., Song, L., Cohen, R., ten Dijke, P. and Chantry, A. (2005). The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGFβ signaling. Oncogene 24, 8080-8084. [DOI] [PubMed] [Google Scholar]
- Wilkes, M., Murphy, S., Garamszegi, N. and Leof, E. (2003). Cell-type-specific activation of PAK2 by TGF-β independent of Smad2 and Smad3. Mol. Cell. Biol. 23, 8878-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotzkey, R., Mehra, A., Sutherland, D., Dobens, L., Liu, X., Dohrmann, C., Attisano, L. and Raftery, L. (1998) Medea is a Drosophila Smad4 homolog that is differentially required to potentiate Dpp responses. Development 125, 1433-1445. [DOI] [PubMed] [Google Scholar]
- Wu, J., Hu, M., Chai, J., Seoane, J., Huse, M., Li, C., Rigotti, D., Kyin, S., Muir, T., Fairman. R. et al. (2001). Crystal structure of a phosphorylated Smad2: recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol. Cell 8, 1277-1289. [DOI] [PubMed] [Google Scholar]
- Xu, L., Kang, Y., Col, S. and Massague, J. (2002). Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10, 271-282. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Kurisaki, A., Yamakawa, N., Minakuchi, K. and Sugino, H. (2006). FKBP12 functions as an adapter of Smad7-Smurf1 on Activin Type I receptor. Mol. Endocrin. 36, 569-579. [DOI] [PubMed] [Google Scholar]
- Yang, S., Galanis, A., Witty, J. and Sharrocks, A. (2006). An extended consensus motif enhances the specificity of substrate modification by Sumo. EMBO J. 25, 5083-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi, Z. and Klipp, E. (2007). Constraint-based modeling and kinetic analysis of the Smad dependent TGF-β signaling pathway. PLoS ONE 2, e936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, W. and Chen, Y. (2009). Specific activation of MAPK by TGF-β receptors in lipid rafts is required for epithelial cell plasticity. Mol. Biol. Cell 20, 1020-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.